Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Retrieval and Proteome Analysis
2.2. Antigenicity and Allergenicity Evaluation
2.3. T Cell Epitope Prediction, Epitope Selection and Vaccine Construction
2.4. Immunoinformatics Analysis of Chimeric Protein
2.4.1. Physicochemical Properties
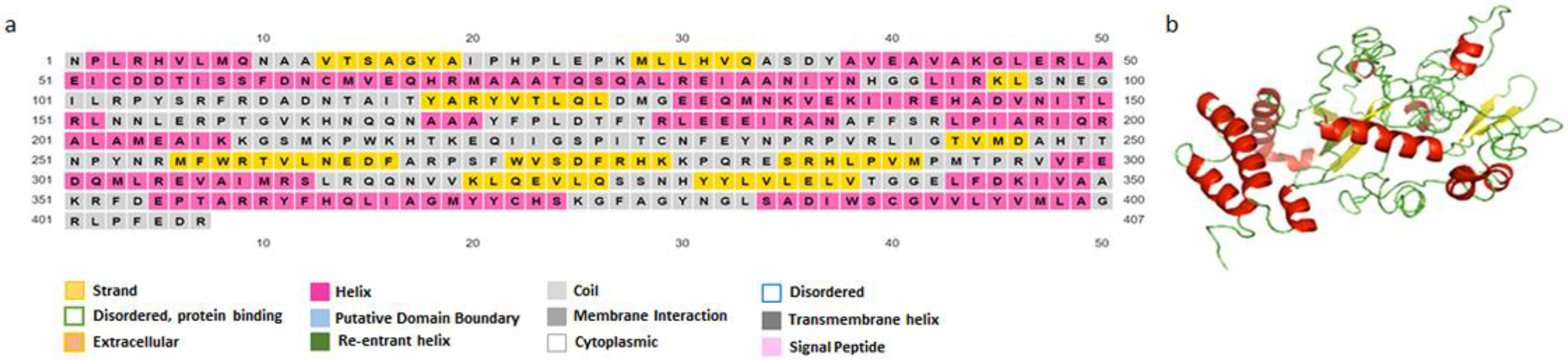
2.4.2. Secondary and Tertiary Structure
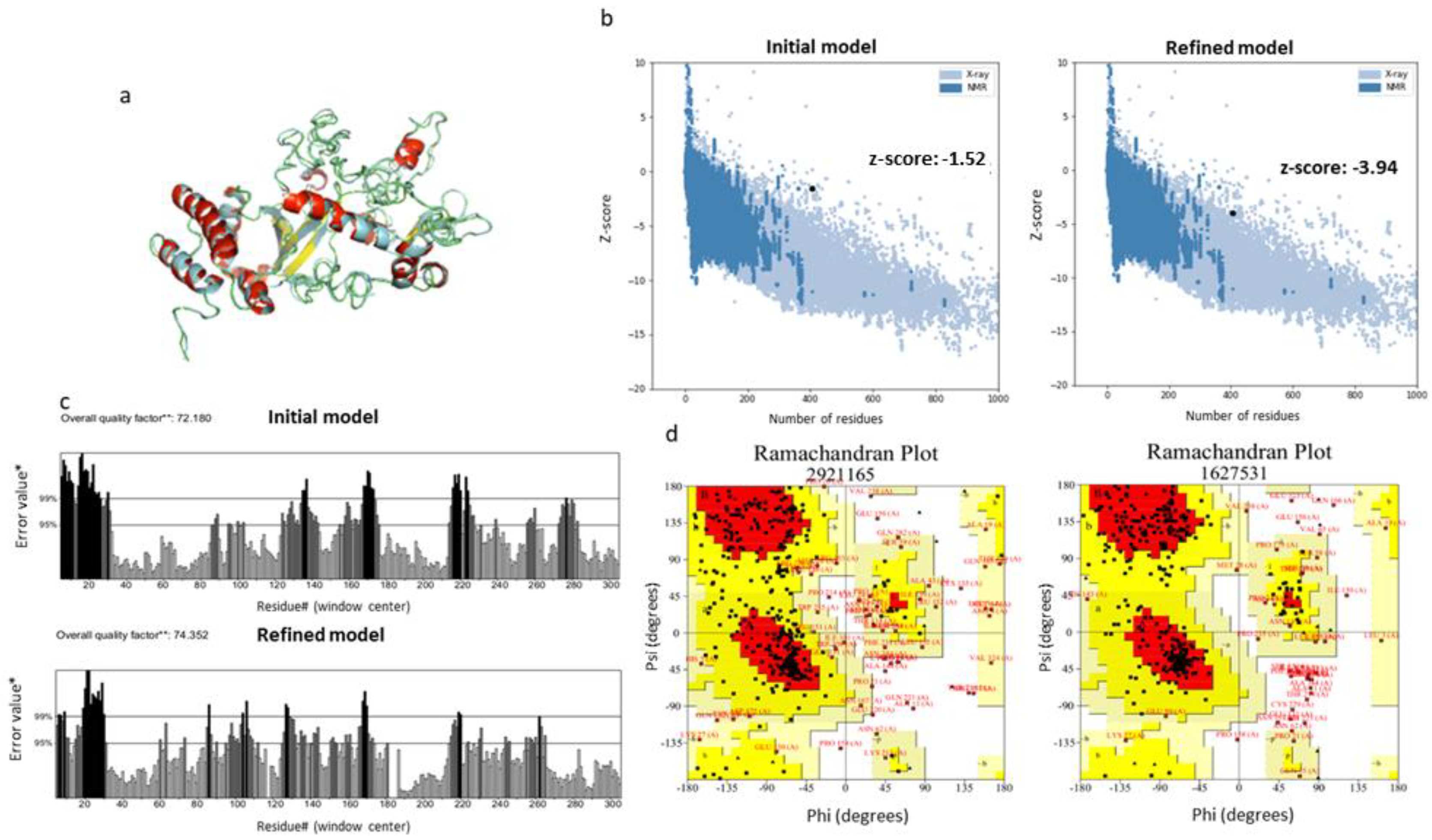
2.4.3. Refinement and Validation of Tertiary Structure Model
2.4.4. In Silico Prediction of LeishChim’s Docking to MHC Class I and II Molecules
2.4.5. Molecular Dynamics (MD) Simulation of LeishChim-MHCI/MHCII Complexes
2.4.6. CTL, HTL, B Cell Linear and Conformational Epitope Prediction
2.4.7. Immune Simulations
2.5. Expression of Chimeric Protein
2.6. PLGA Vaccine Preparation and Characterization
2.6.1. Materials
2.6.2. Preparation and Characterization of PLGA Nanoparticles
2.6.3. Quantification of Antigen Loading
2.6.4. Quantification of MPLA Loading
2.6.5. In Vitro Release Studies
2.6.6. In Vitro Stability
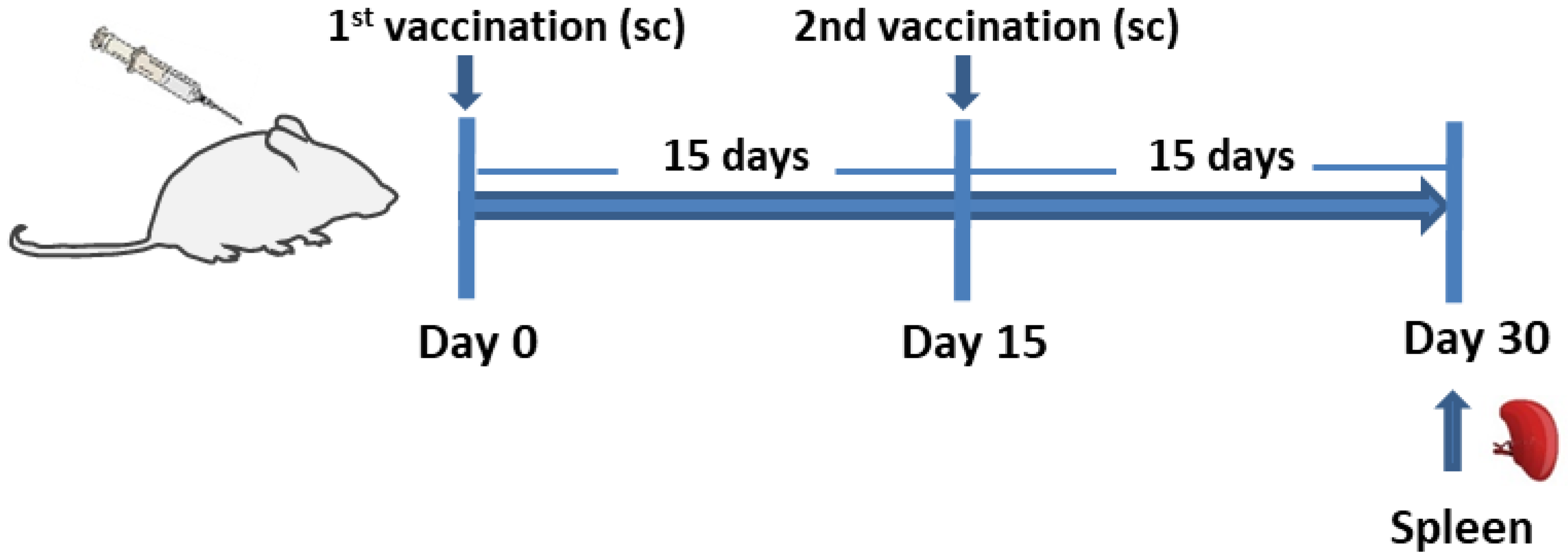
2.7. Experimental Animals
2.8. Vaccination Schedule
2.9. LeishChim-Specific Spleen Cell Proliferation Assay
2.10. Flow Cytometry Analysis for Determination of LeishChim-Specific Memory and Cytokine-Producing T Cells
2.11. Statistical Analysis
3. Results
3.1. Conserved Leishmania Protein Retrieval and Identification of Cellular Localization
3.2. Antigenicity and Allergenicity Evaluation
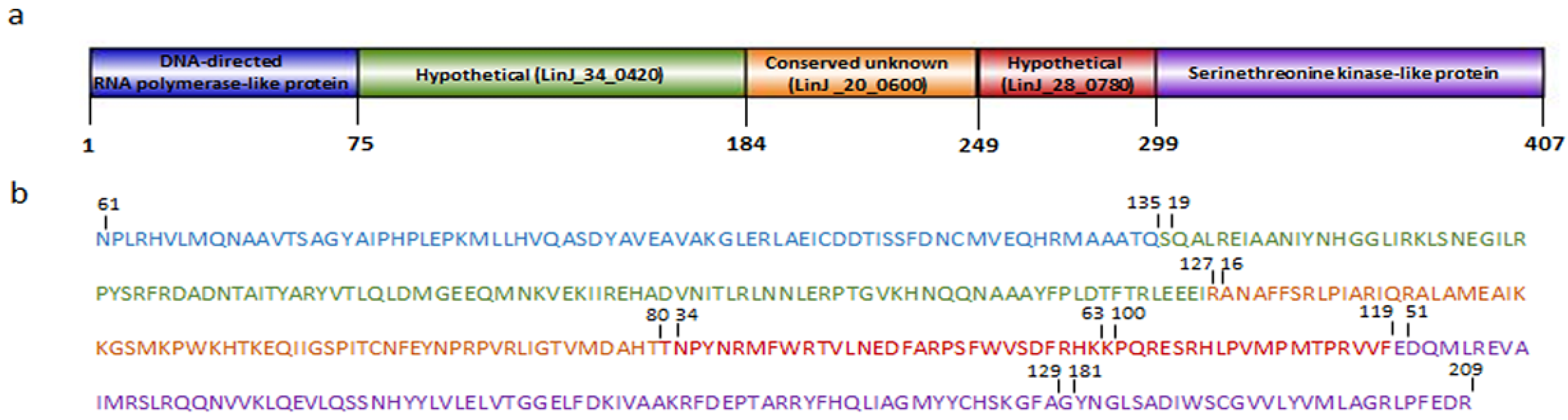
3.3. MHC I- and II-Binding Epitope Identification, Selection and Vaccine Construction
3.4. Immunoinformatics Evaluation of Chimeric Protein
3.4.1. Physicochemical Properties, Antigenicity and Allergenicity Evaluation
3.4.2. Secondary and Tertiary Structure
3.4.3. Refinement and Validation of the Tertiary Structure
3.4.4. In Silico Prediction of LeishChim’s Docking onto MHCI and MHCII Molecules
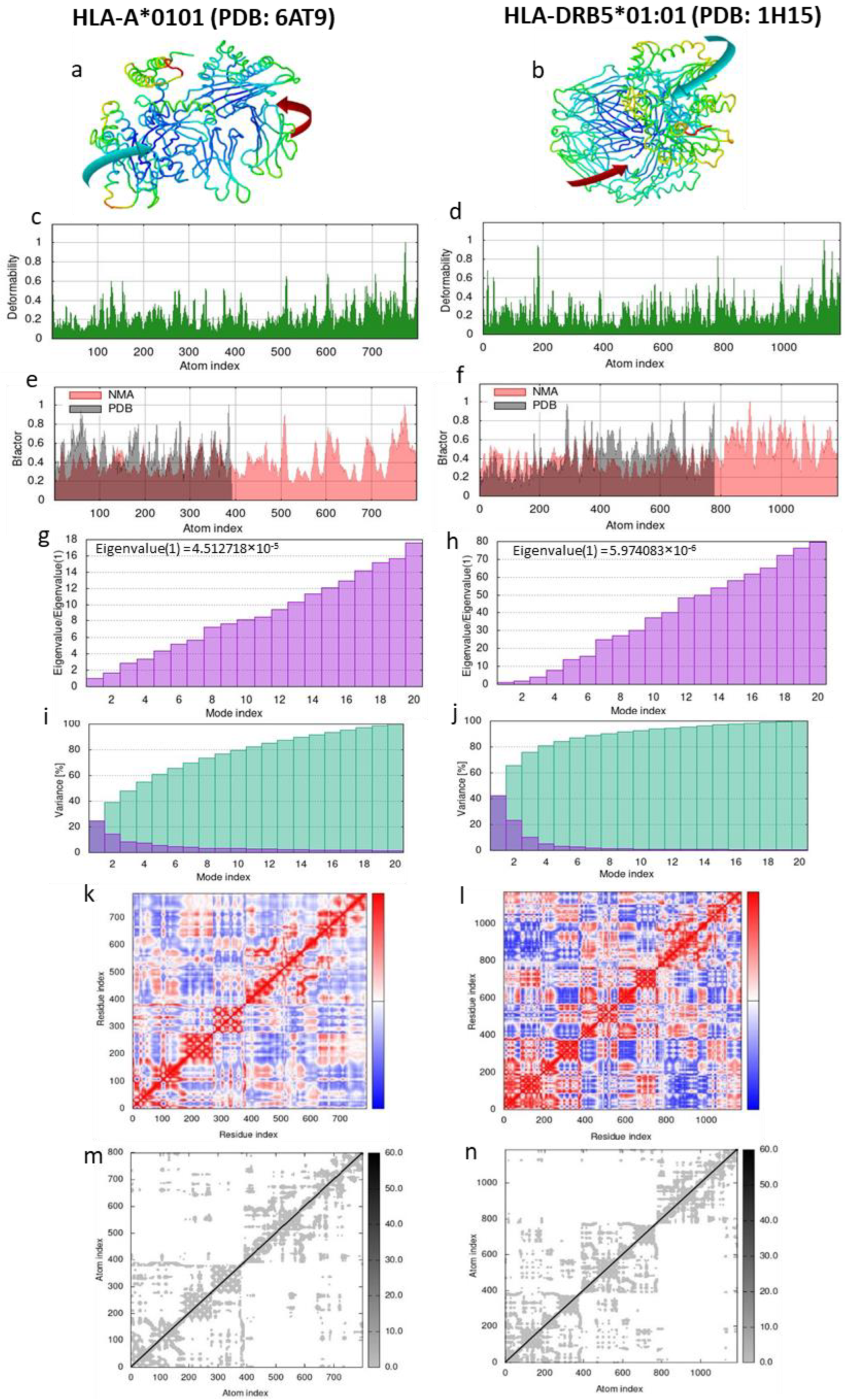
3.4.5. Stability Prediction of the LeishChim and MHCI/MHCII Complexes
3.4.6. B Cell Linear and Conformational Epitope Prediction
3.4.7. CTL and HTL Epitope Prediction
3.4.8. In Silico Immune Simulation Profile of LeishChim
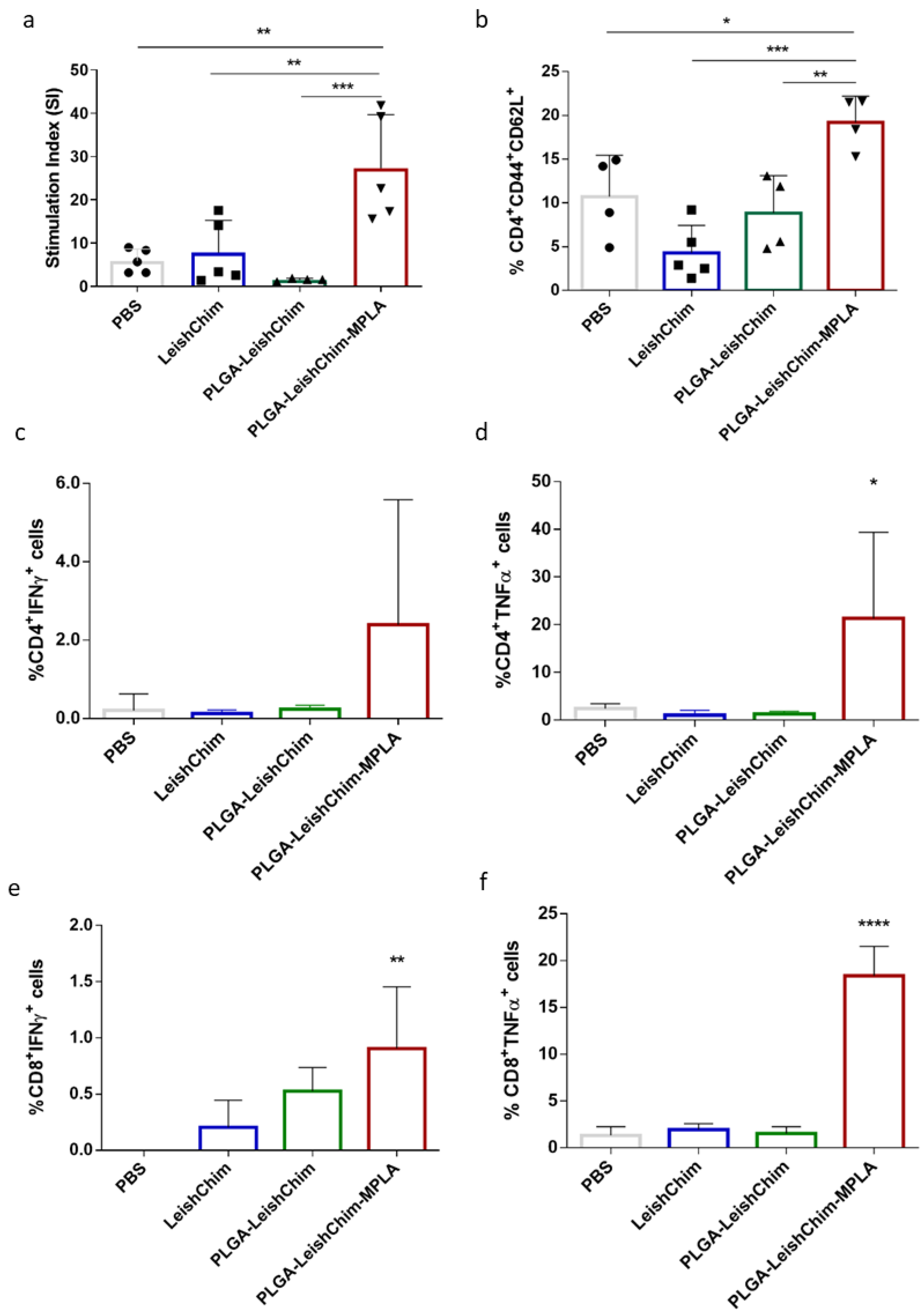
3.5. Immunization with Multi-Epitope Chimeric Protein Encapsulated in PLGA NPs Elicited Antigen-Specific Cellular Immune Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- World Health Organization. Leishmaniasis. Available online: http://www.who.int/leishmaniasis/en/ (accessed on 21 November 2022).
- Chappuis, F.; Sundar, S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: What are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007, 5, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Colombo, F.A.; Odorizzi, R.M.F.N.; Laurenti, M.D.; Galati, E.A.B.; Canavez, F.; Pereira-Chioccola, V.L. Detection of Leishmania (Leishmania) infantum RNA in fleas and ticks collected from naturally infected dogs. Parasitol. Res. 2011, 109, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Didwania, N.; Shadab, M.; Sabur, A.; Ali, N. Alternative to Chemotherapy—The Unmet Demand against Leishmaniasis. Front. Immunol. 2017, 8, 1779. [Google Scholar] [CrossRef]
- Sundar, S.; Singh, A. Recent developments and future prospects in the treatment of visceral leishmaniasis. Ther. Adv. Infect. Dis. 2016, 3, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Das, P. Recent progress in drug targets and inhibitors towards combating leishmaniasis. Acta Trop. 2018, 181, 95–104. [Google Scholar] [CrossRef]
- Greenwood, B. The contribution of vaccination to global health: Past, present and future. Philos. Trans. R. Soc. B 2014, 369, 20130433. [Google Scholar] [CrossRef] [PubMed]
- Duthie, M.S.; Raman, V.S.; Piazza, F.M.; Reed, S.G. The development and clinical evaluation of second-generation leishmaniasis vaccines. Vaccine 2012, 30, 134–141. [Google Scholar] [CrossRef]
- Iborra, S.; Solana, J.C.; Requena, J.M.; Soto, M. Vaccine candidates against leishmania under current research. Expert Rev. Vaccines 2018, 17, 323–334. [Google Scholar] [CrossRef]
- Yasmin, H.; Adhikary, A.; Al-Ahdal, M.N.; Soy, S.; Kishore, U. Host–Pathogen Interaction in Leishmaniasis: Immune Response and Vaccination Strategies. Immuno 2022, 2, 218–254. [Google Scholar] [CrossRef]
- Kaye, P.M.; Cruz, I.; Picado, A.; Van Bocxlaer, K.; Croft, S.L. Leishmaniasis immunopathology—Impact on design and use of vaccines, diagnostics and drugs. Semin. Immunopathol. 2020, 42, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Cecilio, P.; Perez-Cabezas, B.; Fernandez, L.; Moreno, J.; Carrillo, E.; Requena, J.M.; Fichera, E.; Reed, S.G.; Coler, R.N.; Kamhawi, S.; et al. Pre-clinical antigenicity studies of an innovative multivalent vaccine for human visceral leishmaniasis. PLoS Negl. Trop. Dis. 2017, 11, e0005951. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.C.; Lage, D.P.; Martins, V.T.; Costa, L.E.; Lage, L.M.R.; Carvalho, A.M.R.S.; Ludolf, F.; Santos, T.T.O.; Roatt, B.M.; Menezes-Souza, D.; et al. A vaccine combining two Leishmania braziliensis proteins offers heterologous protection against Leishmania infanturn infection. Mol. Immunol. 2016, 76, 70–79. [Google Scholar] [CrossRef] [PubMed]
- De Brito, R.C.F.; Cardoso, J.M.D.O.; Reis, L.E.S.; Vieira, J.F.; Mathias, F.A.S.; Roatt, B.M.; Aguiar-Soares, R.D.D.O.; Ruiz, J.C.; Resende, D.D.; Reis, A.B. Peptide Vaccines for Leishmaniasis. Front. Immunol. 2018, 9, 1043. [Google Scholar] [CrossRef]
- Paninabordignon, P.; Tan, A.; Termijtelen, A.; Demotz, S.; Corradin, G.; Lanzavecchia, A. Universally Immunogenic T-Cell Epitopes—Promiscuous Binding to Human Mhc Class-Ii and Promiscuous Recognition by T-Cells. Eur. J. Immunol. 1989, 19, 2237–2242. [Google Scholar] [CrossRef]
- Hamdy, S.; Haddadi, A.; Hung, R.W.; Lavasanifar, A. Targeting dendritic cells with nano-particulate PLGA cancer vaccine formulations. Adv. Drug Deliv. Rev. 2011, 63, 943–955. [Google Scholar] [CrossRef]
- Jiang, W.L.; Gupta, R.K.; Deshpande, M.C.; Schwendeman, S.P. Biodegradable poly(lactic-co-glycolic acid) microparticles for injectable delivery of vaccine antigens. Adv. Drug Deliv. Rev. 2005, 57, 391–410. [Google Scholar] [CrossRef]
- Ma, W.X.; Chen, M.S.; Kaushal, S.; McElroy, M.; Zhang, Y.; Ozkan, C.; Bouvet, M.; Kruse, C.; Grotjahn, D.; Ichim, T.; et al. PLGA nanoparticle-mediated delivery of tumor antigenic peptides elicits effective immune responses. Int. J. Nanomed. 2012, 7, 1475–1487. [Google Scholar] [CrossRef]
- Pandit, S.; Cevher, E.; Zariwala, M.G.; Somavarapu, S.; Alpar, H.O. Enhancement of immune response of HBsAg loaded poly (L-lactic acid) microspheres against Hepatitis B through incorporation of alum and chitosan. J. Microencapsul. 2007, 24, 539–552. [Google Scholar] [CrossRef]
- Margaroni, M.; Agallou, M.; Athanasiou, E.; Kammona, O.; Kiparissides, C.; Gaitanaki, C.; Karagouni, E. Vaccination with poly(D,L-lactide-co-glycolide) nanoparticles loaded with soluble Leishmania antigens and modified with a TNF alpha-mimicking peptide or monophosphoryl lipid A confers protection against experimental visceral leishmaniasis. Int. J. Nanomed. 2017, 12, 6169–6183. [Google Scholar] [CrossRef]
- Moon, J.J.; Suh, H.; Polhemus, M.E.; Ockenhouse, C.F.; Yadava, A.; Irvine, D.J. Antigen-Displaying Lipid-Enveloped PLGA Nanoparticles as Delivery Agents for a Plasmodium vivax Malaria Vaccine. PLoS ONE 2012, 7, e31472. [Google Scholar] [CrossRef] [PubMed]
- Tafaghodi, M.; Khamesipour, A.; Jaafari, M.R. Immunization against leishmaniasis by PLGA nanospheres encapsulated with autoclaved Leishmania major (ALM) and CpG-ODN. Parasitol. Res. 2011, 108, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2-a server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006, 34, W202–W209. [Google Scholar] [CrossRef]
- Dimitrov, I.; Naneva, L.; Doytchinova, I.; Bangov, I. AllergenFP: Allergenicity prediction by descriptor fingerprints. Bioinformatics 2014, 30, 846–851. [Google Scholar] [CrossRef]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.J.; Peters, B.; Sette, A. The immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef]
- Rammensee, H.G.; Friede, T.; Stevanoviic, S. MHC ligands and peptide motifs: First listing. Immunogenetics 1995, 41, 178–228. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Shin, W.H.; Lee, G.R.; Heo, L.; Lee, H.; Seok, C. Prediction of Protein Structure and Interaction by GALAXY Protein Modeling Programs. BioDesign 2014, 2, 1–11. [Google Scholar]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Macarthur, M.W.; Moss, D.S.; Thornton, J.M. Procheck—A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Brooks, C.L. Symmetry, form, and shape: Guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 115–133. [Google Scholar] [CrossRef]
- Lopez-Blanco, J.R.; Aliaga, J.I.; Quintana-Orti, E.S.; Chacon, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Lopez-Blanco, J.R.; Garzon, J.I.; Chacon, P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics 2011, 27, 2843–2850. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Peters, B.; Bulik, S.; Tampe, R.; Van Endert, P.M.; Holzhutter, H.G. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 2003, 171, 1741–1749. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- Liu, W.Z.; Xie, Y.B.; Ma, J.Y.; Luo, X.T.; Nie, P.; Zuo, Z.X.; Lahrmann, U.; Zhao, Q.; Zheng, Y.Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.I.; Nascimento, I.P.; Barral, A.; Soto, M.; Barral-Netto, M. Challenges and perspectives in vaccination against leishmaniasis. Parasitol. Int. 2009, 58, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; McElrath, M.J.; Lewis, D.J.M.; Del Giudice, G. Challenges and responses in human vaccine development. Curr. Opin. Immunol. 2014, 28, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Oyston, P.; Robinson, K. The current challenges for vaccine development. J. Med. Microbiol. 2012, 61, 889–894. [Google Scholar] [CrossRef]
- Fiuza, J.A.; Gannavaram, S.; Santiago, H.D.; Selvapandiyan, A.; Souza, D.M.; Passos, L.S.A.; de Mendonca, L.Z.; Lemos-Giunchetti, D.D.; Ricci, N.D.; Bartholomeu, D.C.; et al. Vaccination using live attenuated Leishmania donovani centrin deleted parasites induces protection in dogs against Leishmania infantum. Vaccine 2015, 33, 280–288. [Google Scholar] [CrossRef]
- Mugunthan, S.P.; Harish, M.C. Multi-epitope-Based Vaccine Designed by Targeting Cytoadherence Proteins of Mycoplasma gallisepticum. ACS Omega 2021, 6, 13742–13755. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef]
- Das, S.; Freier, A.; Boussoffara, T.; Das, S.; Oswald, D.; Losch, F.O.; Selka, M.; Sacerdoti-Sierra, N.; Schonian, G.; Wiesmuller, K.H.; et al. Modular Multiantigen T Cell Epitope-Enriched DNA Vaccine Against Human Leishmaniasis. Sci. Transl. Med. 2014, 6, 234ra56. [Google Scholar] [CrossRef]
- Amanna, I.J.; Slifka, M.K. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology 2011, 411, 206–215. [Google Scholar] [CrossRef]
- Adu-Bobie, J.; Capecchi, B.; Serruto, D.; Rappuoli, R.; Pizza, M. Two years into reverse vaccinology. Vaccine 2003, 21, 605–610. [Google Scholar] [CrossRef]
- Caro-Gomez, E.; Gazi, M.; Goez, Y.; Valbuena, G. Discovery of novel cross-protective Rickettsia prowazekii T-cell antigens using a combined reverse vaccinology and in vivo screening approach. Vaccine 2014, 32, 4968–4976. [Google Scholar] [CrossRef] [PubMed]
- Mehla, K.; Ramana, J. Identification of epitope-based peptide vaccine candidates against enterotoxigenic Escherichia coli: A comparative genomics and immunoinformatics approach. Mol. Biosyst. 2016, 12, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Nezafat, N.; Eslami, M.; Negahdaripour, M.; Rahbar, M.R.; Ghasemi, Y. Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol. Biosyst. 2017, 13, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Sethi, S.; Krishna, R. Multi-epitope based vaccine design against Staphylococcus epidermidis: A subtractive proteomics and immunoinformatics approach. Microb. Pathog. 2022, 165, 105484. [Google Scholar] [CrossRef] [PubMed]
- Sami, S.A.; Marma, K.K.S.; Mahmud, S.; Khan, M.A.N.; Albogami, S.; El-Shehawi, A.M.; Rakib, A.; Chakraborty, A.; Mohiuddin, M.; Dhama, K.; et al. Designing of a Multi-epitope Vaccine against the Structural Proteins of Marburg Virus Exploiting the Immunoinformatics Approach. ACS Omega 2021, 6, 32043–32071. [Google Scholar] [CrossRef]
- Bhuiyan, M.A.; Quayum, S.T.; Ahammad, F.; Alam, R.; Samad, A.; Nain, Z. Discovery of potential immune epitopes and peptide vaccine design—A prophylactic strategy against Rift Valley fever virus. F1000Research 2020, 9, 999. [Google Scholar] [CrossRef]
- Ul Qamar, M.T.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.T.; Fatima, I.; Shahid, F.; Chen, L.L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef]
- Delany, I.; Rappuoli, R.; Seib, K.L. Vaccines, Reverse Vaccinology, and Bacterial Pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a012476. [Google Scholar] [CrossRef]
- Chakravarty, J.; Kumar, S.; Trivedi, S.; Rai, V.K.; Singh, A.; Ashman, J.A.; Laughlin, E.M.; Coler, R.N.; Kahn, S.J.; Beckmann, A.M.; et al. A clinical trial to evaluate the safety and immunogenicity of the LEISH-F1+MPL-SE vaccine for use in the prevention of visceral leishmaniasis. Vaccine 2011, 29, 3531–3537. [Google Scholar] [CrossRef]
- Coler, R.N.; Goto, Y.; Bogatzki, L.; Raman, V.; Reed, S.G. Leish-111f, a recombinant polyprotein vaccine that protects against visceral leishmaniasis by elicitation of CD4(+) T cells. Infect. Immun. 2007, 75, 4648–4654. [Google Scholar] [CrossRef]
- Goto, Y.; Bhatia, A.; Raman, V.S.; Liang, H.; Mohamath, R.; Picone, A.F.; Vidal, S.E.; Vedvick, T.S.; Howard, R.F.; Reed, S.G. KSAC, the first defined polyprotein vaccine candidate for visceral leishmaniasis. Clin. Vaccine Immunol. 2011, 18, 1118–1124. [Google Scholar] [CrossRef]
- Molano, I.; Alonso, M.G.; Miron, C.; Redondo, E.; Requena, J.M.; Soto, M.; Nieto, C.G.; Alonso, C. A Leishmania infantum multi-component antigenic protein mixed with live BCG confers protection to dogs experimentally infected with L-infantum. Vet. Immunol. Immunopathol. 2003, 92, 1–13. [Google Scholar] [CrossRef]
- Joshi, S.; Rawat, K.; Yadav, N.K.; Kumar, V.; Siddiqi, M.I.; Dube, A. Visceral leisnmaniasis: Advancements in vaccine development via classical and molecular approaches. Front. Immunol. 2014, 5, 380. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.D.E.; Ferreira, L.F.G.R.; Hernandes, M.Z.; de Brito, M.E.F.; de Oliveira, B.C.; da Silva, A.A.; de-Melo-Neto, O.P.; Rezende, A.M.; Pereira, V.R.A. Combination of In Silico Methods in the Search for Potential CD4(+) and CD8(+) T Cell Epitopes in the Proteome of Leishmania braziliensis. Front. Immunol. 2016, 7, 327. [Google Scholar] [CrossRef]
- Khatoon, N.; Pandey, R.K.; Ojha, R.; Aathmanathan, V.S.; Krishnan, M.; Prajapati, V.K. Exploratory algorithm to devise multi-epitope subunit vaccine by investigating Leishmania donovani membrane proteins. J. Biomol. Struct. Dyn. 2019, 37, 2381–2393. [Google Scholar] [CrossRef]
- Khatoon, N.; Pandey, R.K.; Prajapati, V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017, 7, 8285. [Google Scholar] [CrossRef]
- Onile, O.S.; Musaigwa, F.; Ayawei, N.; Omoboyede, V.; Onile, T.A.; Oghenevovwero, E.; Aruleba, R.T. Immunoinformatics Studies and Design of a Potential Multi-Epitope Peptide Vaccine to Combat the Fatal Visceral Leishmaniasis. Vaccines 2022, 10, 1598. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Vashishtha, S.; Kundu, B.; Ghosh, M. In-silico design of an immunoinformatics based multi-epitope vaccine against Leishmania donovani. BMC Bioinform. 2022, 23, 319. [Google Scholar] [CrossRef] [PubMed]
- Vakili, B.; Eslami, M.; Hatam, G.R.; Zare, B.; Erfani, N.; Nezafat, N.; Ghasemi, Y. Immunoinformatics-aided design of a potential multi-epitope peptide vaccine against Leishmania infantum. Int. J. Biol. Macromol. 2018, 120, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Vakili, B.; Nezafat, N.; Hatam, G.R.; Zare, B.; Erfani, N.; Ghasemi, Y. Proteome-scale identification of Leishmania infantum for novel vaccine candidates: A hierarchical subtractive approach. Comput. Biol. Chem. 2018, 72, 16–25. [Google Scholar] [CrossRef]
- Bhowmick, S.; Ali, N. Identification of Novel Leishmania donovani Antigens that Help Define Correlates of Vaccine-Mediated Protection in Visceral Leishmaniasis. PLoS ONE 2009, 4, e5820. [Google Scholar] [CrossRef] [PubMed]
- Iborra, S.; Parody, N.; Abanades, D.R.; Bonay, P.; Prates, D.; Novais, F.O.; Barral-Netto, M.; Alonso, C.; Soto, M. Vaccination with the Leishmania major ribosomal proteins plus CpG oligodeoxynucleotides induces protection against experimental cutaneous leishmaniasis in mice. Microbes Infect. 2008, 10, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.W.; Santos, D.M.; Fukutani, K.F.; Clarencio, J.; Miranda, J.C.; Brodskyn, C.; Barral, A.; Barral-Netto, M.; Soto, M.; de Oliveira, C.I. Vaccination with L. infantum chagasi Nucleosomal Histones Confers Protection against New World Cutaneous Leishmaniasis Caused by Leishmania braziliensis. PLoS ONE 2012, 7, e52296. [Google Scholar] [CrossRef]
- Santarem, N.; Silvestre, R.; Tavares, J.; Silva, M.; Cabral, S.; Maciel, J.; Cordeiro-da-Silva, A. Immune response regulation by Leishmania secreted and nonsecreted antigens. J. Biomed. Biotechnol. 2007, 2007, 085154. [Google Scholar] [CrossRef]
- Clark, E.A.; Ledbetter, J.A. How B-Cells and T-Cells Talk to Each Other. Nature 1994, 367, 425–428. [Google Scholar] [CrossRef]
- Paulnock, D.M. Macrophage Activation by T-Cells. Curr. Opin. Immunol. 1992, 4, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Tscharke, D.C.; Croft, N.P.; Doherty, P.C.; La Gruta, N.L. Sizing up the key determinants of the CD8(+) T cell response. Nat. Rev. Immunol. 2015, 15, 705–716. [Google Scholar] [CrossRef]
- Rothbard, J.B.; Taylor, W.R. A Sequence Pattern Common to T-Cell Epitopes. EMBO J. 1988, 7, 93–100. [Google Scholar] [CrossRef]
- Dias, D.S.; Ribeiro, P.A.F.; Martins, V.T.; Lage, D.P.; Costa, L.E.; Chavez-Fumagalli, M.A.; Ramos, F.F.; Santos, T.T.O.; Ludolf, F.; Oliveira, J.S.; et al. Vaccination with a CD4(+) and CD8(+) T-cell epitopes-based recombinant chimeric protein derived from Leishmania infantum proteins confers protective immunity against visceral leishmaniasis. Transl. Res. 2018, 200, 18–34. [Google Scholar] [CrossRef]
- Khan, M.A.A.; Ami, J.Q.; Faisal, K.; Chowdhury, R.; Ghosh, P.; Hossain, F.; Abd El Wahed, A.; Mondal, D. An immunoinformatic approach driven by experimental proteomics: In silico design of a subunit candidate vaccine targeting secretory proteins of Leishmania donovani amastigotes. Parasite Vector 2020, 13, 196. [Google Scholar] [CrossRef]
- Ribeiro, P.A.F.; Dias, D.S.; Lage, D.P.; Martins, V.T.; Costa, L.E.; Santos, T.T.O.; Ramos, F.F.; Tavares, G.S.V.; Mendonca, D.V.C.; Ludolf, F.; et al. Immunogenicity and protective efficacy of a new Leishmania hypothetical protein applied as a DNA vaccine or in a recombinant form against Leishmania infantum infection. Mol. Immunol. 2019, 106, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, P.A.F.; Dias, D.S.; Novais, M.V.M.; Lage, D.P.; Tavares, G.S.V.; Mendonca, D.V.C.; Oliveira, J.S.; Chavez-Fumagalli, M.A.; Roatt, B.M.; Duarte, M.C.; et al. A Leishmania hypothetical protein-containing liposome-based formulation is highly immunogenic and induces protection against visceral leishmaniasis. Cytokine 2018, 111, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Vakili, B.; Nezafat, N.; Zare, B.; Erfani, N.; Akbari, M.; Ghasemi, Y.; Rahbar, M.R.; Hatam, G.R. A new multi-epitope peptide vaccine induces immune responses and protection against Leishmania infantum in BALB/c mice. Med. Microbiol. Immunol. 2020, 209, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Prakash, J.; Shukla, H.; Das, K.C.; Tripathi, T.; Dubey, V.K. Design a multi-epitope subunit vaccine for immune-protection against Leishmania parasite. Pathog. Glob. Health 2020, 114, 471–481. [Google Scholar] [CrossRef]
- Martins, V.T.; Duarte, M.C.; Chavez-Fumagalli, M.A.; Menezes-Souza, D.; Coelho, C.S.P.; de Magalhaes-Soares, D.F.; Fernandes, A.P.; Soto, M.; Tavares, C.A.P.; Coelho, E.A.F. A Leishmania-specific hypothetical protein expressed in both promastigote and amastigote stages of Leishmania infantum employed for the serodiagnosis of, and as a vaccine candidate against, visceral leishmaniasis. Parasite Vector 2015, 8, 363. [Google Scholar] [CrossRef]
- Efstathiou, A.; Smirlis, D. Leishmania Protein Kinases: Important Regulators of the Parasite Life Cycle and Molecular Targets for Treating Leishmaniasis. Microorganisms 2021, 9, 691. [Google Scholar] [CrossRef]
- Kumari, D.; Mahajan, S.; Kour, P.; Singh, K. Virulence factors of Leishmania parasite: Their paramount importance in unraveling novel vaccine candidates and therapeutic targets. Life Sci. 2022, 306, 120829. [Google Scholar] [CrossRef] [PubMed]
- Seyed, N.; Taheri, T.; Rafati, S. Post-Genomics and Vaccine Improvement for Leishmania. Front. Microbiol. 2016, 7, 467. [Google Scholar] [CrossRef]
- World Health Organization. WHO Technical Report Series 962. WHO Expert Committee on Biological Standardization, 57th Report. Available online: https://www.who.int/publications/i/item/9789241209625 (accessed on 5 December 2022).
- Pandey, R.K.; Bhatt, T.K.; Prajapati, V.K. Novel Immunoinformatics Approaches to Design Multi-epitope Subunit Vaccine for Malaria by Investigating Anopheles Salivary Protein. Sci. Rep. 2018, 8, 1125. [Google Scholar] [CrossRef]
- Chen, X.Y.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Cunningham, A.C. Parasitic adaptive mechanisms in infection by Leishmania. Exp. Mol. Pathol. 2002, 72, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Murray, H.W.; Cartelli, D.M. Killing of Intracellular Leishmania-Donovani by Human Mononuclear Phagocytes—Evidence for Oxygen-Dependent and Oxygen-Independent Leishmanicidal Activity. J. Clin. Investig. 1983, 72, 32–44. [Google Scholar] [CrossRef]
- Basu, A.; Chakrabarti, G.; Saha, A.; Bandyopadhyay, S. Modulation of CD11C(+) splenic dendritic cell functions in murine visceral leishmaniasis: Correlation with parasite replication in the spleen. Immunology 2000, 99, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Engwerda, C.R.; Kaye, P.M. Organ-specific immune responses associated with infectious disease. Immunol. Today 2000, 21, 73–78. [Google Scholar] [CrossRef]
- Bodas, M.; Jain, N.; Awasthi, A.; Martin, S.; Loka, R.K.P.; Dandekar, D.; Mitra, D.; Saha, B. Inhibition of IL-2 induced IL-10 production as a principle of phase-specific immunotherapy. J. Immunol. 2006, 177, 4636–4643. [Google Scholar] [CrossRef]
- Pippa, N.; Gazouli, M.; Pispas, S. Recent Advances and Future Perspectives in Polymer-Based Nanovaccines. Vaccines 2021, 9, 558. [Google Scholar] [CrossRef]
- Agallou, M.; Margaroni, M.; Athanasiou, E.; Toubanaki, D.K.; Kontonikola, K.; Karidi, K.; Kammona, O.; Kiparissides, C.; Karagouni, E. Identification of BALB/c Immune Markers Correlated with a Partial Protection to Leishmania infantum after Vaccination with a Rationally Designed Multi-epitope Cysteine Protease A Peptide-Based Nanovaccine. PLoS Neglect. Trop. Dis. 2017, 11, e0005311. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, E.; Agallou, M.; Tastsoglou, S.; Kammona, O.; Hatzigeorgiou, A.; Kiparissides, C.; Karagouni, E. A Poly(Lactic-co-Glycolic) Acid Nanovaccine Based on Chimeric Peptides from Different Leishmania infantum Proteins Induces Dendritic Cells Maturation and Promotes Peptide-Specific IFN gamma-Producing CD8(+) T Cells Essential for the Protection against Experimental Visceral Leishmaniasis. Front. Immunol. 2017, 8, 684. [Google Scholar] [CrossRef]
- Santos, D.M.; Carneiro, M.W.; de Moura, T.R.; Fukutani, K.; Clarencio, J.; Soto, M.; Espuelas, S.; Brodskyn, C.; Barral, A.; Barral-Netto, M.; et al. Towards development of novel immunization strategies against leishmaniasis using PLGA nanoparticles loaded with kinetoplastid membrane protein-11. Int. J. Nanomed. 2012, 7, 2115–2127. [Google Scholar] [CrossRef]
- Tafaghodi, M.; Eskandari, M.; Kharazizadeh, M.; Khamesipour, A.; Jaafari, M.R. Immunization against leishmaniasis by PLGA nanospheres loaded with an experimental autoclaved Leishmania major (ALM) and Quillaja saponins. Trop. Biomed. 2010, 27, 639–650. [Google Scholar]
- De Brito, R.C.F.; Ruiz, J.C.; Cardoso, J.M.D.; Ostolin, T.L.V.D.; Reis, L.E.S.; Mathias, F.A.S.; Aguiar-Soares, R.D.D.; Roatt, B.M.; Correa-Oliveira, R.; Resende, D.D.; et al. Chimeric Vaccines Designed by Immunoinformatics-Activated Polyfunctional and Memory T cells that Trigger Protection against Experimental Visceral Leishmaniasis. Vaccines 2020, 8, 252. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.; Claro, M.; Alexandre-Pires, G.; Santos-Mateus, D.; Martins, C.; Valerio-Bolas, A.; Rafael-Fernandes, M.; Pereira, M.A.; da Fonseca, I.P.; Tomas, A.M.; et al. Leishmania infanturn antigens modulate memory cell subsets of liver resident T lymphocyte. Immunobiology 2017, 222, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Sabur, A.; Bhowmick, S.; Chhajer, R.; Ejazi, S.A.; Didwania, N.; Asad, M.; Bhattacharyya, A.; Sinha, U.; Ali, N. Liposomal Elongation Factor-1 alpha Triggers Effector CD4 and CD8 T Cells for Induction of Long-Lasting Protective Immunity against Visceral Leishmaniasis. Front. Immunol. 2018, 9, 18. [Google Scholar] [CrossRef]
- Hofmeyer, K.A.; Duthie, M.S.; Laurance, J.D.; Favila, M.A.; Van Hoeven, N.; Coler, R.N.; Reed, S.G. Optimizing Immunization Strategies for the Induction of Antigen-Specific CD4 and CD8 T Cell Responses for Protection against Intracellular Parasites. Clin. Vaccine Immunol. 2016, 23, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Hohman, L.S.; Peters, N.C. CD4(+) T Cell-Mediated Immunity against the Phagosomal Pathogen Leishmania: Implications for Vaccination. Trends Parasitol. 2019, 35, 423–435. [Google Scholar] [CrossRef]
- Dey, R.; Dagur, P.K.; Selvapandiyan, A.; McCoy, J.P.; Salotra, P.; Duncan, R.; Nakhasi, H.L. Live Attenuated Leishmania donovani p27 Gene Knockout Parasites Are Nonpathogenic and Elicit Long-Term Protective Immunity in BALB/c Mice. J. Immunol. 2013, 190, 2138–2149. [Google Scholar] [CrossRef]
- Agallou, M.; Margaroni, M.; Kotsakis, S.D.; Karagouni, E. A Canine-Directed Chimeric Multi-Epitope Vaccine Induced Protective Immune Responses in BALB/c Mice Infected with Leishmania Infantum. Vaccines 2020, 8, 350. [Google Scholar] [CrossRef]
- Basu, R.; Roy, S.; Walden, P. HLA class I-restricted T cell epitopes of the kinetoplastid membrane protein-11 presented by Leishmania donovani-infected human macrophages. J. Infect. Dis. 2007, 195, 1373–1380. [Google Scholar] [CrossRef]
- Stager, S.; Alexander, J.; Kirby, A.C.; Botto, M.; Van Rooijen, N.; Smith, D.F.; Brombacher, F.; Kaye, P.M. Natural antibodies and complement are endogenous adjuvants for vaccine-induced CD8(+) T-cell responses. Nat. Med. 2003, 9, 1287–1292. [Google Scholar] [CrossRef]
- Heit, A.; Schmitz, F.; Haas, T.; Busch, D.H.; Wagner, H. Antigen co-encapsulated with adjuvants efficiently drive protective T cell immunity. Eur. J. Immunol. 2007, 37, 2063–2074. [Google Scholar] [CrossRef]
- Silva, A.L.; Rosalia, R.A.; Varypataki, E.; Sibuea, S.; Ossendorp, F.; Jiskoot, W. Poly-(lactic-co-glycolic-acid)-based particulate vaccines: Particle uptake by dendritic cells is a key parameter for immune activation. Vaccine 2015, 33, 847–854. [Google Scholar] [CrossRef] [PubMed]

| No | Sequence ID | Function | Cellular Localization 1 | Antigenicity (VaxiJen 2, ANTIGENPro 3) |
|---|---|---|---|---|
| 1 | LinJ.35.1030 | casein kinase, putative | Cytoplasm | 0.5737/0.836 |
| 2 | LinJ.35.1540 | rieske iron-sulfuric protein, mitochondrion precursor | Cytoplasm | 0.5906/0.785 |
| 3 | LinJ.08.1290 | beta tubulin | Cytoplasm | 0.5436/0.764 |
| 4 | LinJ.21.2240 | beta tubulin | Cytoplasm | 0.5349/0.780 |
| 5 | LinJ.36.5660 | branch point binding protein, putative | Cytoplasm | 0.7933/0.917 |
| 6 | LinJ.08.1280 | beta tubulin | Cytoplasm | 0.5334/0.784 |
| 7 | LinJ.28.0780 | hypothetical protein | Cytoplasm | 0.5545/0.652 |
| 8 | LinJ.28.2940 | receptor for activated C kinase 1 | Extracellular | 0.6190/0.814 |
| 9 | LinJ.22.1300 | cyclophilin 6, putative | Extracellular | 0.5135/0.775 |
| 10 | LinJ.16.0470 | 60S ribosomal protein L21, putative | Mitochondria | 0.5437/0.665 |
| 11 | LinJ.20.0600 | conserved protein, unknown function | Mitochondria | 0.6737/0.735 |
| 12 | LinJ.34.0420 | hypothetical protein, conserved | Mitochondria | 0.5455/0.779 |
| 13 | LinJ.34.3620 | ribosomal protein L14, putative | Mitochondria | 0.8221/0.624 |
| 14 | LinJ.35.3810 | 60S ribosomal protein L27A/L29, putative | Mitochondria | 0.7317/0.754 |
| 15 | LinJ.24.0040 | 60S ribosomal protein L17, putative | Mitochondria | 0.6032/0.850 |
| 16 | LinJ.20.0250 | transmembrane protein, putative | Nucleus | 0.5086/0.576 |
| 17 | LinJ.18.0650 | serine/threonine kinase-like protein, putative | Nucleus | 0.5320/0.546 |
| 18 | LinJ.33.1560 | RNA-binding protein, putative | Nucleus | 0.5448/0.809 |
| 19 | LinJ.28.2200 | DNA-directed RNA polymerase-like protein | Nucleus | 0.5213/0.918 |
| 20 | LinJ.10.0050 | ribosomal protein l35a, putative | Nucleus | 0.6450/0.869 |
| 21 | LinJ.28.0210 | Histone H2B variant V | Nucleus | 0.5420/0.616 |
| 22 | LinJ.32.0930 | 60S ribosomal protein L18a, putative | Nucleus | 0.5872/0.509 |
| 23 | LinJ.33.3340 | small nuclear ribonucleoprotein SmD2 | Nucleus | 0.5678/0.534 |
| 24 | LinJ.36.6680 | 40S ribosomal protein S8, putative | Nucleus | 0.8525/0.616 |
| 25 | LinJ.21.0440 | Protein of unknown function (DUF667) | Nucleus | 0.5903/0.840 |
| 26 | LinJ.27.1450 | DNA-directed RNA polymerase II-like protein | Nucleus | 0.5001/0.731 |
| No | Protein (Code) | No of MHCI Epitopes | No of MHCII Epitopes | ||
|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | ||
| 1 | LinJ.08.1290 | 2 | 13 | 4 | 38 |
| 2 | LinJ.08.1280 | 2 | 13 | 3 | 38 |
| 3 | LinJ.21.2240 | 2 | 13 | 3 | 40 |
| 4 | LinJ.22.1300 | 2 | 6 | 4 | 18 |
| 5 | LinJ.21.0440 | 3 | 6 | 5 | 24 |
| 6 | LinJ.36.6680 | 3 | 6 | 3 | 61 |
| 7 | LinJ.16.0470 | 0 | 5 | 0 | 2 |
| 8 | LinJ.32.0930 | 1 | 5 | 0 | 5 |
| 9 | LinJ.24.0040 | 2 | 4 | 0 | 9 |
| 10 | LinJ.34.0420 | 2 | 4 | 1 | 29 |
| 11 | LinJ.18.0650 | 0 | 4 | 2 | 35 |
| 12 | LinJ.28.2200 | 3 | 3 | 0 | 21 |
| 13 | LinJ.28.0780 | 1 | 3 | 0 | 50 |
| 14 | LinJ.35.1540 | 6 | 3 | 2 | 64 |
| 15 | LinJ.20.0250 | 5 | 3 | 1 | 37 |
| 16 | LinJ.36.5660 | 4 | 2 | 3 | 38 |
| 17 | LinJ.34.3620 | 1 | 2 | 3 | 55 |
| 18 | LinJ.33.1560 | 3 | 2 | 1 | 10 |
| 19 | LinJ.33.3340 | 1 | 2 | 5 | 3 |
| 20 | LinJ.35.3810 | 1 | 1 | 3 | 3 |
| 21 | LinJ.28.0210 | 1 | 1 | 4 | 44 |
| 22 | LinJ.10.0050 | 4 | 1 | 0 | 39 |
| 23 | LinJ.20.0600 | 0 | 1 | 6 | 25 |
| 24 | LinJ.27.1450 | 3 | 0 | 0 | 6 |
| 25 | LinJ.35.1030 | 2 | 11 | 4 | 26 |
| 26 | LinJ.28.2940 | 2 | 5 | 0 | 35 |
| No | Protein (Code) | Number of MHCI Epitopes | Number of MHCII Epitopes | (%) Coverage of Amino Acid Sequence | Homology Human/Mouse |
|---|---|---|---|---|---|
| 1 | LinJ.34.0420 | 4 | 2 | 30.8% | no |
| 2 | LinJ.18.0650 | 4 | 9 | 23.6% | 41.29/42.97 |
| 3 | LinJ.28.2200 | 3 | 4 | 46.7% | 40.26/29.92 |
| 4 | LinJ.28.0780 | 3 | 50 | 81.1% | no |
| 5 | LinJ.20.0600 | 1 | 14 | 38.2% | no |
| Physicochemical Characteristic | Amino Acid Number | Molecular Weight | Instability Index | GRAVY | Half Life | Aliphatic Index | Theoretical pI |
|---|---|---|---|---|---|---|---|
| Score | 407 | 46,757.76 Da | 45.65 | −0.338 | >10 h (E. coli, in vivo) | 85.11 | 9.05 |
| Protein | HLA Alleles (PDB ID) | Global Energy 1 | aVdW 2 | rVdW 3 | ACE 4 | HB 5 |
|---|---|---|---|---|---|---|
| LeishChim | HLA-A*0201 (PDB: 1I4F) | 2.37 | −4.43 | 0.79 | −0.01 | −0.22 |
| HLA-A*0101 (PDB: 6AT9) | −12.54 | −19.50 | 10.78 | 3.91 | −2.85 | |
| HLA-B*0702 (PDB: 5EO0) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| HLA-B*3501 (PDB:2FYY) | 5.58 | −21.45 | 13.08 | 11.14 | −0.52 | |
| HLA-DRB1*03:01 (PDB: 1A6A) | −22.06 | −40.07 | 21.56 | 11.65 | −3.75 | |
| HLA-DRB5*01:01 (PDB: 1H15) | −2.69 | −34.44 | 38.98 | 1.97 | −4.19 | |
| HLA-DRB1*01:01 (PDB: 2FSE) | −17.35 | −26.01 | 11.39 | 6.73 | −0.35 | |
| HLA-DRB3*02:02 (PDB: 3C5J) | 0.43 | −6.65 | 3.13 | 2.47 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margaroni, M.; Agallou, M.; Tsanaktsidou, E.; Kammona, O.; Kiparissides, C.; Karagouni, E. Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses. Vaccines 2023, 11, 304. https://doi.org/10.3390/vaccines11020304
Margaroni M, Agallou M, Tsanaktsidou E, Kammona O, Kiparissides C, Karagouni E. Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses. Vaccines. 2023; 11(2):304. https://doi.org/10.3390/vaccines11020304
Chicago/Turabian StyleMargaroni, Maritsa, Maria Agallou, Evgenia Tsanaktsidou, Olga Kammona, Costas Kiparissides, and Evdokia Karagouni. 2023. "Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses" Vaccines 11, no. 2: 304. https://doi.org/10.3390/vaccines11020304
APA StyleMargaroni, M., Agallou, M., Tsanaktsidou, E., Kammona, O., Kiparissides, C., & Karagouni, E. (2023). Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses. Vaccines, 11(2), 304. https://doi.org/10.3390/vaccines11020304







