Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vaccines and Challenge Viruses
2.2. Characterization of NanoS100–SwIAV or Nano11–SwIAV
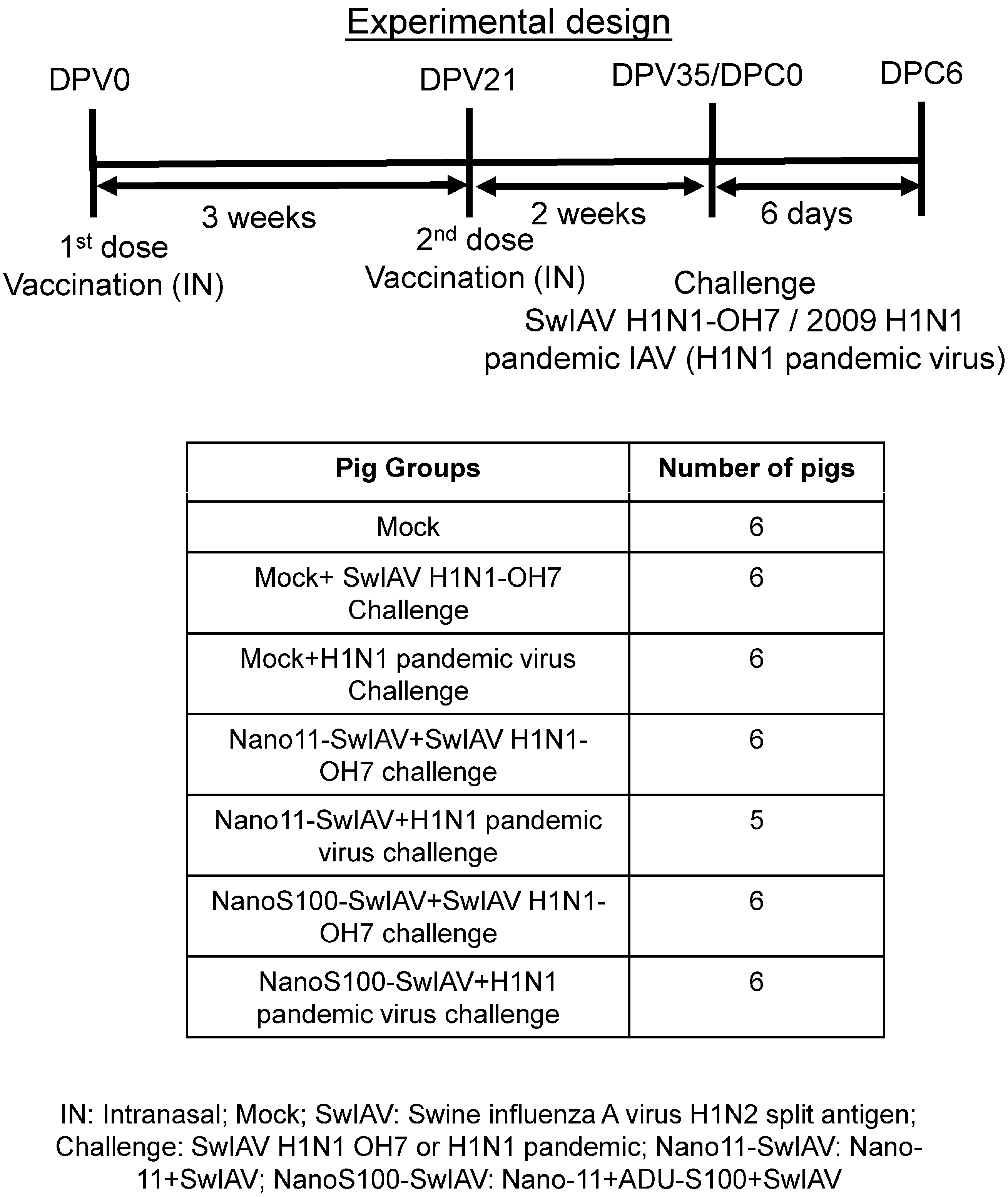
2.3. Animal Studies
2.4. Surface and Intracellular Cytokine Labeling for Flow Cytometry
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Virus Titration
2.7. Statistical Analysis
3. Results
3.1. Characterization of the Vaccine Formulations
3.2. NanoS100–SwIAV Vaccine Partially Decreased the Challenge Virus Titers in the Respiratory Tract of Conventional Pigs
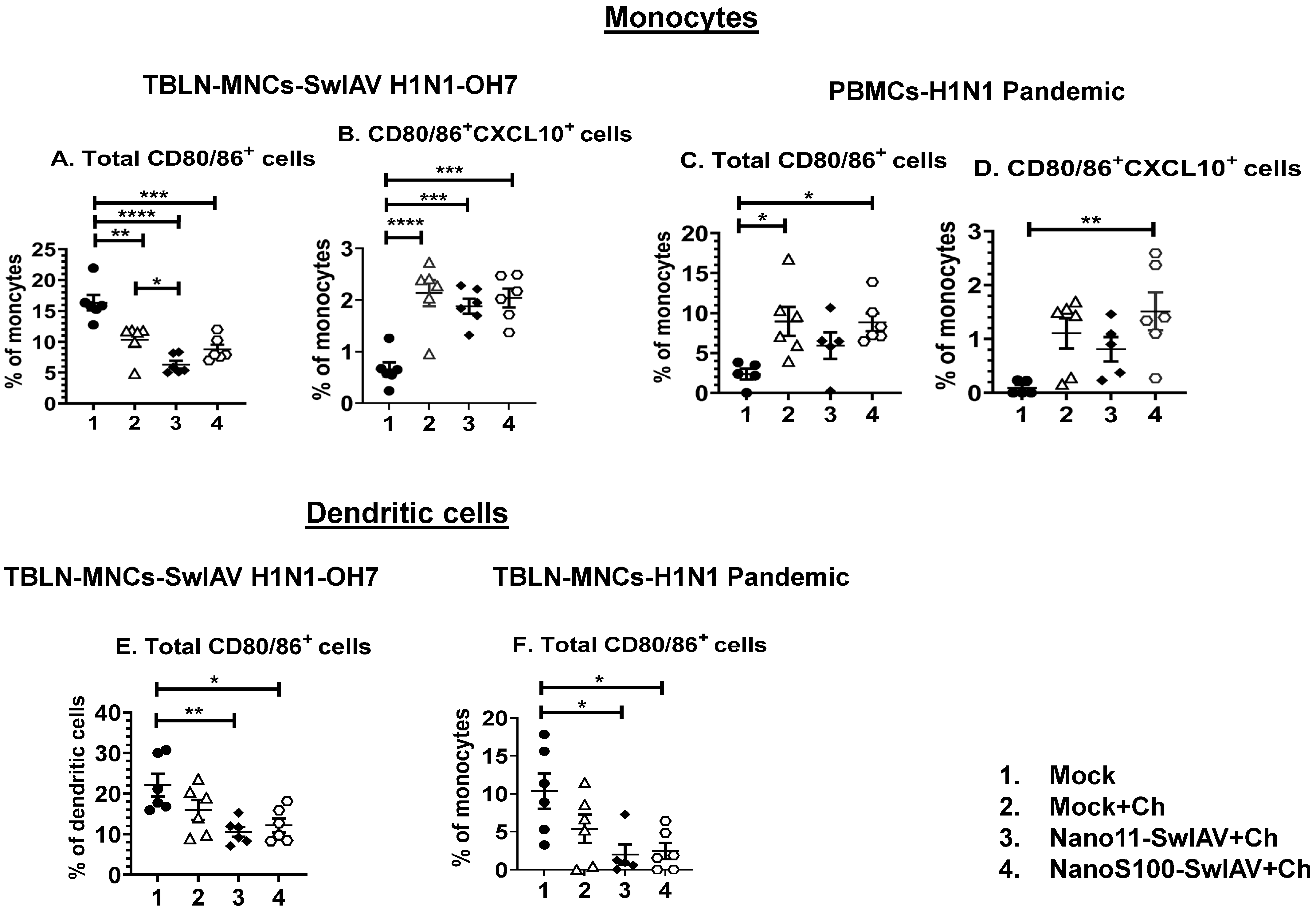
3.3. Both Nano11–SwIAV and NanoS100–SwIAV Vaccines Downregulated the Frequencies of Activated Dendritic Cells (CD3−CD172a+SynCAM+CD80/86+) and Enhanced the Activated Monocytes (CD3−CD172a+SynCAM−CXCL10+CD80/86+) in Both Mucosal and Systemic Compartments of Vaccinates at DPC6
3.4. Both the Candidate Nano11-Based Vaccines Elicited Strong Virus-Specific IL-17A and IFNγ Secreting Recall CTL Responses in The Lung Draining TBLN of Vaccinates
3.5. Both of the Candidate Nano11 Vaccines Elicited the Challenge Virus-Specific IFNγ Secreting T Cell Responses in PBMCs of Vaccinated Pigs
3.6. Both of the Candidate Vaccines in SwIAV-H1N1-OH7-Challenged Pigs Elicited Cross-Reactive Virus-Specific Mucosal Antibody Response in Vaccinates at DPC6
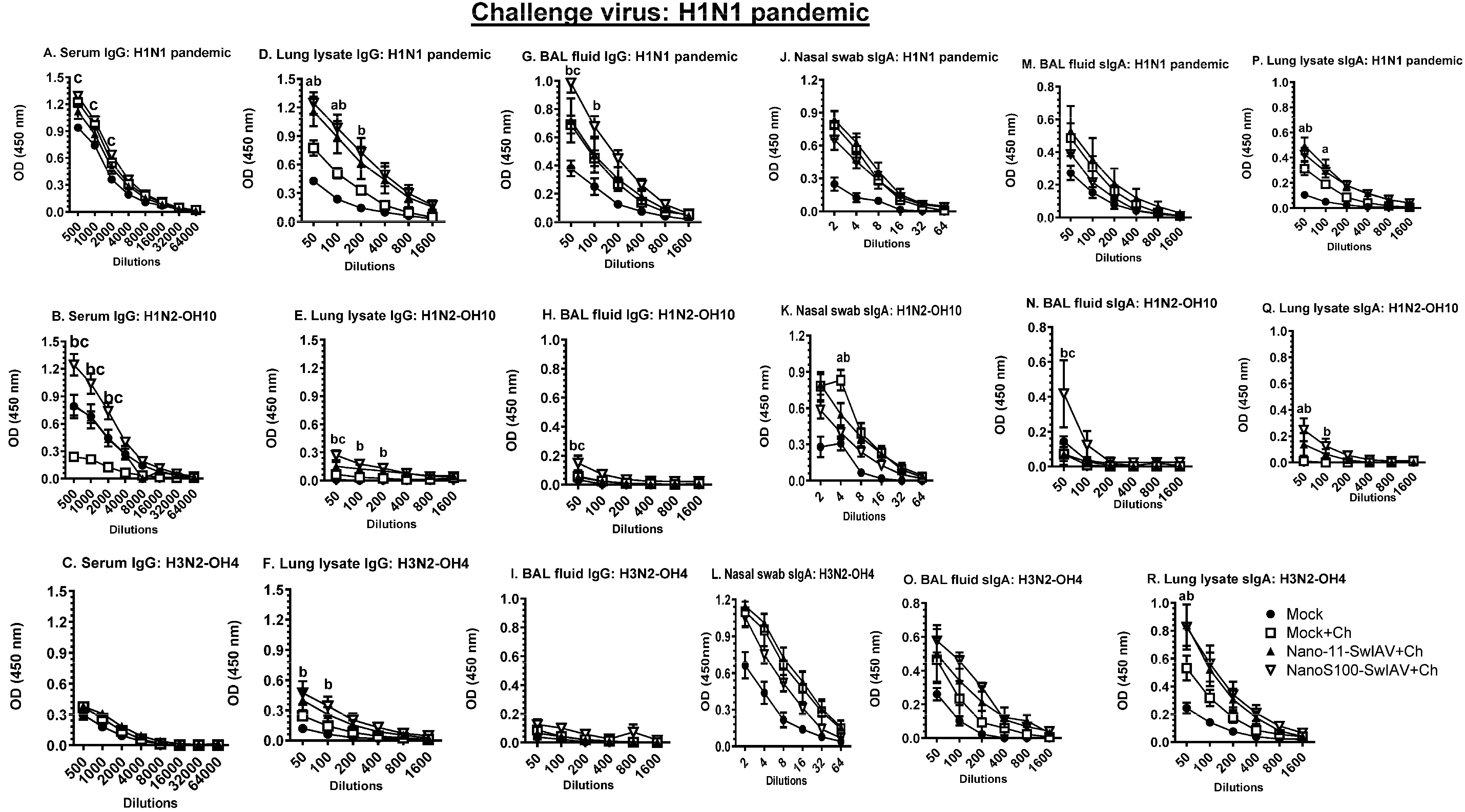
3.7. NanoS100–SwIAV Vaccine Elicited Both Cross-Reactive H1N1 Pandemic Virus-Specific Lung and Systemic IgG and Lung sIgA Responses in Vaccinates at DPC6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, T.K.; Chang, J.; Arendsee, Z.W.; Venkatesh, D.; Souza, C.K.; Kimble, J.B.; Lewis, N.S.; Davis, C.T.; Vincent, A.L. Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb. Perspect. Med. 2021, 11, a038737. [Google Scholar] [CrossRef]
- Ma, W. Swine influenza virus: Current status and challenge. Virus Res. 2020, 288, 198118. [Google Scholar] [CrossRef]
- Hayward, A.C.; Wang, L.; Goonetilleke, N.; Fragaszy, E.B.; Bermingham, A.; Copas, A.; Dukes, O.; Millett, E.R.; Nazareth, I.; Nguyen-Van-Tam, J.S.; et al. Natural T Cell-mediated Protection against Seasonal and Pandemic Influenza. Results of the Flu Watch Cohort Study. Am. J. Respir. Crit. Care Med. 2015, 191, 1422–1431. [Google Scholar] [CrossRef]
- Gras, S.; Kedzierski, L.; Valkenburg, S.A.; Laurie, K.; Liu, Y.C.; Denholm, J.T.; Richards, M.J.; Rimmelzwaan, G.F.; Kelso, A.; Doherty, P.C.; et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc. Natl. Acad. Sci. USA 2010, 107, 12599–12604. [Google Scholar] [CrossRef]
- Christensen, J.P.; Doherty, P.C.; Branum, K.C.; Riberdy, J.M. Profound protection against respiratory challenge with a lethal H7N7 influenza A virus by increasing the magnitude of CD8(+) T-cell memory. J. Virol. 2000, 74, 11690–11696. [Google Scholar] [CrossRef]
- Epstein, S.L. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: An experiment of nature. J. Infect. Dis. 2006, 193, 49–53. [Google Scholar] [CrossRef]
- Kreijtz, J.H.; de Mutsert, G.; van Baalen, C.A.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J. Virol. 2008, 82, 5161–5166. [Google Scholar] [CrossRef]
- Lee, L.Y.; Ha, D.L.A.; Simmons, C.; de Jong, M.D.; Chau, N.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar] [CrossRef]
- Morgan, S.B.; Hemmink, J.D.; Porter, E.; Harley, R.; Shelton, H.; Aramouni, M.; Everett, H.E.; Brookes, S.M.; Bailey, M.; Townsend, A.M.; et al. Aerosol Delivery of a Candidate Universal Influenza Vaccine Reduces Viral Load in Pigs Challenged with Pandemic H1N1 Virus. J. Immunol. 2016, 196, 5014–5023. [Google Scholar] [CrossRef]
- Dhakal, S.; Renukaradhya, G.J. Nanoparticle-based vaccine development and evaluation against viral infections in pigs. Vet. Res. 2019, 50, 90. [Google Scholar] [CrossRef]
- Chan, Y.; Ng, S.W.; Singh, S.K.; Gulati, M.; Gupta, G.; Chaudhary, S.K.; Hing, G.B.; Collet, T.; MacLoughlin, R.; Lobenberg, R.; et al. Revolutionizing polymer-based nanoparticle-linked vaccines for targeting respiratory viruses: A perspective. Life Sci. 2021, 280, 119744. [Google Scholar] [CrossRef]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines Against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef]
- Patil, V.; Hernandez-Franco, J.F.; HogenEsch, H.; Renukaradhya, G.J. Alpha-D-glucan-based vaccine adjuvants: Current status and future perspectives. Front. Immunol. 2022, 13, 858321. [Google Scholar] [CrossRef]
- Verschoor, C.P.; Haynes, L.; Pawelec, G.; Loeb, M.; Andrew, M.K.; Kuchel, G.A.; McElhaney, J.E. Corrigendum: Key Determinants of Cell-Mediated Immune Responses: A Randomized Trial of High Dose vs. Standard Dose Split-Virus Influenza Vaccine in Older Adults. Front. Aging 2021, 2, 718966. [Google Scholar] [CrossRef]
- Wagar, L.E.; Rosella, L.; Crowcroft, N.; Lowcock, B.; Drohomyrecky, P.C.; Foisy, J.; Gubbay, J.; Rebbapragada, A.; Winter, A.L.; Achonu, C.; et al. Humoral and cell-mediated immunity to pandemic H1N1 influenza in a Canadian cohort one year post-pandemic: Implications for vaccination. PLoS ONE 2011, 6, e28063. [Google Scholar] [CrossRef]
- Chotpitayasunondh, T.; Thisyakorn, U.; Pancharoen, C.; Pepin, S.; Nougarede, N. Safety, humoral and cell mediated immune responses to two formulations of an inactivated, split-virion influenza A/H5N1 vaccine in children. PLoS ONE 2008, 3, e4028. [Google Scholar] [CrossRef]
- Patil, V.; Hernandez-Franco, J.F.; Yadagiri, G.; Bugybayeva, D.; Dolatyabi, S.; Feliciano-Ruiz, N.; Schrock, J.; Hanson, J.; Ngunjiri, J.; HogenEsch, H.; et al. A split influenza vaccine formulated with a combination adjuvant composed of alpha-D-glucan nanoparticles and a STING agonist elicits cross-protective immunity in pigs. J. Nanobiotechnol. 2022, 20, 477. [Google Scholar] [CrossRef]
- Stephenson, I.; Nicholson, K.G.; Gluck, R.; Mischler, R.; Newman, R.W.; Palache, A.M.; Verlander, N.Q.; Warburton, F.; Wood, J.M.; Zambon, M.C. Safety and antigenicity of whole virus and subunit influenza A/Hong Kong/1073/99 (H9N2) vaccine in healthy adults: Phase I randomised trial. Lancet 2003, 362, 1959–1966. [Google Scholar] [CrossRef]
- O’Gorman, W.E.; Huang, H.; Wei, Y.L.; Davis, K.L.; Leipold, M.D.; Bendall, S.C.; Kidd, B.A.; Dekker, C.L.; Maecker, H.T.; Chien, Y.H.; et al. The Split Virus Influenza Vaccine rapidly activates immune cells through Fcgamma receptors. Vaccine 2014, 32, 5989–5997. [Google Scholar] [CrossRef]
- Ali, A.; Khatri, M.; Wang, L.; Saif, Y.M.; Lee, C.W. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet. Microbiol. 2012, 158, 60–68. [Google Scholar] [CrossRef]
- Yassine, H.M.; Khatri, M.; Zhang, Y.J.; Lee, C.W.; Byrum, B.A.; O’Quin, J.; Smith, K.A.; Saif, Y.M. Characterization of triple reassortant H1N1 influenza A viruses from swine in Ohio. Vet. Microbiol. 2009, 139, 132–139. [Google Scholar] [CrossRef]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef]
- Yassine, H.M.; Al-Natour, M.Q.; Lee, C.W.; Saif, Y.M. Interspecies and intraspecies transmission of triple reassortant H3N2 influenza A viruses. Virol. J. 2007, 4, 129. [Google Scholar] [CrossRef]
- Hernandez-Franco, J.F.; Mosley, Y.C.; Franco, J.; Ragland, D.; Yao, Y.; HogenEsch, H. Effective and Safe Stimulation of Humoral and Cell-Mediated Immunity by Intradermal Immunization with a Cyclic Dinucleotide/Nanoparticle Combination Adjuvant. J. Immunol. 2021, 206, 700–711. [Google Scholar] [CrossRef]
- Dhakal, S.; Hiremath, J.; Bondra, K.; Lakshmanappa, Y.S.; Shyu, D.L.; Ouyang, K.; Kang, K.I.; Binjawadagi, B.; Goodman, J.; Tabynov, K.; et al. Biodegradable nanoparticle delivery of inactivated swine influenza virus vaccine provides heterologous cell-mediated immune response in pigs. J. Control Release 2017, 247, 194–205. [Google Scholar] [CrossRef]
- Auray, G.; Keller, I.; Python, S.; Gerber, M.; Bruggmann, R.; Ruggli, N.; Summerfield, A. Characterization and Transcriptomic Analysis of Porcine Blood Conventional and Plasmacytoid Dendritic Cells Reveals Striking Species-Specific Differences. J. Immunol. 2016, 197, 4791–4806. [Google Scholar] [CrossRef]
- Li, Y.; Puebla-Clark, L.; Hernandez, J.; Diaz, I.; Mateu, E. Development of Pig Conventional Dendritic Cells From Bone Marrow Hematopoietic Cells in vitro. Front. Immunol. 2020, 11, 553859. [Google Scholar] [CrossRef]
- Arriens, M.A.; Summerfield, A.; McCullough, K.C. Differential adhesion molecule expression on porcine mononuclear cell populations. Scand. J. Immunol. 1998, 47, 487–495. [Google Scholar] [CrossRef]
- Pruvot, M.; Denstedt, E.; Latinne, A.; Porco, A.; Montecino-Latorre, D.; Khammavong, K.; Milavong, P.; Phouangsouvanh, S.; Sisavanh, M.; Nga, N.T.T.; et al. WildHealthNet: Supporting the development of sustainable wildlife health surveillance networks in Southeast Asia. Sci. Total. Environ. 2022, 863, 160748. [Google Scholar] [CrossRef]
- Lee, E.Y.; Lee, Z.H.; Song, Y.W. CXCL10 and autoimmune diseases. Autoimmun. Rev. 2009, 8, 379–383. [Google Scholar] [CrossRef]
- Trifilo, M.J.; Montalto-Morrison, C.; Stiles, L.N.; Hurst, K.R.; Hardison, J.L.; Manning, J.E.; Masters, P.S.; Lane, T.E. CXC chemokine ligand 10 controls viral infection in the central nervous system: Evidence for a role in innate immune response through recruitment and activation of natural killer cells. J. Virol. 2004, 78, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Embgenbroich, M.; Burgdorf, S. Current Concepts of Antigen Cross-Presentation. Front. Immunol. 2018, 9, 1643. [Google Scholar] [CrossRef]
- Khatri, M.; Dwivedi, V.; Krakowka, S.; Manickam, C.; Ali, A.; Wang, L.; Qin, Z.; Renukaradhya, G.J.; Lee, C.W. Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: A potential animal model for human H1N1 influenza virus. J. Virol. 2010, 84, 11210–11218. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; de Boer-Luijtze, E.A.; Bianchi, A.T.J. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 2001, 82, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Lange, E.; Kalthoff, D.; Blohm, U.; Teifke, J.P.; Breithaupt, A.; Maresch, C.; Starick, E.; Fereidouni, S.; Hoffmann, B.; Mettenleiter, T.C.; et al. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 2009, 90, 2119–2123. [Google Scholar] [CrossRef]
- Talker, S.C.; Stadler, M.; Koinig, H.C.; Mair, K.H.; Rodriguez-Gomez, I.M.; Graage, R.; Zell, R.; Durrwald, R.; Starick, E.; Harder, T.; et al. Influenza A Virus Infection in Pigs Attracts Multifunctional and Cross-Reactive T Cells to the Lung. J. Virol. 2016, 90, 9364–9382. [Google Scholar] [CrossRef]
- Hemann, E.A.; Kang, S.M.; Legge, K.L. Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J. Immunol. 2013, 191, 2486–2494. [Google Scholar] [CrossRef]
- Lee, Y.N.; Lee, Y.T.; Kim, M.C.; Gewirtz, A.T.; Kang, S.M. A Novel Vaccination Strategy Mediating the Induction of Lung-Resident Memory CD8 T Cells Confers Heterosubtypic Immunity against Future Pandemic Influenza Virus. J. Immunol. 2016, 196, 2637–2645. [Google Scholar] [CrossRef]
- Reber, A.J.; Music, N.; Kim, J.H.; Gansebom, S.; Chen, J.; York, I. Extensive T cell cross-reactivity between diverse seasonal influenza strains in the ferret model. Sci. Rep. 2018, 8, 6112. [Google Scholar] [CrossRef]
- Hayashida, K.; Shimaoka, Y.; Ochi, T.; Lipsky, P.E. Rheumatoid arthritis synovial stromal cells inhibit apoptosis and up-regulate Bcl-xL expression by B cells in a CD49/CD29-CD106-dependent mechanism. J. Immunol. 2000, 164, 1110–1116. [Google Scholar] [CrossRef]
- Koopman, G.; Keehnen, R.M.; Lindhout, E.; Newman, W.; Shimizu, Y.; van Seventer, G.A.; de Groot, C.; Pals, S.T. Adhesion through the LFA-1 (CD11a/CD18)-ICAM-1 (CD54) and the VLA-4 (CD49d)-VCAM-1 (CD106) pathways prevents apoptosis of germinal center B cells. J. Immunol. 1994, 152, 3760–3767. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.J.; Iavarone, A.T.; Portnoy, D.A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010, 328, 1703–1705. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Hill, K.K.; Filak, H.; Mogan, J.; Knowles, H.; Zhang, B.; Perraud, A.L.; Cambier, J.C.; Lenz, L.L. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 2011, 187, 2595–2601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, H.; Guo, P.; Hu, L.; Wang, Z.; Wang, J.; Ju, Y.; Meng, S. Broadly Protective CD8(+) T Cell Immunity to Highly Conserved Epitopes Elicited by Heat Shock Protein gp96-Adjuvanted Influenza Monovalent Split Vaccine. J. Virol. 2021, 95, 12. [Google Scholar] [CrossRef]
- Falkeborn, T.; Brave, A.; Larsson, M.; Akerlind, B.; Schroder, U.; Hinkula, J. Endocine, N3OA and N3OASq; three mucosal adjuvants that enhance the immune response to nasal influenza vaccination. PLoS ONE 2013, 8, e70527. [Google Scholar] [CrossRef]
- Rosenkrands, I.; Vingsbo-Lundberg, C.; Bundgaard, T.J.; Lindenstrom, T.; Enouf, V.; van der Werf, S.; Andersen, P.; Agger, E.M. Enhanced humoral and cell-mediated immune responses after immunization with trivalent influenza vaccine adjuvanted with cationic liposomes. Vaccine 2011, 29, 6283–6291. [Google Scholar] [CrossRef]
- Moliva, J.I.; Hossfeld, A.P.; Sidiki, S.; Canan, C.H.; Dwivedi, V.; Beamer, G.; Turner, J.; Torrelles, J.B. Selective delipidation of Mycobacterium bovis BCG enables direct pulmonary vaccination and enhances protection against Mycobacterium tuberculosis. Mucosal Immunol. 2019, 12, 805–815. [Google Scholar] [CrossRef]
- Wright, A.K.; Bangert, M.; Gritzfeld, J.F.; Ferreira, D.M.; Jambo, K.C.; Wright, A.D.; Collins, A.M.; Gordon, S.B. Experimental human pneumococcal carriage augments IL-17A-dependent T-cell defence of the lung. PLoS Pathog. 2013, 9, e1003274. [Google Scholar] [CrossRef]
- Luo, Y.; Van Nguyen, U.; de la Fe Rodriguez, P.Y.; Devriendt, B.; Cox, E. F4+ ETEC infection and oral immunization with F4 fimbriae elicits an IL-17-dominated immune response. Vet. Res. 2015, 46, 121. [Google Scholar] [CrossRef]
- Vemula, S.V.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Vaccine approaches conferring cross-protection against influenza viruses. Expert. Rev. Vaccines 2017, 16, 1141–1154. [Google Scholar] [CrossRef]
- Gianchecchi, E.; Torelli, A.; Montomoli, E. The use of cell-mediated immunity for the evaluation of influenza vaccines: An upcoming necessity. Human Vaccines Immunother. 2019, 15, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Subiza, J.L.; Palomares, O.; Quinti, I.; Sanchez-Ramon, S. Editorial: Trained Immunity-Based Vaccines. Front. Immunol. 2021, 12, 716296. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef] [PubMed]




| Phenotypes of Ivmohoid and Maeloid Cells T-Helper/Memory Cells [CD3+CD4+CD8α+CD8β−] CTLs [CD3+CD4−CD8α+CD8β+] Monocytes [CD3−CD17a+SynCAM−] | Vaccine Type | Challenge: SwlAVH1N1-OH7 | Challenge: H1N1 Pandemic |
|---|---|---|---|
| TBLN MNCs: CXCL10+CD80/86+ activated monocytes | NanoS100-SwlAV | *** p < 0.001 | ** p < 0.01 |
| Nano11-SwlAV | *** p < 0.001 | NS | |
| TBLN MNCs: CD49d+ antigen responsive T-helper/memory cells | NanoS100-SwlAV | - | - |
| Nano11-SwlAV | - | * p < 0.05 | |
| TBLN MNCs: IFNγ+ antigen activated CTLs | NanoS100-SwlAV | *** p < 0.05 | - |
| Nano11-SwlAV | ** p < 0.01 | - | |
| TBLN MNCs: CD49d+IFNγ+ antigen responsive CTLs | NanoS100-SwlAV | ** p < 0.01 | - |
| Nano11-SwlAV | * p < 0.05 | - | |
| TBLN MNCs: IL-17A+ antigen activated CTLs | NanoS100-SwlAV | NS | ** p < 0.01 |
| Nano11-SwlAV | * p < 0.05 | * p < 0.05 | |
| TBLN MNCs: CD49d+IL-17A+ antigen responsive CTLs | NanoS100-SwlAV | - | ** p < 0.01 |
| Nano11-SwlAV | * p < 0.05 | ** p < 0.01 | |
| PBMCs: IFNγ+ antigen activated CTLs | NanoS 100-SwlAV | - | ** p < 0.01 |
| Nano11-SwlAV | - | * p < 0.05 | |
| PBMCs: CD49d+IFNY+ antigen responsive CTLs | NanoS100-SwlAV | - | ** p < 0.01 |
| Nano11-SwlAV | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, V.; Hernandez-Franco, J.F.; Yadagiri, G.; Bugybayeva, D.; Dolatyabi, S.; Feliciano-Ruiz, N.; Schrock, J.; Suresh, R.; Hanson, J.; Yassine, H.; et al. Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs. Vaccines 2023, 11, 1707. https://doi.org/10.3390/vaccines11111707
Patil V, Hernandez-Franco JF, Yadagiri G, Bugybayeva D, Dolatyabi S, Feliciano-Ruiz N, Schrock J, Suresh R, Hanson J, Yassine H, et al. Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs. Vaccines. 2023; 11(11):1707. https://doi.org/10.3390/vaccines11111707
Chicago/Turabian StylePatil, Veerupaxagouda, Juan F. Hernandez-Franco, Ganesh Yadagiri, Dina Bugybayeva, Sara Dolatyabi, Ninoshkaly Feliciano-Ruiz, Jennifer Schrock, Raksha Suresh, Juliette Hanson, Hadi Yassine, and et al. 2023. "Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs" Vaccines 11, no. 11: 1707. https://doi.org/10.3390/vaccines11111707
APA StylePatil, V., Hernandez-Franco, J. F., Yadagiri, G., Bugybayeva, D., Dolatyabi, S., Feliciano-Ruiz, N., Schrock, J., Suresh, R., Hanson, J., Yassine, H., HogenEsch, H., & Renukaradhya, G. J. (2023). Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs. Vaccines, 11(11), 1707. https://doi.org/10.3390/vaccines11111707








