Integrative Bioinformatics Approaches Indicate a Particular Pattern of Some SARS-CoV-2 and Non-SARS-CoV-2 Proteins
Abstract
1. Introduction
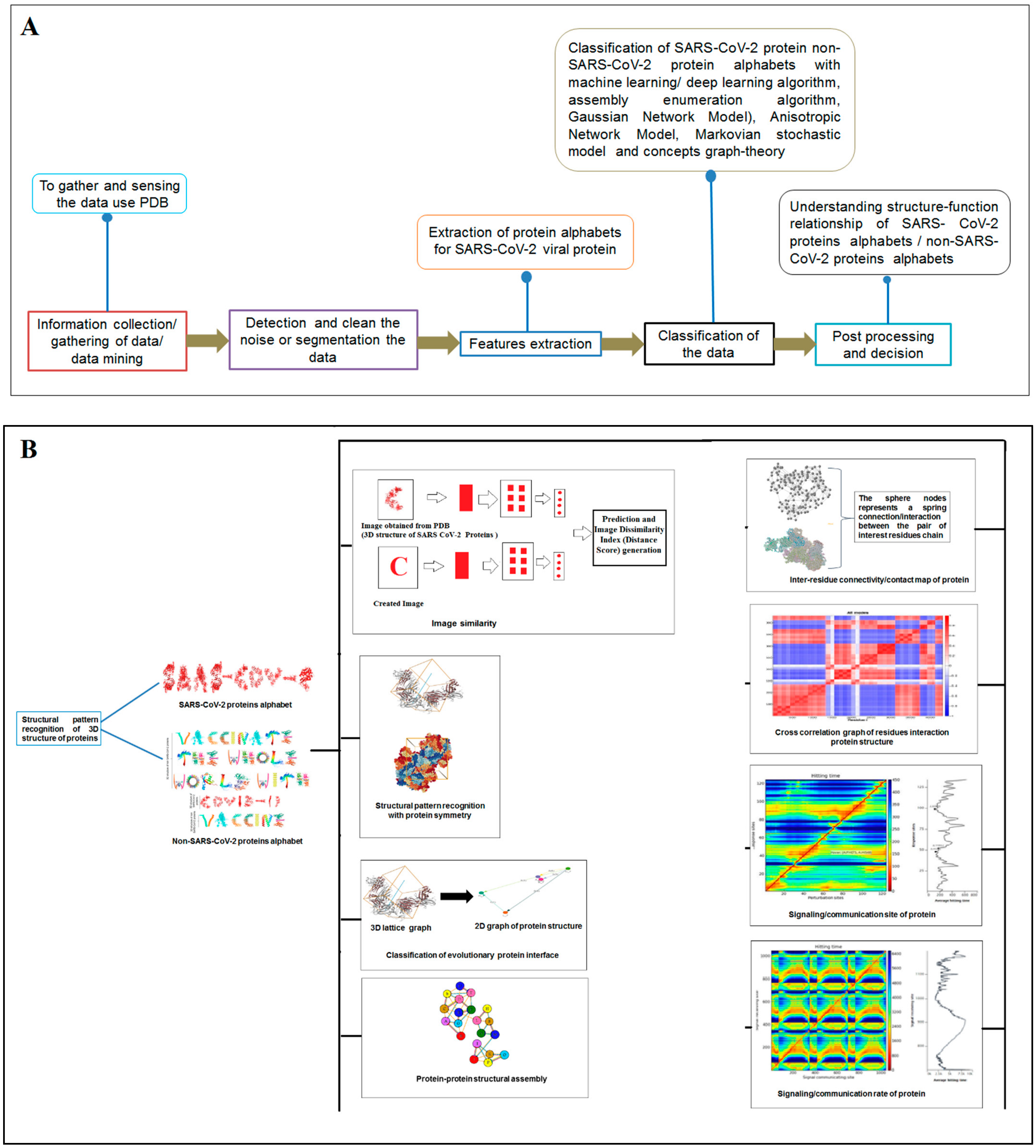
2. Materials and Methods
2.1. Data Mining Using PDB and Collection of Proteins as Alphabets from SARS-CoV-2 Proteins and Non-SARS-CoV-2 Proteins
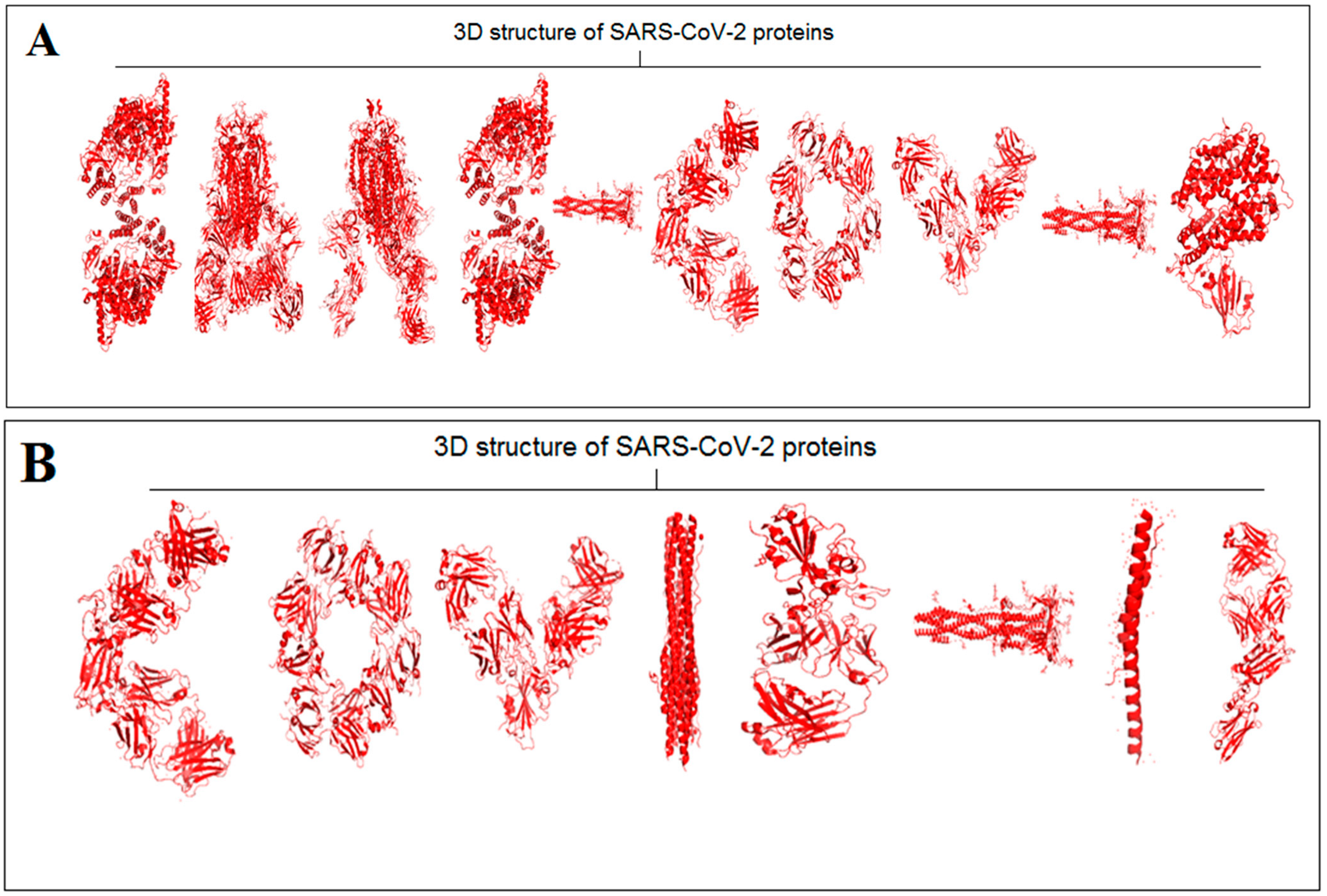
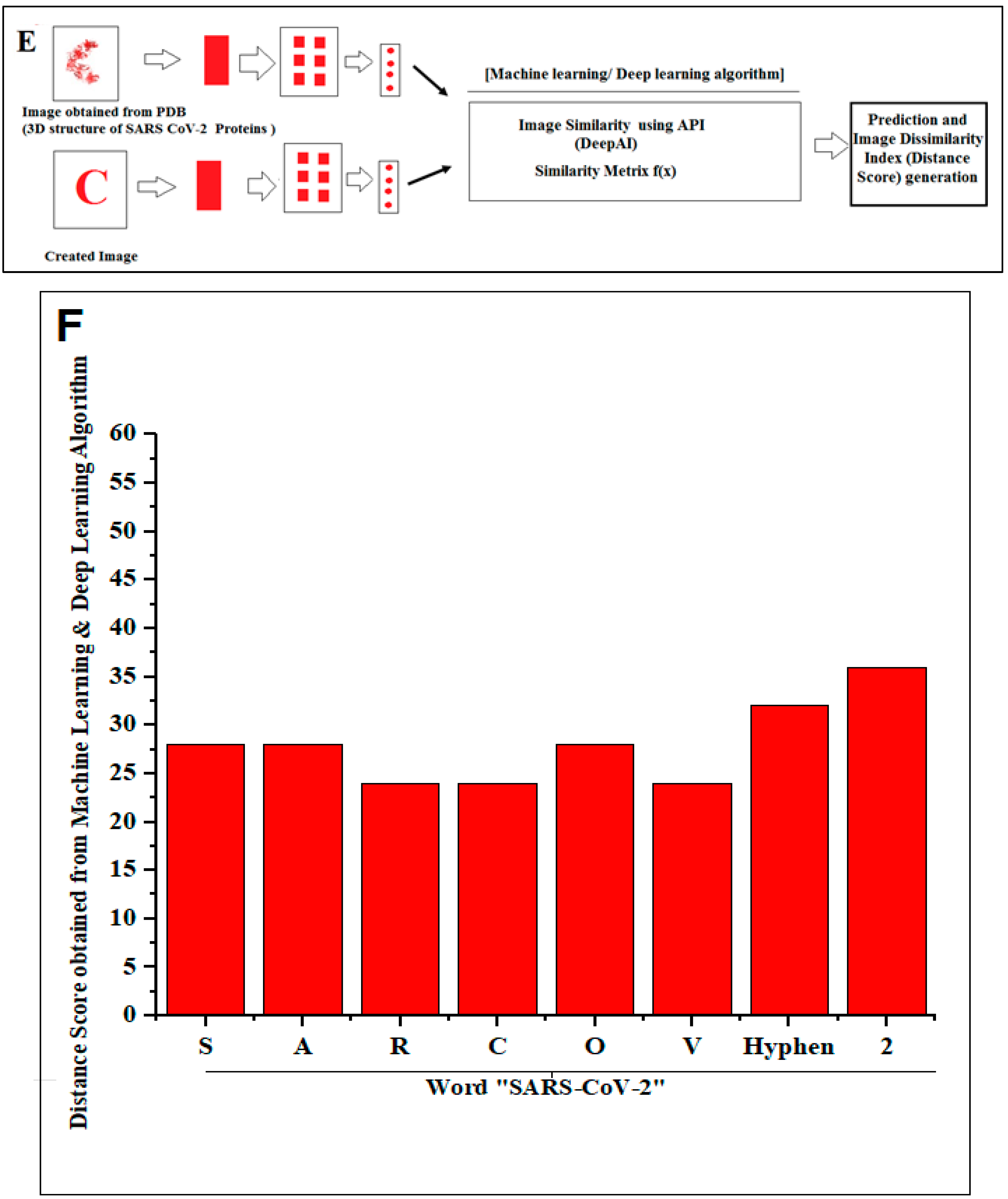
2.2. Pattern Recognition of 3D Structures of SARS-CoV-2 and Non-SARS-CoV-2 Proteins
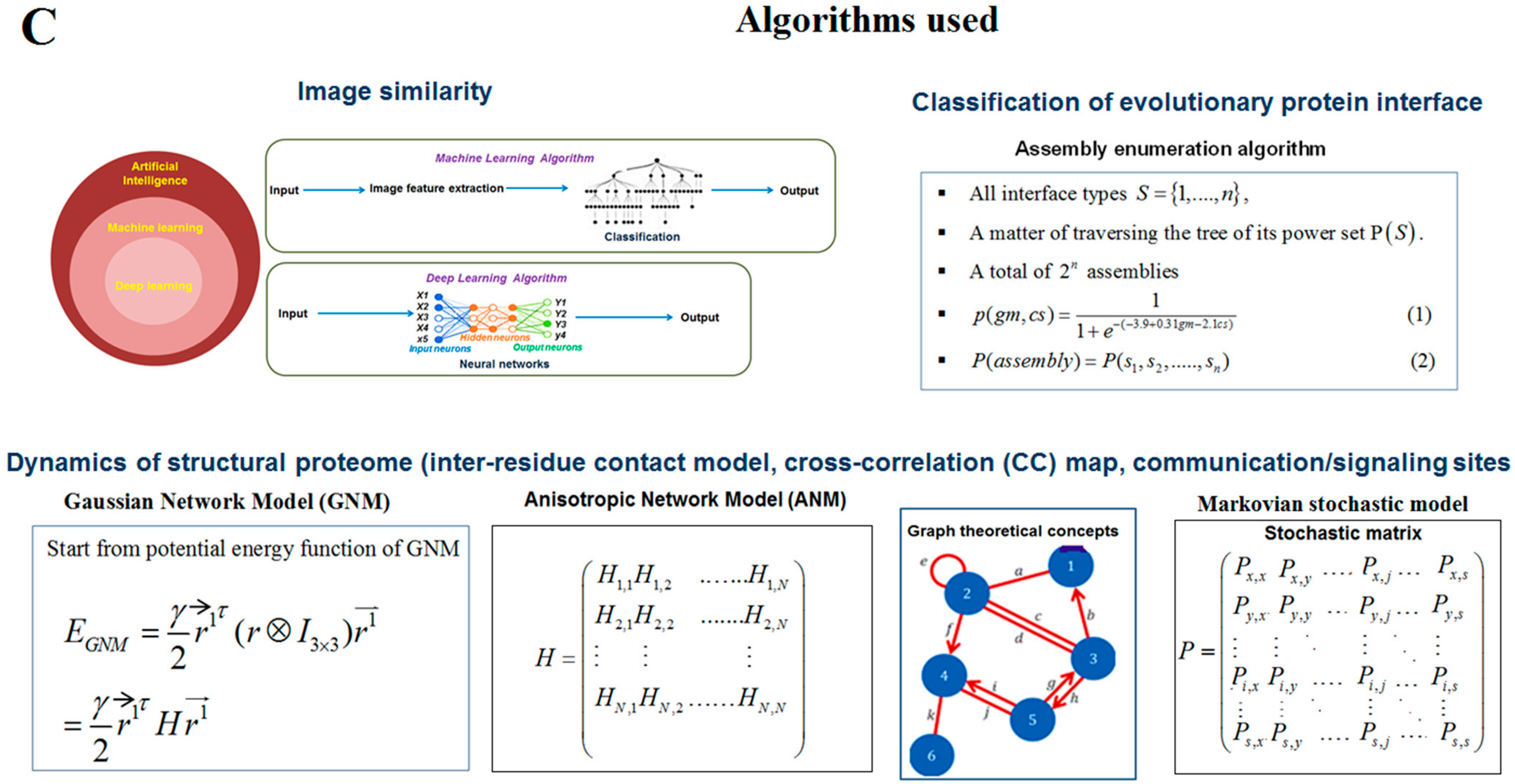
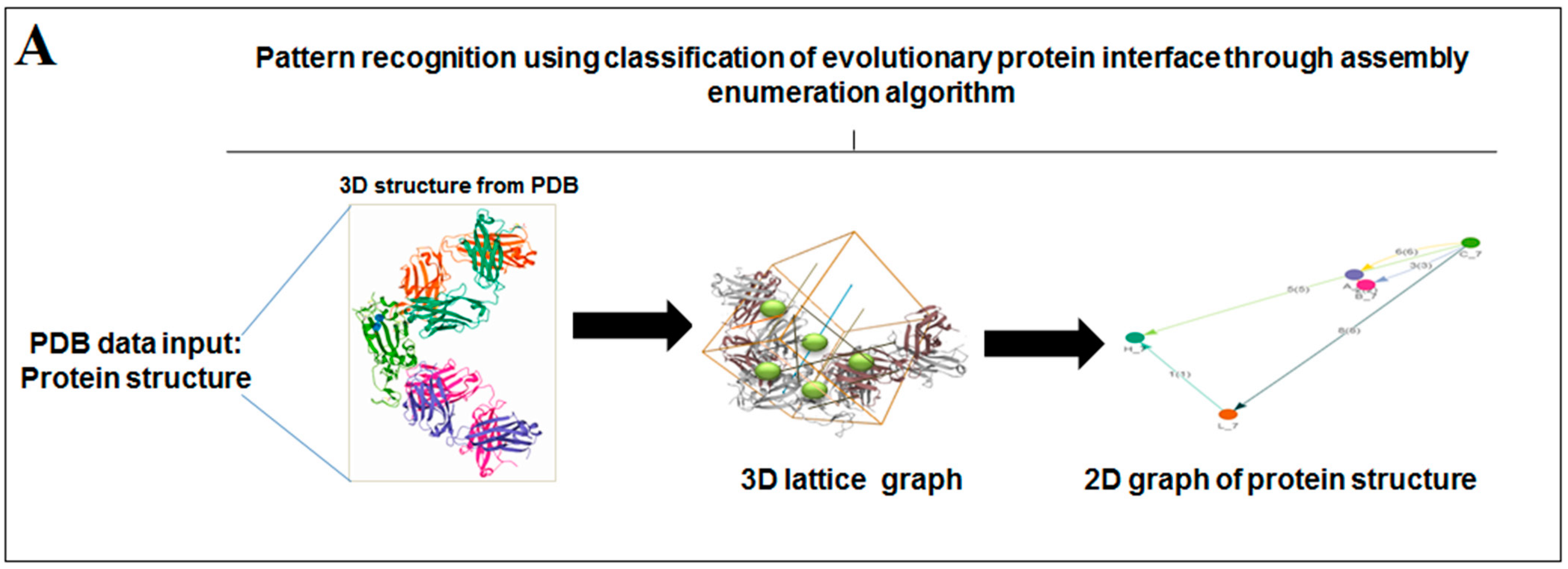
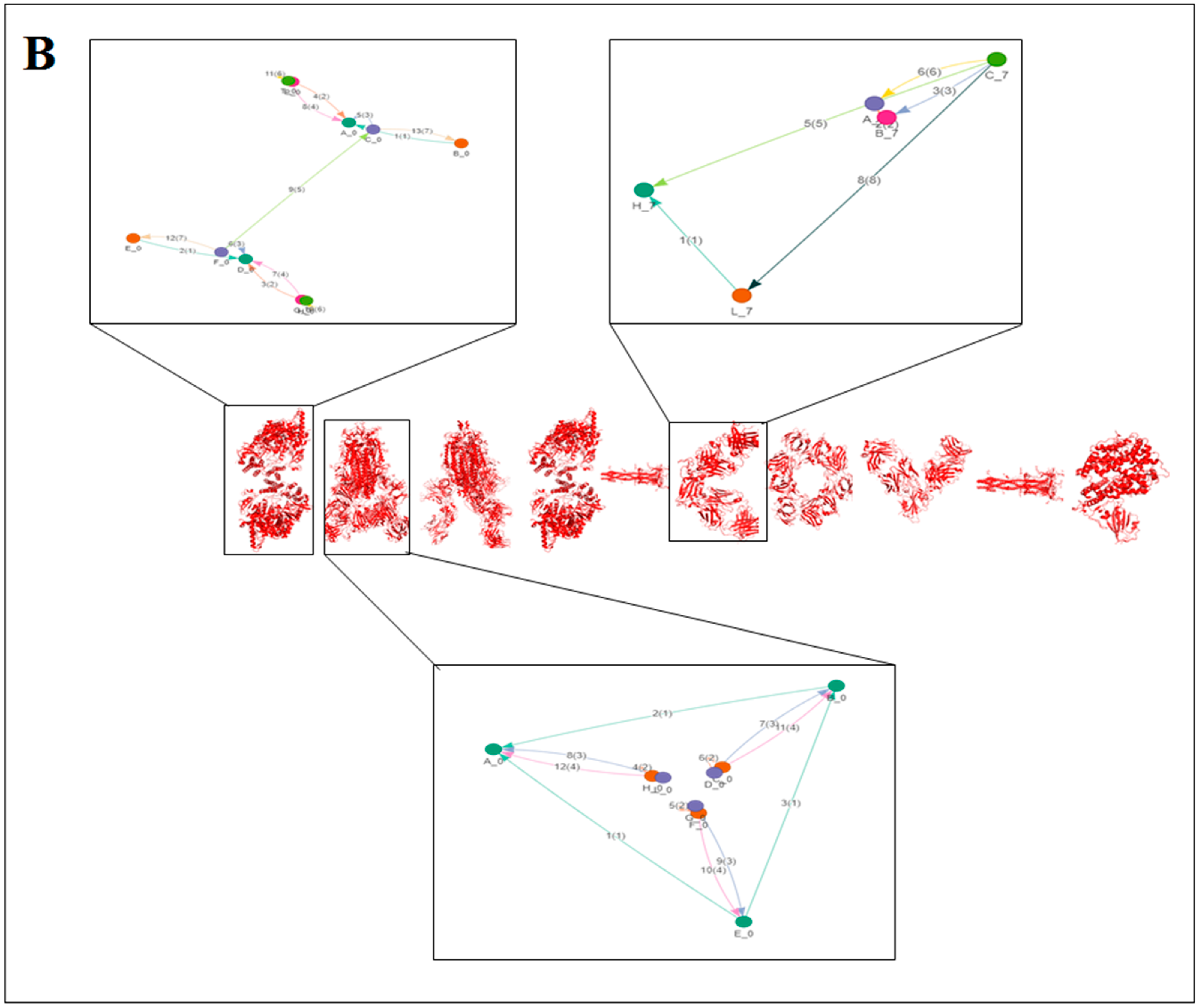
2.3. Pattern Recognition Using the Classification of Evolutionary Protein Interface through Assembly Enumeration Algorithm
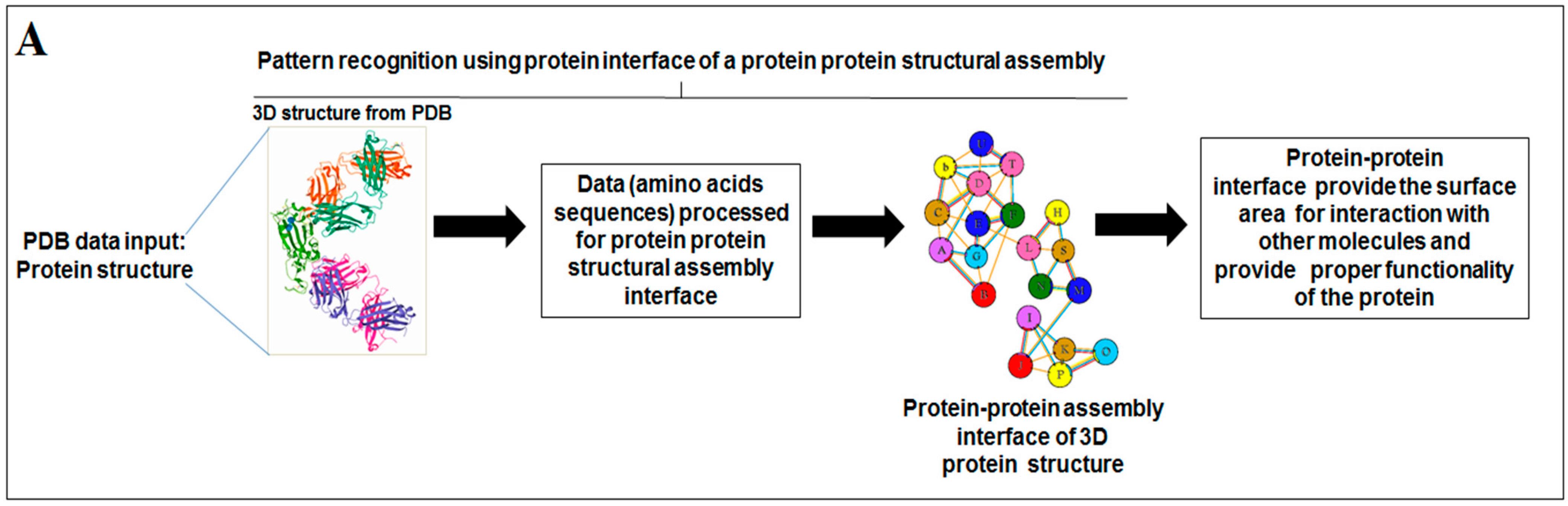
2.4. Pattern Recognition Using the Protein–Protein Interface of 3D Structures of SARS-CoV-2 and Non-SARS-CoV-2
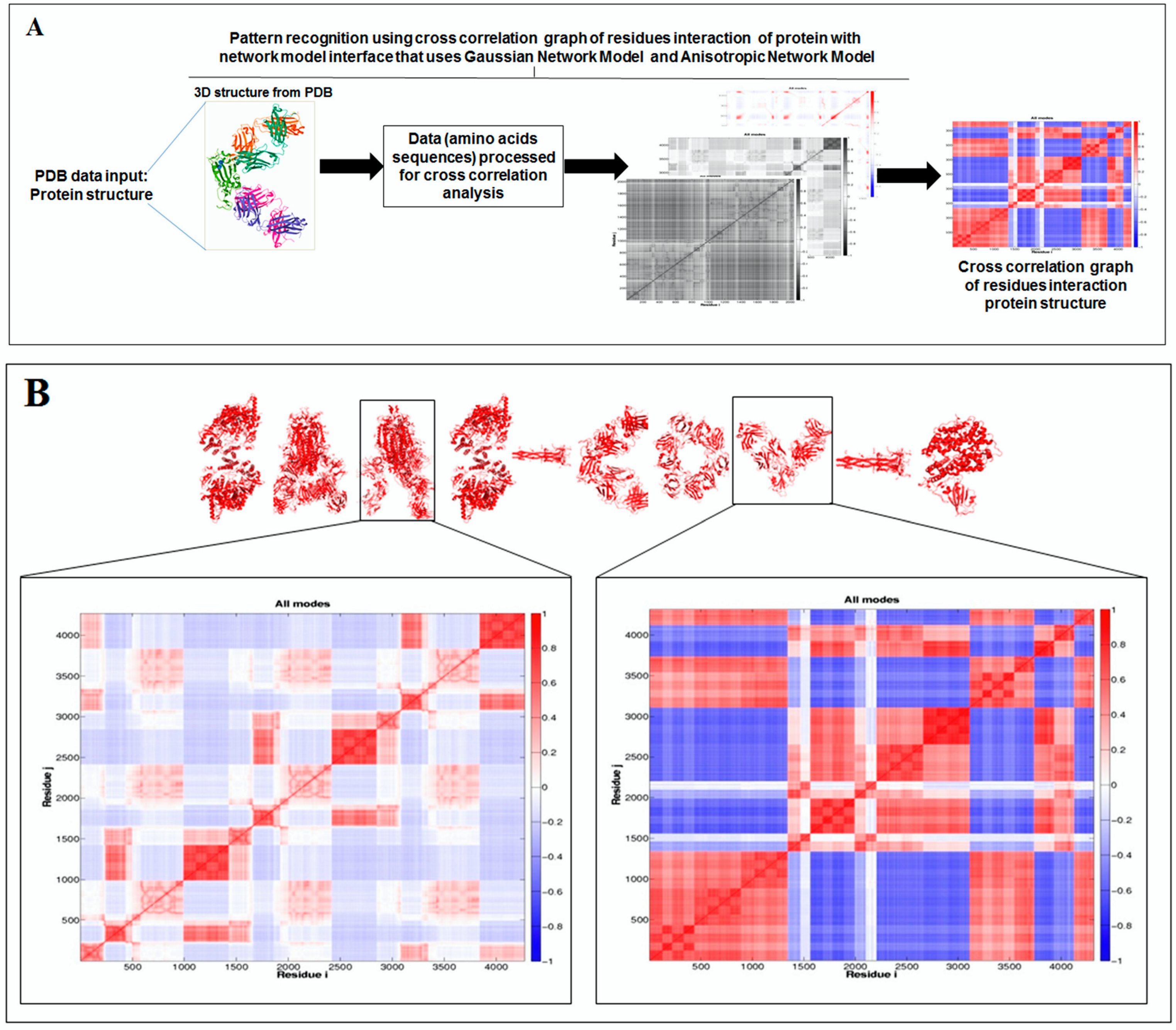
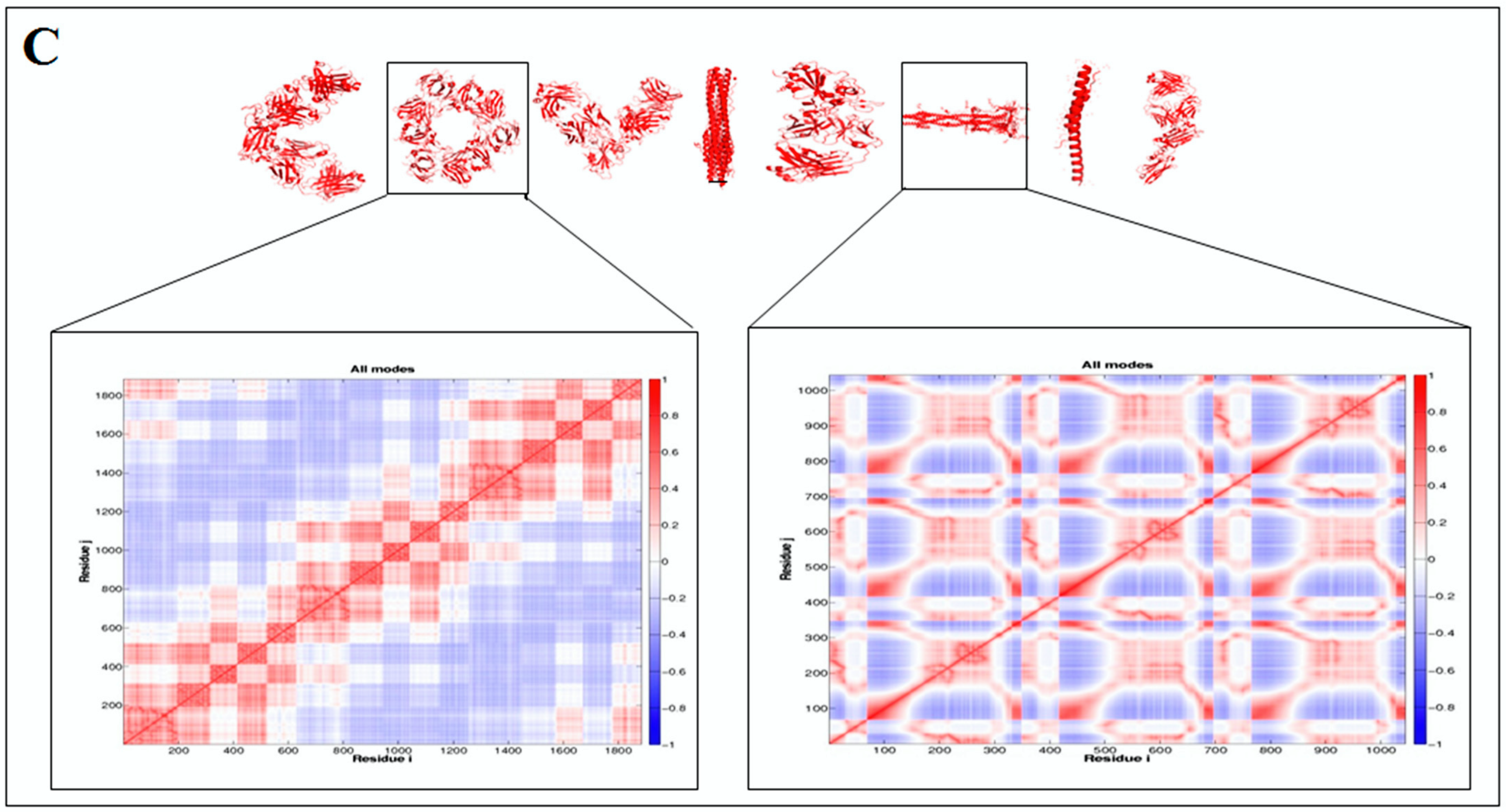
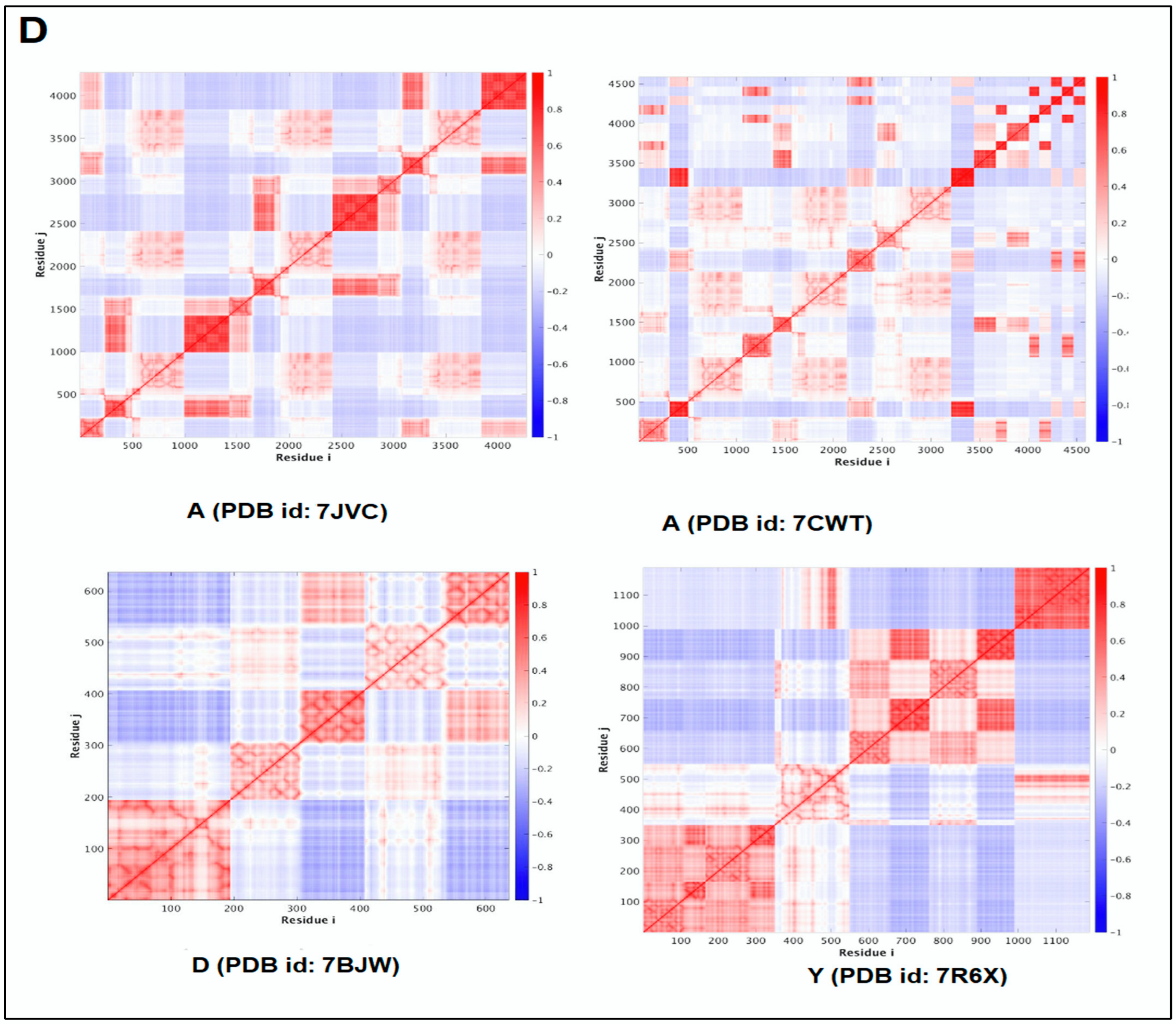
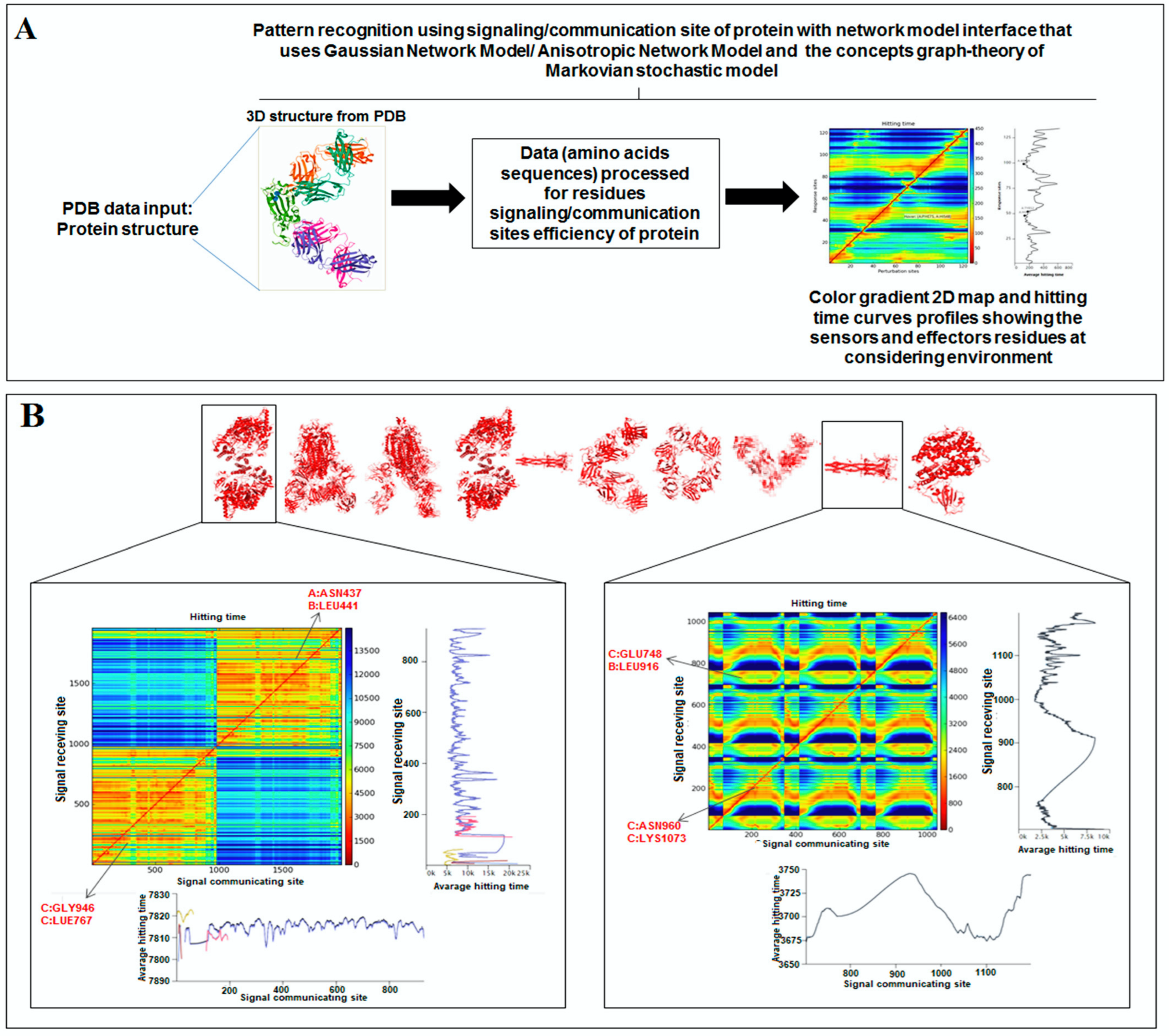
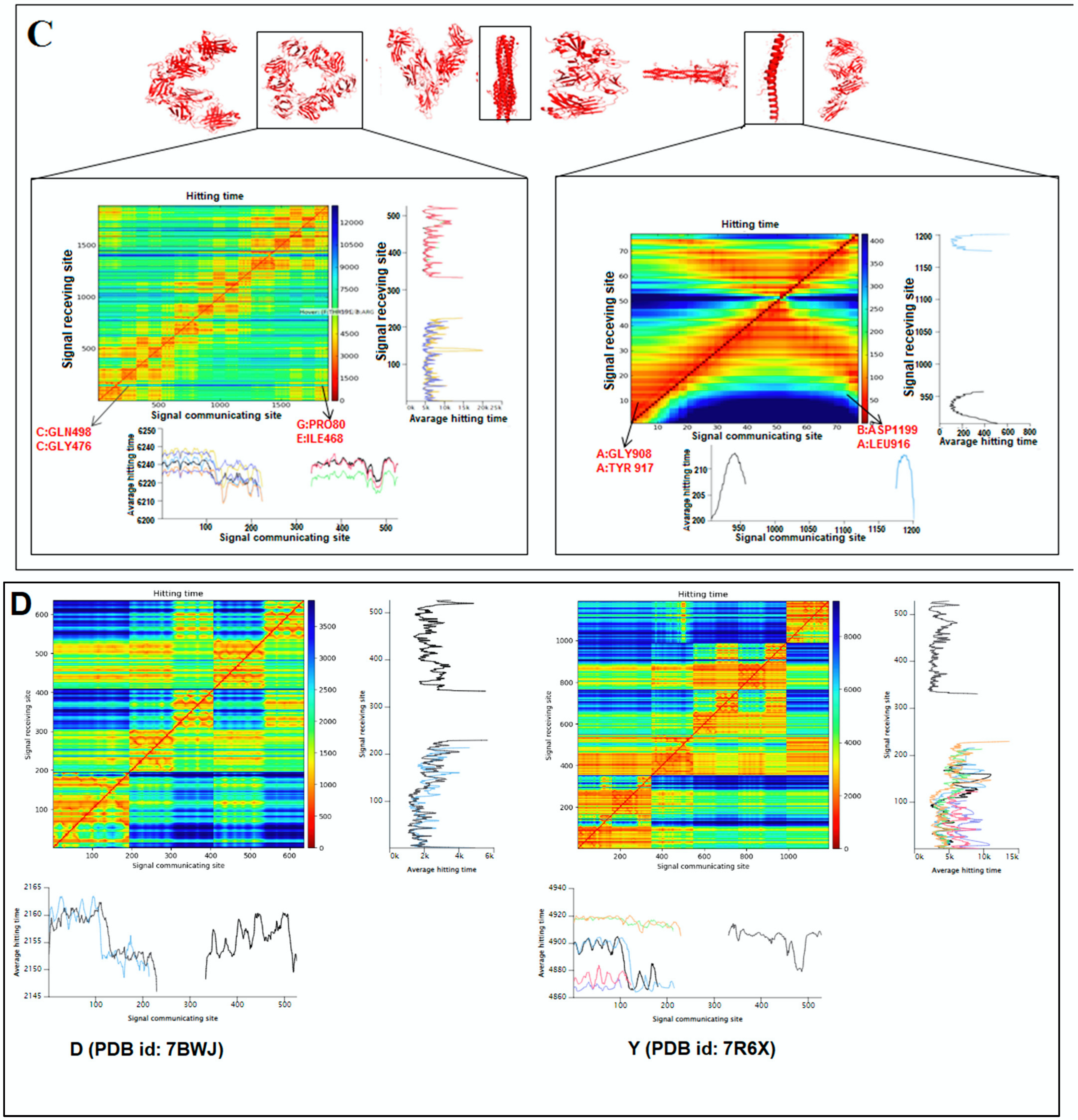
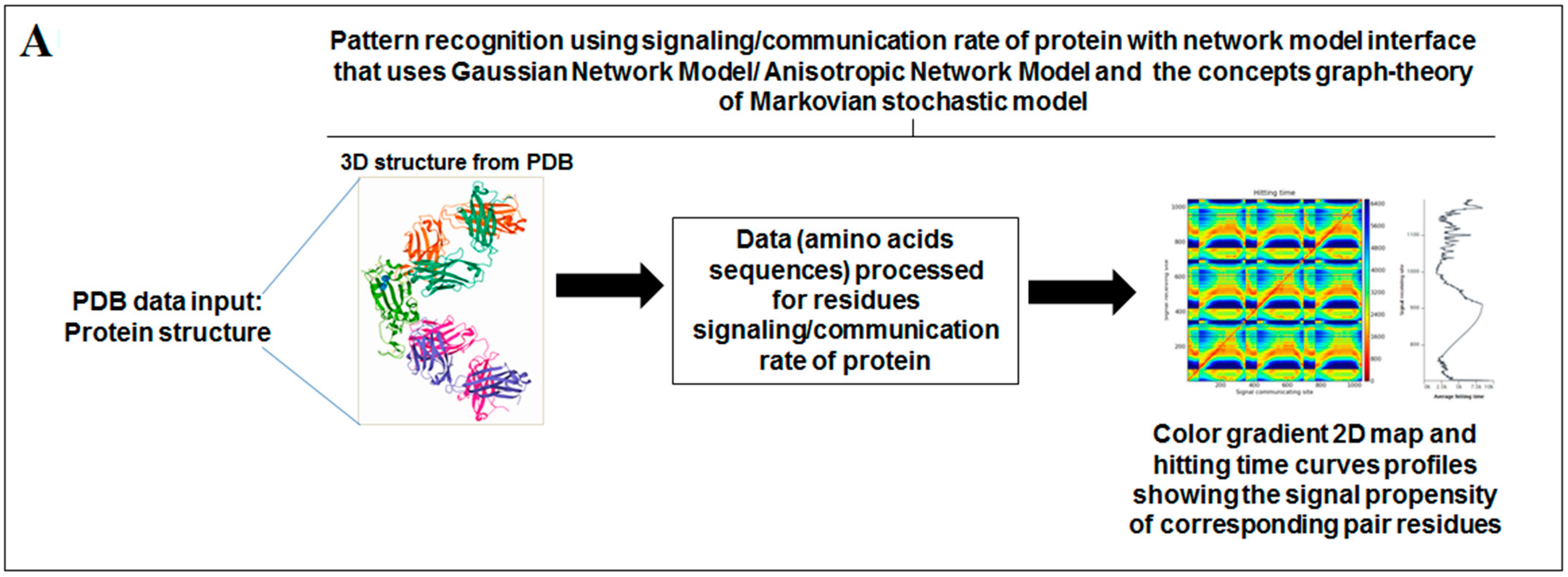
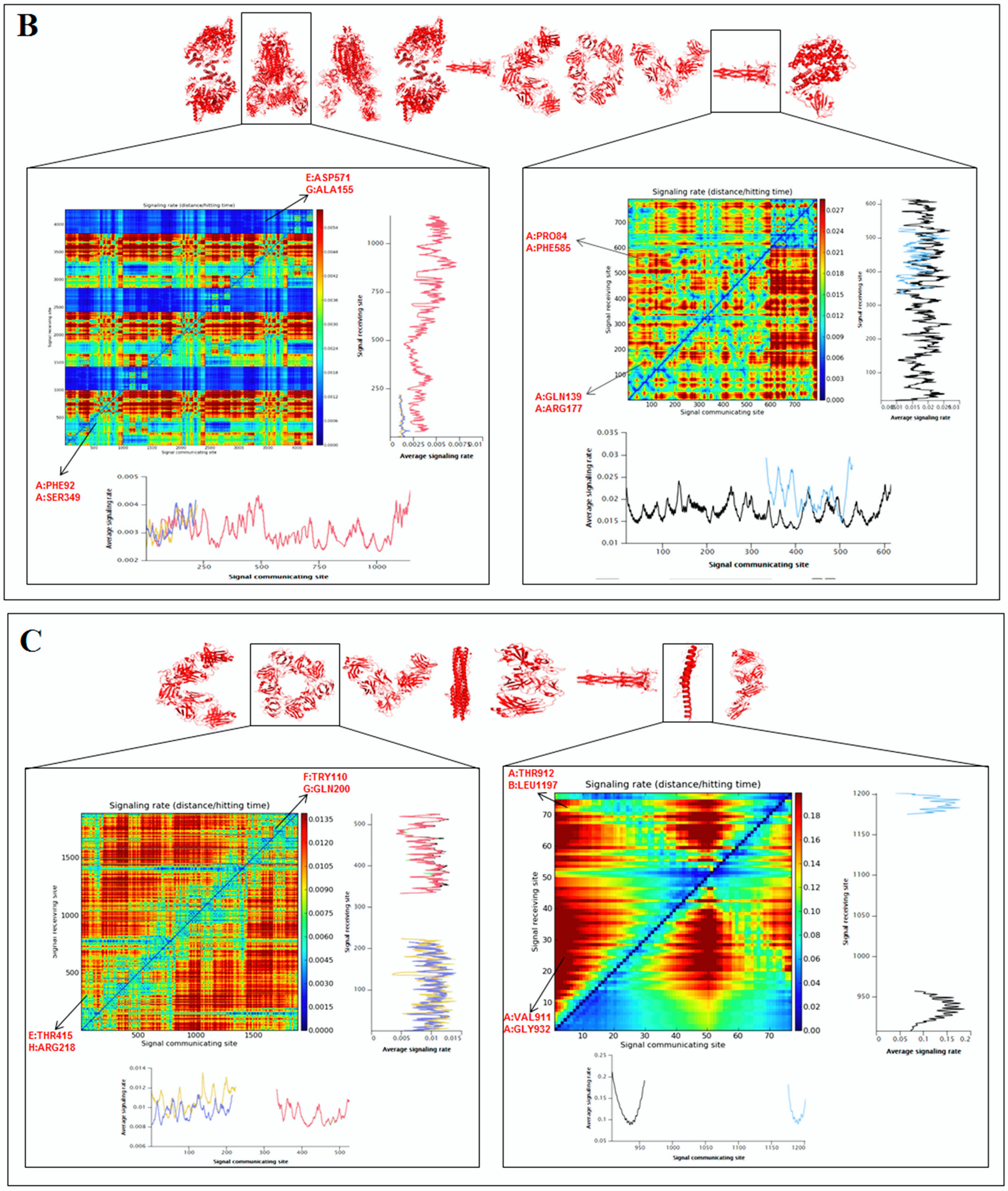
2.5. Pattern Recognition with Dynamics of Structural Proteome
2.6. Post-Processing and Decision
3. Result
3.1. Data Mining Using PDB and Collection of Proteins as Alphabets from SARS-CoV-2 and Non-SARS-CoV-2 Proteins
3.2. Structural Pattern Recognition of 3D Structures of SARS-CoV-2 and Non-SARS-CoV-2 Proteins
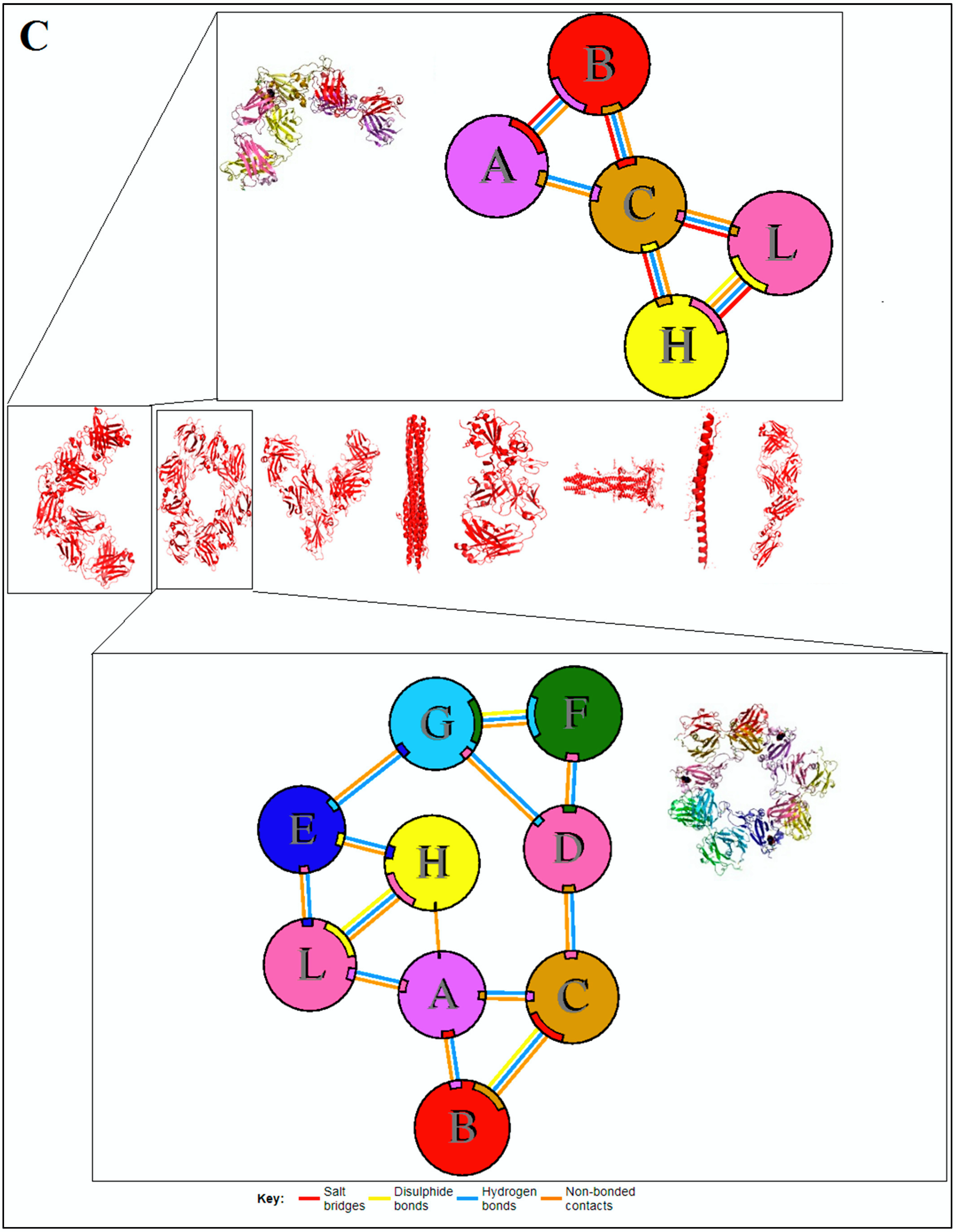
3.3. Pattern Recognition Using the Classification of Evolutionary Protein Interface through Assembly Enumeration Algorithm
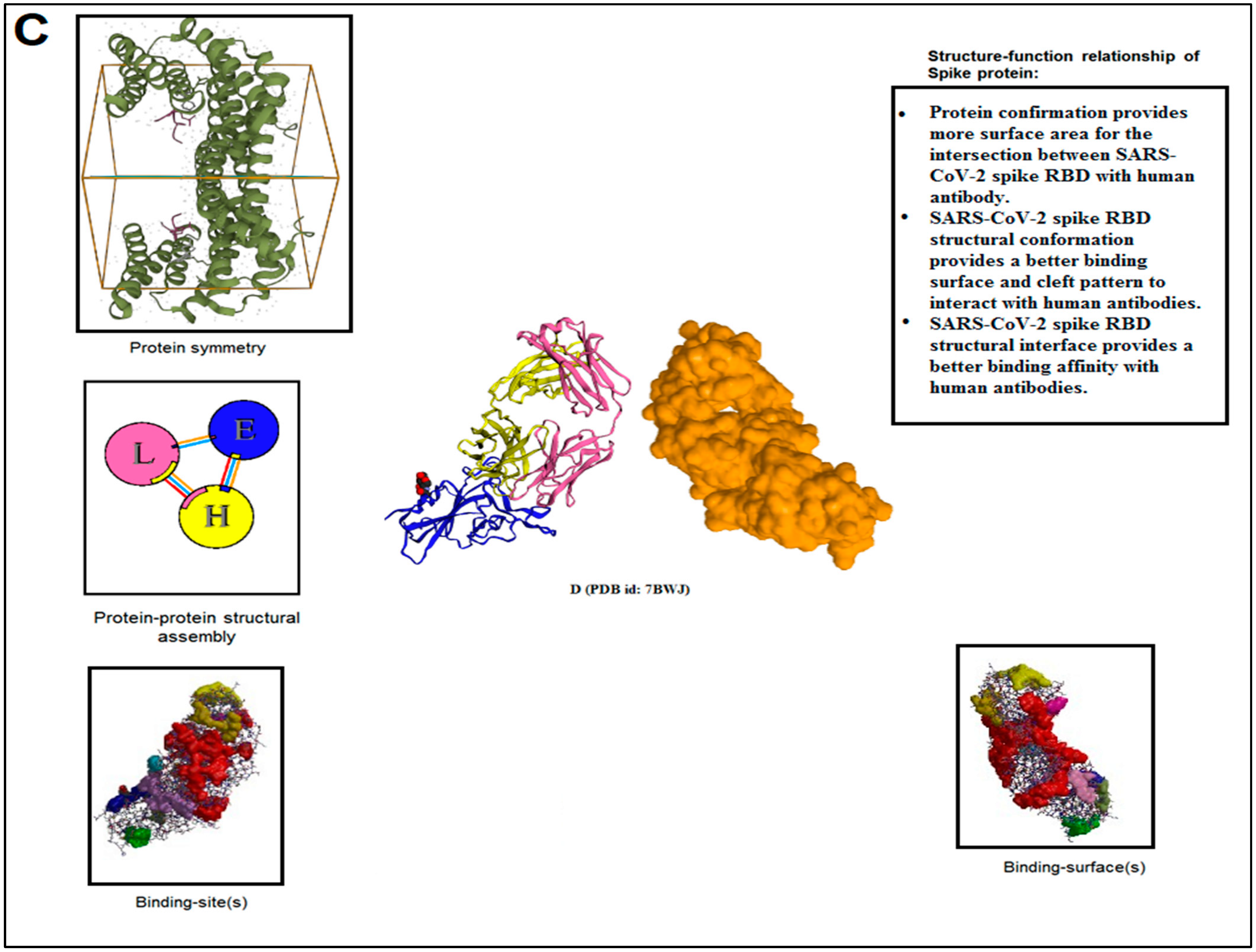
3.4. Pattern Recognition Using Protein–Protein Interface 3D Structures of SARS-CoV-2 and Non-SARS-CoV-2
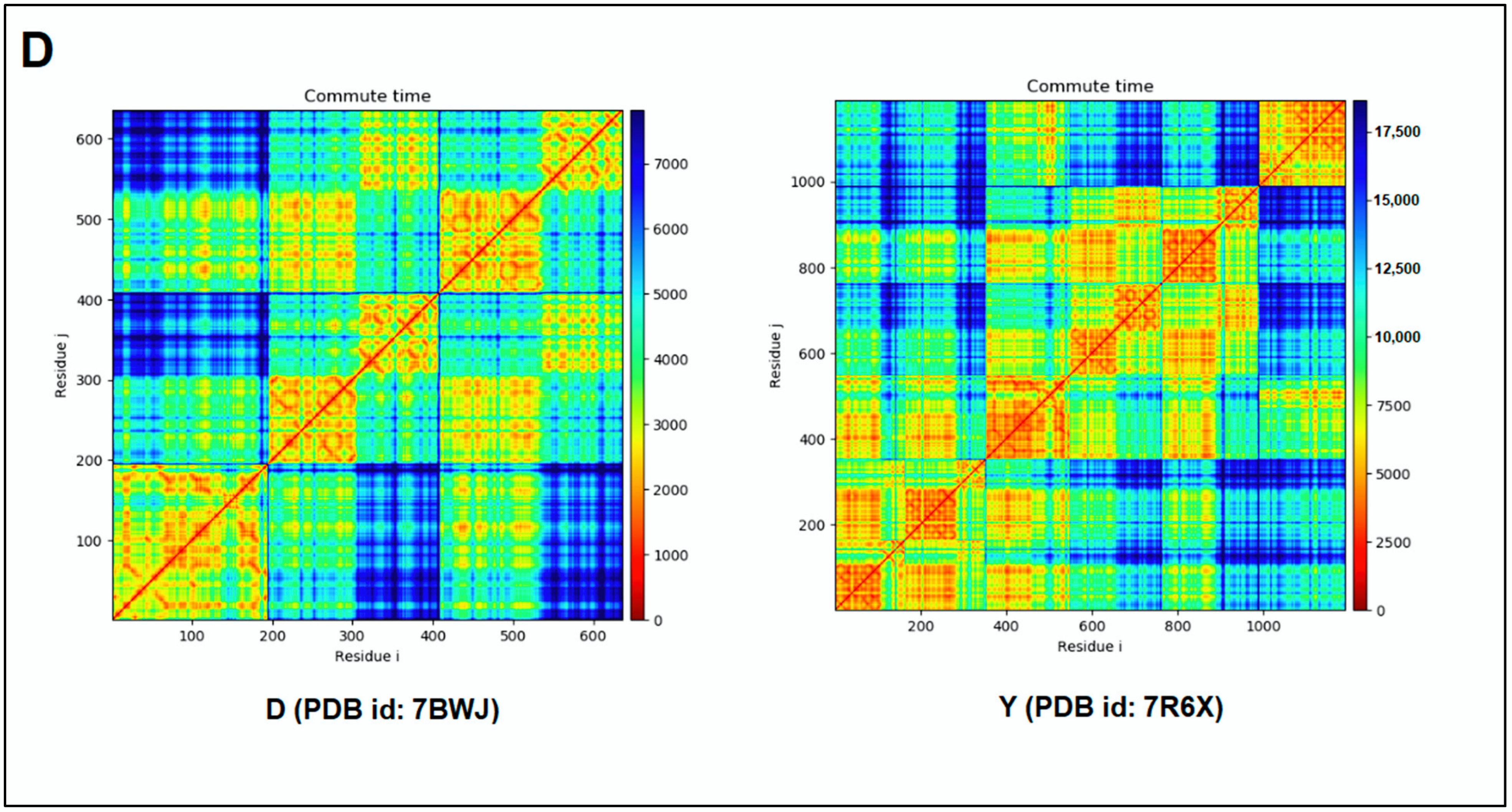
3.5. Pattern Recognition with Dynamics of Structural Proteome
3.6. Post-Processing and Decision
4. Discussion
5. Limitation of the Study
6. Conclusions
7. Perspectives
- (i)
- Importance of the field. Pattern recognition is a rapidly developing field with enormous applicability in biological sciences. This study tried to understand the pattern identification of SARS-CoV-2 proteins. Finally, the study presents new information on the pattern identification of SARS-CoV-2 proteins.
- (ii)
- A summary of the current thinking. We have searched for protein-like alphabets involving 3D structure of SARS-CoV-2 from PDB and created two words, “SARS- CoV-2” and “COVID-19”. We have also developed two slogans using non-SARS-CoV-2 proteins, and the slogans are “Vaccinate the world against COVID-19” and “Say no to SARS-CoV-2”. We have used 12 SARS-CoV-2 proteins and 14 non-SARS-CoV-2 proteins to design those words and slogans. We have performed image comparison with protein-like alphabets with English alphabets using the deep AI model. The structural symmetry analysis indicates alphabet-shaped symmetric proteins, such as C, O, I, Hyphen (-), 1(One), S, and A. To determine the dynamics of the structural proteome, we evaluated the inter-residue contact by developing inter-residue contact models with both residue and chain and illustrated the cross-correlations between residues through a cross-correlation (CC) map. In order to understand the residue functionality of proteins, we analyzed the communication/signaling sites of protein residue and signal communication/signal receiving rate of protein alphabets. The assembly enumeration algorithm, anisotropic network model, Gaussian network model, Markovian stochastic model, and other integrative bioinformatics approaches, and tools were used to depict the structural and functional relationships of the protein alphabets of SARS-CoV-2 and COVID-19. After image comparison of protein-like alphabets, the distance score of “I” was the lowest with 22, and “9” was the highest with 40. For post-processing and decision, two protein alphabets were evaluated, protein alphabet “C” (PDB ID: 6XC3) and alphabet “S” (PDB ID: 7OYG), and we understood the structural, functional, and evolutionary relationships using modeling approaches.
- (iii)
- Future directions. This study sheds further light on the uniqueness in the functionality of SARS-CoV-2 proteins. The evolutionary process appears to enhance the protein structure smoothly to provide suitable functionality shaped by natural selection. The computational approach may assist in solving patterns related to the structural aspects of other proteins and help to decipher the riddles and puzzles involving the complex structure–function relationships of proteins, which is an important area of modern biology. It has a great promise for capturing the evolutionary information of proteins and the potential for success in future work. It might help to understand the therapeutic target protein pattern, which will be beneficial as a potential therapeutic target discovery.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schalkoff, R.J. Pattern recognition. In Wiley Encyclopedia of Computer Science and Engineering; Wah, B.W., Ed.; John Wiley & Sons: Berkeley, CA, USA, 2007. [Google Scholar] [CrossRef]
- Dougherty, G. Pattern Recognition and Classification: An Introduction; Springer Science & Business Media: New York, NY, USA, 2012; ISBN 978-1-4614-5322-2. [Google Scholar] [CrossRef]
- Sverrisson, F.; Feydy, J.; Correia, B.E.; Bronstein, M.M. Fast end-to-end learning on protein surfaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual Conference, 19–25 June 2021; pp. 15272–15281. [Google Scholar]
- Jain, A.K.; Duin, R.P.W.; Mao, J. Statistical pattern recognition: A review. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 4–37. [Google Scholar] [CrossRef]
- Webb, A.R. Statistical Pattern Recognition, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2003; pp. 1–668. ISBN 978-1-119-95295-4. [Google Scholar]
- Bishop, C.M. Pattern recognition. In Machine Learning; Springer: New York, NY, USA, 2006; Volume 1, p. 738. [Google Scholar]
- Oussous, A.; Benjelloun, F.-Z.; Lahcen, A.A.; Belfkih, S. Big Data technologies: A survey. J. King Saud Univ. -Comput. Inf. Sci. 2018, 30, 431–448. [Google Scholar] [CrossRef]
- Choy, C.; Lee, J.; Ranftl, R.; Park, J.; Koltun, V. High-dimensional convolutional networks for geometric pattern recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 16–18 June 2020; pp. 11227–11236. [Google Scholar]
- Pal, S.K.; Mitra, P. Pattern Recognition Algorithms for Data Mining; CRC Press: Boca Raton, FL, USA, 2004; pp. 1–280. ISBN 9780367394240. [Google Scholar]
- Dhall, D.; Kaur, R.; Juneja, M. Machine learning: A review of the algorithms and its applications. In Proceedings of the ICRIC 2019; Lecture Notes in Electrical Engineering; Singh, P., Kar, A., Singh, Y., Kolekar, M., Tanwar, S., Eds.; Springer: Cham, Switzerland, 2020; Volume 597, pp. 47–63. [Google Scholar]
- Saranya, A.; Kottilingam, K. A Survey on Bone Fracture Identification Techniques using Quantitative and Learning Based Algorithms. In Proceedings of the 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS), Tamilnadu, India, 25–27 March 2021; pp. 241–248. [Google Scholar]
- Paolanti, M.; Frontoni, E. Multidisciplinary pattern recognition applications: A review. Comput. Sci. Rev. 2020, 37, 100276. [Google Scholar] [CrossRef]
- AlQuraishi, M. Machine learning in protein structure prediction. Curr. Opin. Chem. Biol. 2021, 1, 65. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Mahajan, S.P.; Sulam, J.; Gray, J.J. Deep learning in protein structural modeling and design. Patterns 2020, 1, 100142. [Google Scholar] [CrossRef] [PubMed]
- Guehairia, O.; Dornaika, F.; Ouamane, A.; Taleb-Ahmed, A. Facial age estimation using tensor based subspace learning and deep random forests. Inf. Sci. 2022, 609, 1309–1317. [Google Scholar] [CrossRef]
- de Ridder, D.; de Ridder, J.; Reinders, M.J. Pattern recognition in bioinformatics. Brief. Bioinform. 2013, 14, 633–647. [Google Scholar] [CrossRef]
- Sarkar, B.; Chakraborty, C. DNA pattern recognition using canonical correlation algorithm. J. Biosci. 2015, 40, 709–719. [Google Scholar] [CrossRef]
- Chhabra, M.; Gujral, R.K. Image pattern recognition for an intelligent healthcare system: An application area of machine learning and big data. J. Comput. Theor. Nanosci. 2019, 16, 3932–3937. [Google Scholar] [CrossRef]
- Kinch, L.N.; Grishin, N.V. Evolution of protein structures and functions. Curr. Opin. Struct. Biol. 2002, 12, 400–408. [Google Scholar] [CrossRef]
- Sikosek, T.; Chan, H.S. Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interface 2014, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. The sprint to solve coronavirus protein structures—And disarm them with drugs. J. Comput. Theor. Nanosci. 2020, 7808, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Lubin, J.H.; Zardecki, C.; Dolan, E.M.; Lu, C.; Shen, Z.; Dutta, S.; Westbrook, J.D.; Hudson, B.P.; Goodsell, D.S.; Williams, J.K.; et al. Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic. Proteins: Struct. Funct. Bioinform. 2022, 90, 1054–1080. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Agoramoorthy, G.; Lee, S.-S. Evolution, Mode of Transmission, and Mutational Landscape of Newly Emerging SARS-CoV-2 Variants. mBio 2021, 12, e01140-21. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Dhama, K.; Lee, S.S. The rapid emergence of multiple sublineages of Omicron (B. 1.1. 529) variant: Dynamic profiling via molecular phylogenetics and mutational landscape studies. J. Infect. Public Health 2022, 15, 1234–1258. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Dhama, K.; Agoramoorthy, G. A comprehensive analysis of the mutational landscape of the newly emerging Omicron (B. 1.1. 529) variant and comparison of mutations with VOCs and VOIs. GeroScience 2022, 22, 1–33. [Google Scholar]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Dhama, K.; Lee, S.S. Continent-wide evolutionary trends of emerging SARS-CoV-2 variants: Dynamic profiles from Alpha to Omicron. GeroScience 2022, 13, 1–22. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Mallik, B. Omicron (B. 1.1. 529)—A new heavily mutated variant: Mapped location and probable properties of its mutations with an emphasis on S-glycoprotein. Int. J. Biol. Macromol. 2022, 31, 980–997. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Chatterjee, S.; Sharma, A.R.; Lee, S.S.; Chakraborty, C. Delta variant (B. 1.617. 2) of SARS-CoV-2: Current understanding of infection, transmission, immune escape, and mutational landscape. Folia Microbiol. 2022, 12, 1–2. [Google Scholar]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Mohapatra, R.K.; Chakraborty, S.; Pal, S.; Dhama, K. Immediate need for next-generation and mutation-proof vaccine to protect against current emerging Omicron sublineages and future SARS-CoV-2 variants: An urgent call for researchers and vaccine companies—Correspondence. Int. J. Surg. 2022, 106, 106903. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 2021, 4, e2270. [Google Scholar] [CrossRef]
- Riesen, K. Structural pattern recognition with graph edit distance. In Advances in Computer Vision and Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Jia, F.; Shi, C.; He, K.; Wang, C.; Xiao, B. Degraded document image binarization using structural symmetry of strokes. Pattern Recognit. 2018, 74, 225–240. [Google Scholar] [CrossRef]
- Del Carpio-Munloz, C.A. Folding pattern recognition in proteins using spectral analysis methods. Genome Inform. 2002, 13, 163–172. [Google Scholar]
- Youkharibache, P. Protodomains: Symmetry-related supersecondary structures in proteins and self-complementarity. In Protein Supersecondary Structures; Springer: Berlin/Heidelberg, Germany, 2019; pp. 187–219. [Google Scholar]
- André, I.; Bradley, P.; Wang, C.; Baker, D. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. USA 2007, 104, 17656–17661. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Alam, S.L.; Fricke, T.; Zadrozny, K.; Sedzicki, J.; Taylor, A.B.; Demeler, B.; Pornillos, O.; Ganser-Pornillos, B.K.; Diaz-Griffero, F. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625–18630. [Google Scholar] [CrossRef] [PubMed]
- Clegg, J. Properties and metabolism of the aqueous cytoplasm and its boundaries. Am. J. Physiol. -Regul. Integr. Comp. Physiol. 1984, 246, R133–R151. [Google Scholar] [CrossRef]
- Pagès, G.; Grudinin, S. Analytical symmetry detection in protein assemblies. II. Dihedral and cubic symmetries. J. Struct. Biol. 2018, 203, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 4, e02098. [Google Scholar] [CrossRef]
- Bliven, S.; Lafita, A.; Parker, A.; Capitani, G.; Duarte, J.M. Automated evaluation of quaternary structures from protein crystals. PLoS Comput. Biol. 2018, 14, e1006104. [Google Scholar] [CrossRef]
- Matsumoto, S.; Ishida, S.; Araki, M.; Kato, T.; Terayama, K.; Okuno, Y. Extraction of protein dynamics information from cryo-EM maps using deep learning. Nat. Mach. Intell. 2021, 3, 153–160. [Google Scholar] [CrossRef]
- Afify, H.M.; Abdelhalim, M.B.; Mabrouk, M.S.; Sayed, A.Y. Protein secondary structure prediction (PSSP) using different machine algorithms. Egypt. J. Med. Hum. Genet. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Li, H.; Chang, Y.-Y.; Lee, J.Y.; Bahar, I.; Yang, L.-W. DynOmics: Dynamics of structural proteome and beyond. Nucleic Acids Res. 2017, 45, W374–W380. [Google Scholar] [CrossRef] [PubMed]
- Chennubhotla, C.; Bahar, I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS Comput. Biol. 2007, 3, e172. [Google Scholar]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- DeepAI. Image Similarity API. 2019. Available online: https://deepai.org/machine-learning-model/image-similarity (accessed on 15 July 2022).
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2009, 37, D355–D359. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 2022, 183, 1024–1042. [Google Scholar] [CrossRef]
- Wang, N.; Sun, Y.; Feng, R.; Wang, Y.; Guo, Y.; Zhang, L.; Deng, Y.Q.; Wang, L.; Cui, Z.; Cao, L.; et al. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021, 31, 101–103. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Starr, T.N.; Czudnochowski, N.; Liu, Z.; Zatta, F.; Park, Y.J.; Addetia, A.; Pinto, D.; Beltramello, M.; Hernandez, P.; Greaney, A.J.; et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef]
- Wodehouse, P. Bioinformatics and pattern recognition come together. J. Pattern Recognit. Res. 2006, 1, 37–41. [Google Scholar]
- Grandgenett, D.P. Symmetrical recognition of cellular DNA target sequences during retroviral integration. Proc. Natl. Acad. Sci. USA 2005, 102, 5903–5904. [Google Scholar] [CrossRef] [PubMed]
- Eck, R.V. Genetic code: Emergence of a symmetrical pattern. Science 1963, 140, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, A.R.; Firtel, R.A. Breaking symmetries: Regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr. Opin. Genet. Dev. 2004, 14, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Howarth, M. Say it with proteins: An alphabet of crystal structures. Nat. Struct. Mol. Biol. 2015, 22, 349. [Google Scholar] [CrossRef] [PubMed]
- Bongini, P.; Cicaloni, V.; Pasqui, A.; Bianchini, M.; Niccolai, N. A Bioinformatics approach to investigate structural and non-structural proteins in human coronaviruses. Front. Genet. 2022, 14, 1303. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. The shape and structure of proteins. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Taujale, R.; Venkat, A.; Huang, L.-C.; Zhou, Z.; Yeung, W.; Rasheed, K.M.; Li, S.; Edison, A.S.; Moremen, K.W.; Kannan, N. Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases. eLife 2020, 9, e54532. [Google Scholar] [CrossRef]
- Hvidsten, T.R.; Lægreid, A.; Kryshtafovych, A.; Andersson, G.; Fidelis, K.; Komorowski, J. A comprehensive analysis of the structure-function relationship in proteins based on local structure similarity. PLoS ONE 2009, 4, e6266. [Google Scholar] [CrossRef]
- Taylor, G.K.; Stoddard, B.L. Structural, functional and evolutionary relationships between homing endonucleases and proteins from their host organisms. Nucleic Acids Res. 2012, 40, 5189–5200. [Google Scholar] [CrossRef][Green Version]


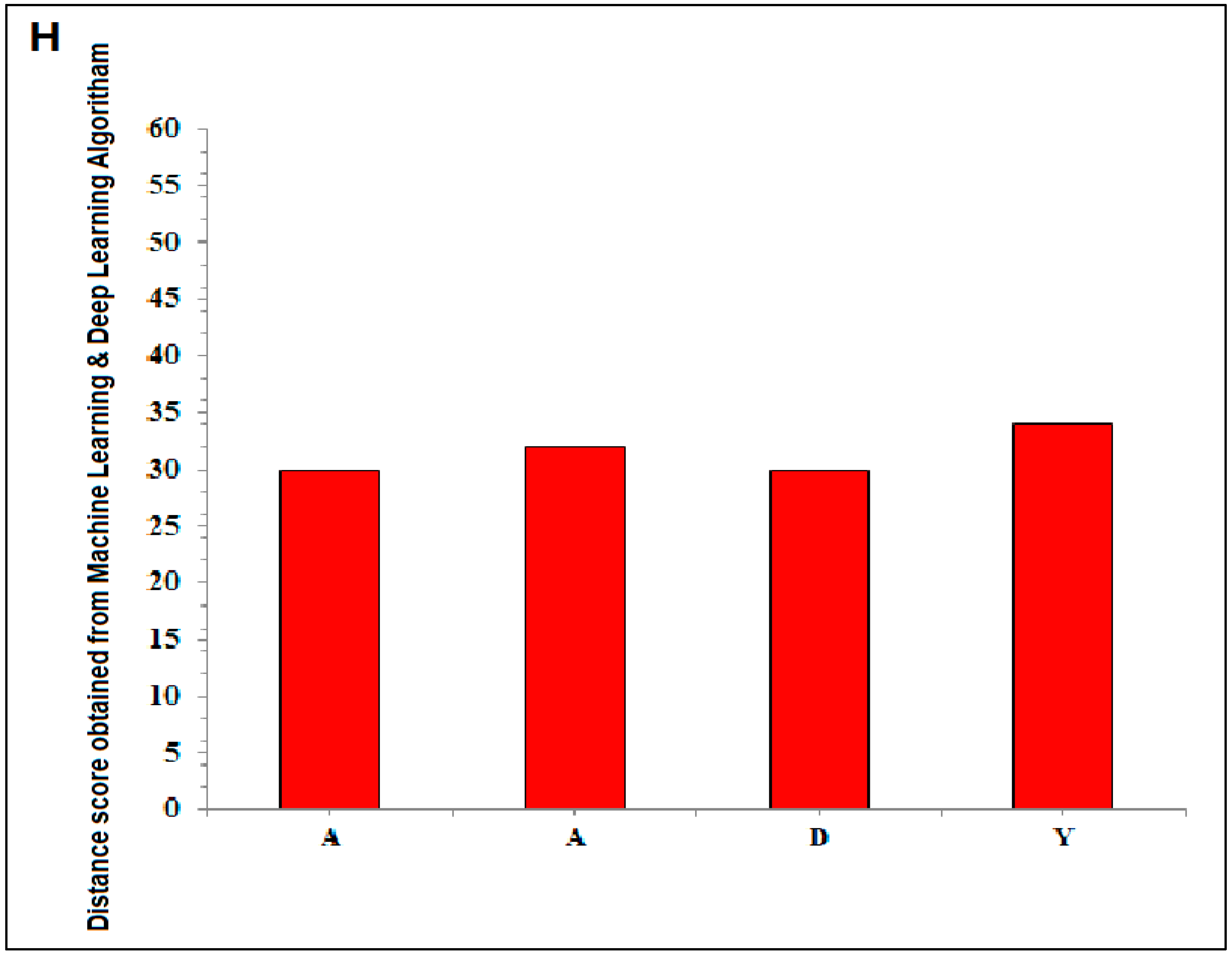

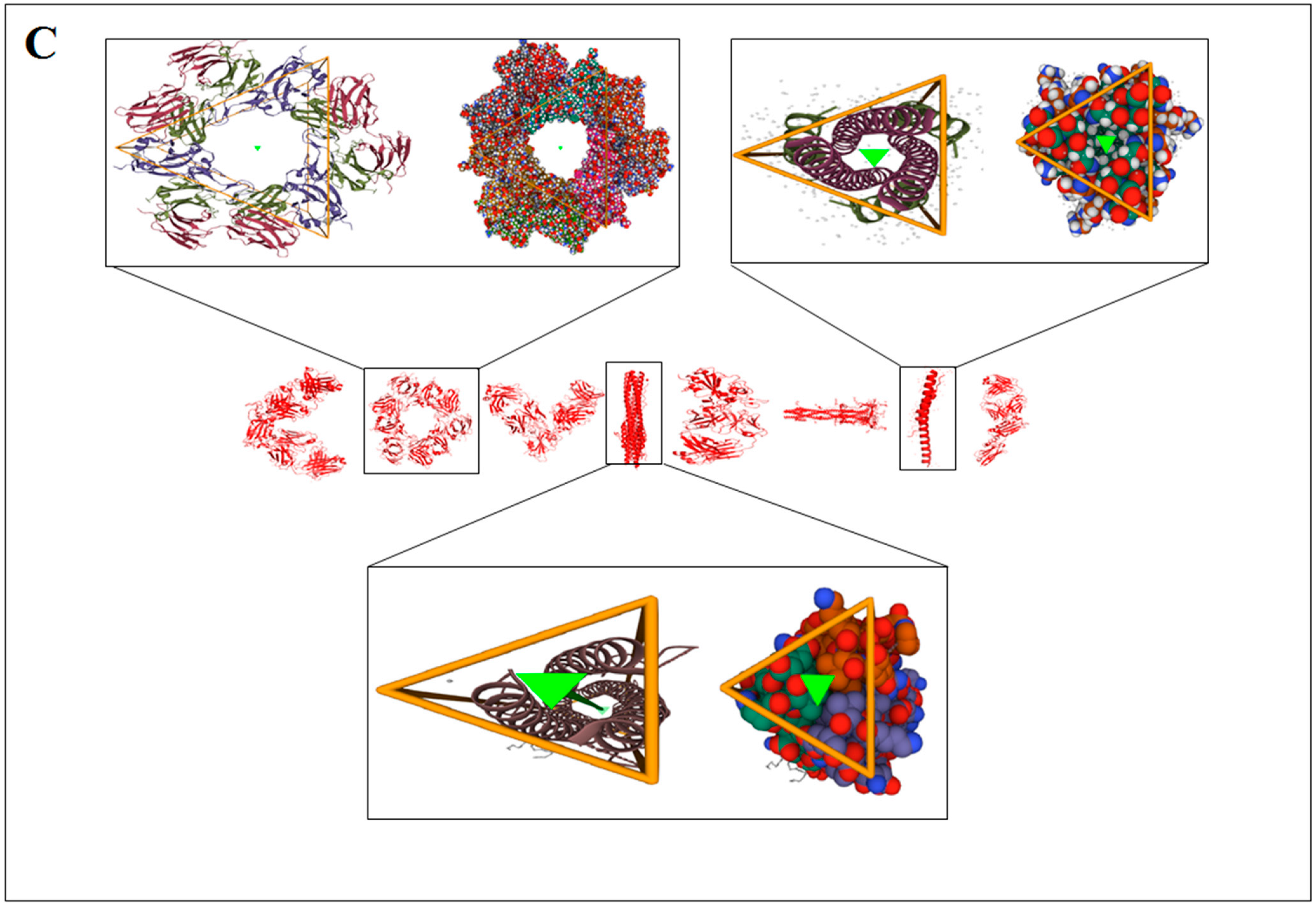
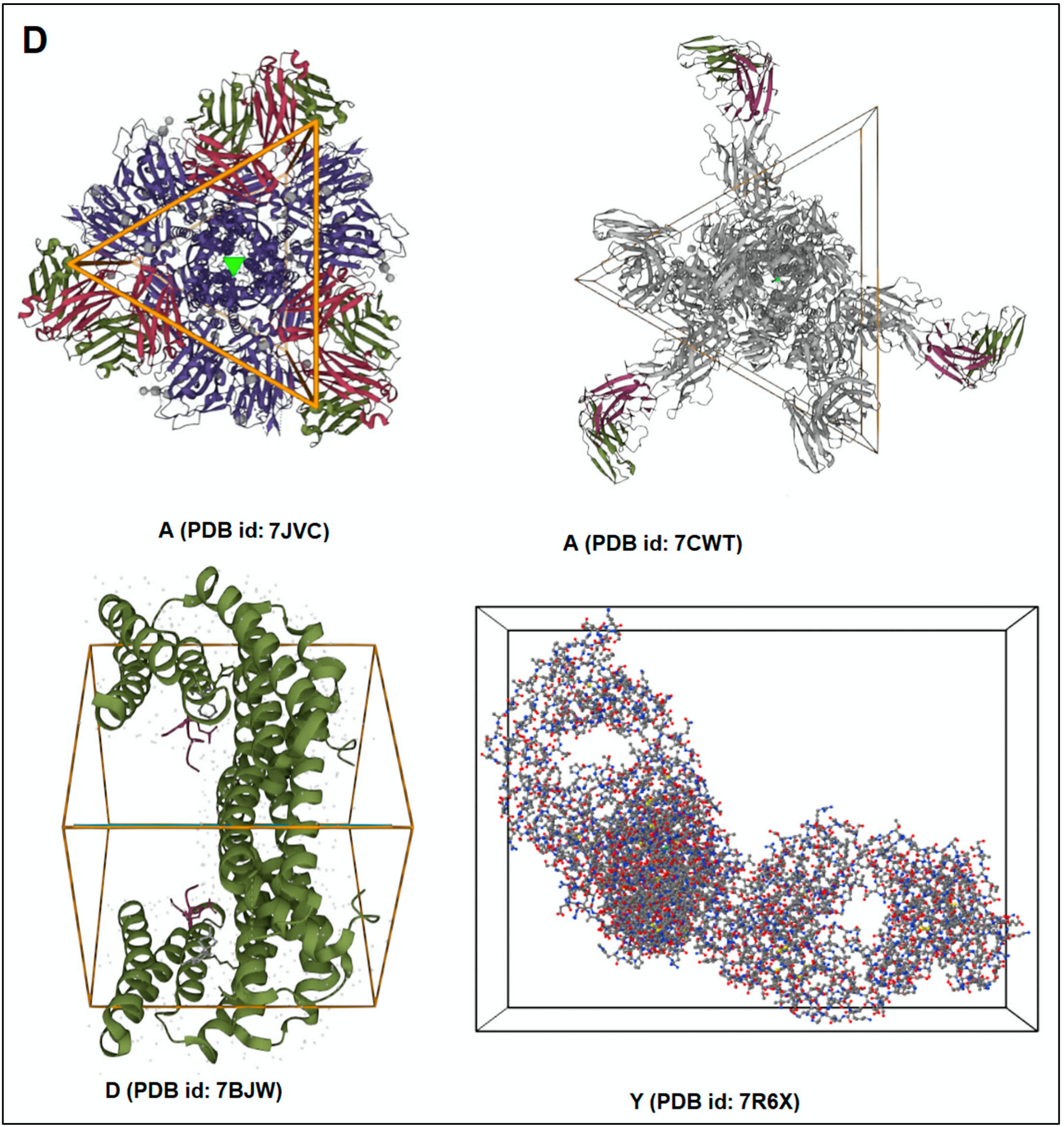


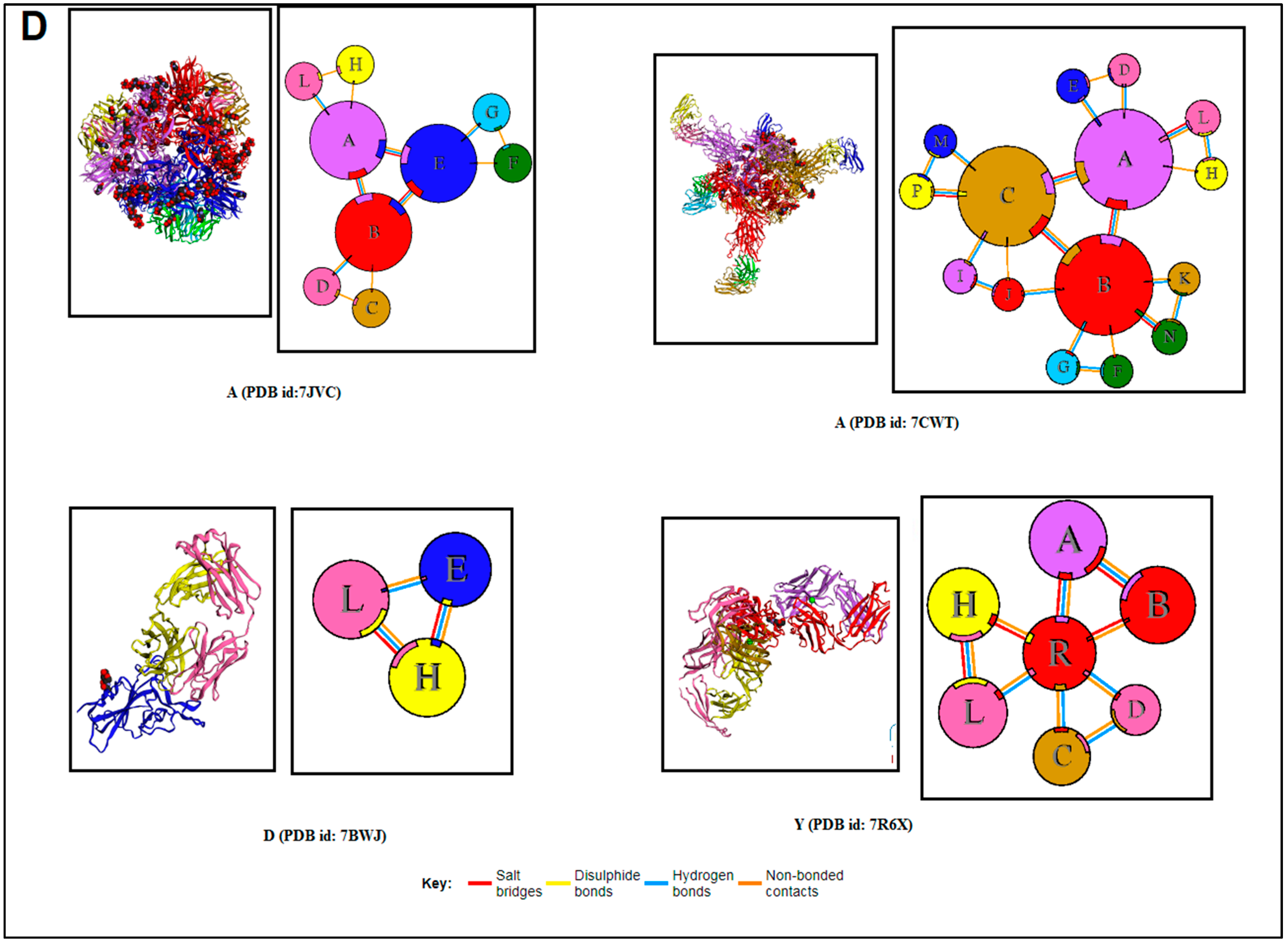




| Sl. No | 3D Structure of the Protein Alphabet Compared with English Alphabet | PDB ID | Remarks | Reference |
|---|---|---|---|---|
| 1. | A | 7JVC | SARS-CoV-2 spike RBD immunodominant sites in complex with the S2A4 neutralizing antibody Fab fragment | [49] |
| 2. | A | 7CWT | Human antibody cocktails (hb27 and fc05 Fab) protein complex with SARS-CoV-2 spike protein | [50] |
| 3. | D | 7BWJ | SARS-CoV-2 spike protein (S1 domain) attached with human antibody (heavy and light chain of Ab) | [51] |
| 4. | Y | 7R6X | Complex structure of SARS-CoV-2 RBD protein complex with S2E12 Fab, S309 Fab, and S304 Fab domain of Ab | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, C.; Bhattacharya, M.; Chatterjee, S.; Sharma, A.R.; Saha, R.P.; Dhama, K.; Agoramoorthy, G. Integrative Bioinformatics Approaches Indicate a Particular Pattern of Some SARS-CoV-2 and Non-SARS-CoV-2 Proteins. Vaccines 2023, 11, 38. https://doi.org/10.3390/vaccines11010038
Chakraborty C, Bhattacharya M, Chatterjee S, Sharma AR, Saha RP, Dhama K, Agoramoorthy G. Integrative Bioinformatics Approaches Indicate a Particular Pattern of Some SARS-CoV-2 and Non-SARS-CoV-2 Proteins. Vaccines. 2023; 11(1):38. https://doi.org/10.3390/vaccines11010038
Chicago/Turabian StyleChakraborty, Chiranjib, Manojit Bhattacharya, Srijan Chatterjee, Ashish Ranjan Sharma, Rudra P. Saha, Kuldeep Dhama, and Govindasamy Agoramoorthy. 2023. "Integrative Bioinformatics Approaches Indicate a Particular Pattern of Some SARS-CoV-2 and Non-SARS-CoV-2 Proteins" Vaccines 11, no. 1: 38. https://doi.org/10.3390/vaccines11010038
APA StyleChakraborty, C., Bhattacharya, M., Chatterjee, S., Sharma, A. R., Saha, R. P., Dhama, K., & Agoramoorthy, G. (2023). Integrative Bioinformatics Approaches Indicate a Particular Pattern of Some SARS-CoV-2 and Non-SARS-CoV-2 Proteins. Vaccines, 11(1), 38. https://doi.org/10.3390/vaccines11010038








