In Vitro Stimulation with Live SARS-CoV-2 Suggests Th17 Dominance In Virus-Specific CD4+ T Cell Response after COVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Viruses
2.3. PBMC Isolation and Stimulation with SARS-CoV-2 and Influenza Viruses
2.4. Activation Induced Cell Marker Assay and Flow Cytometry
2.5. Statistical Analysis
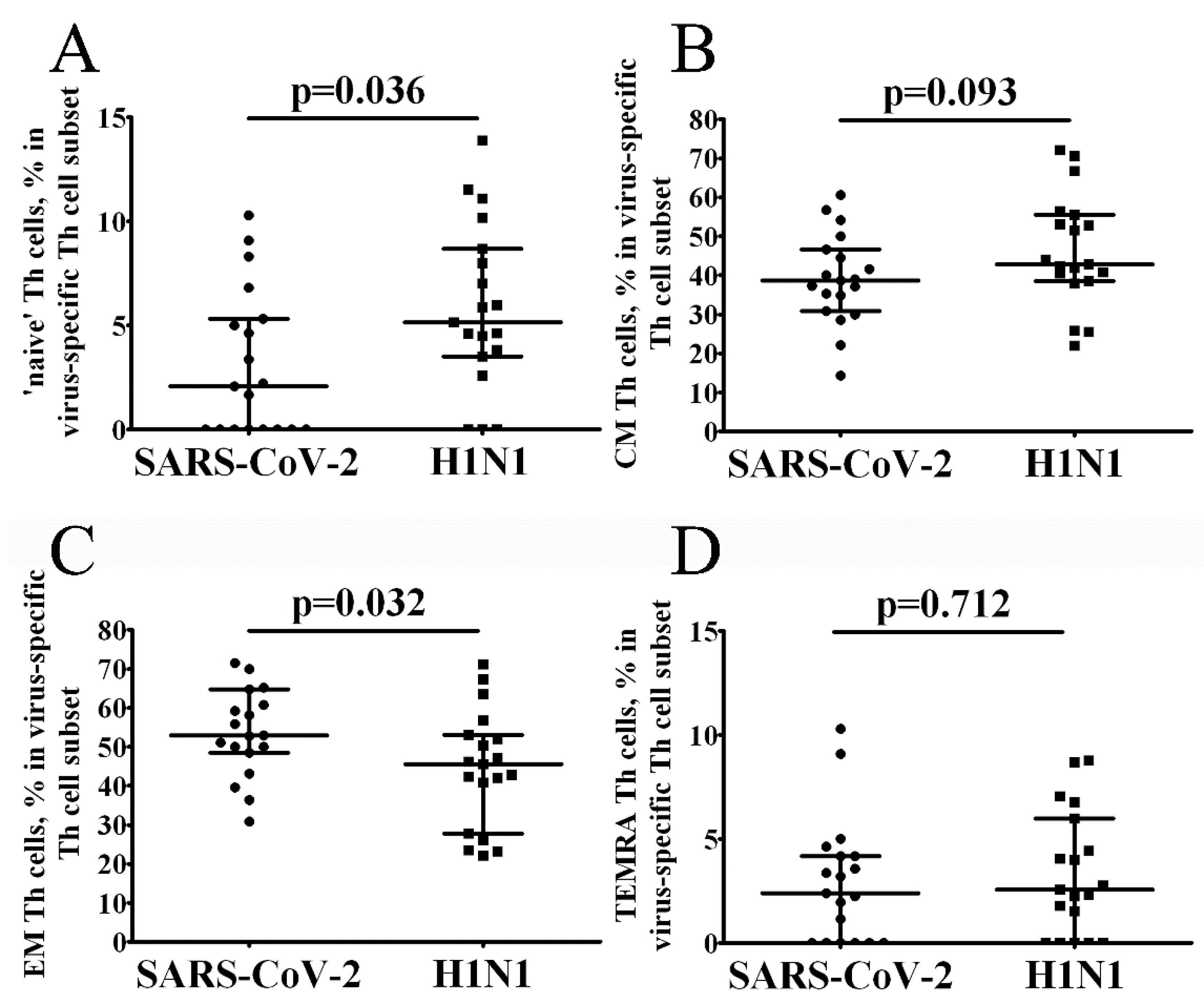
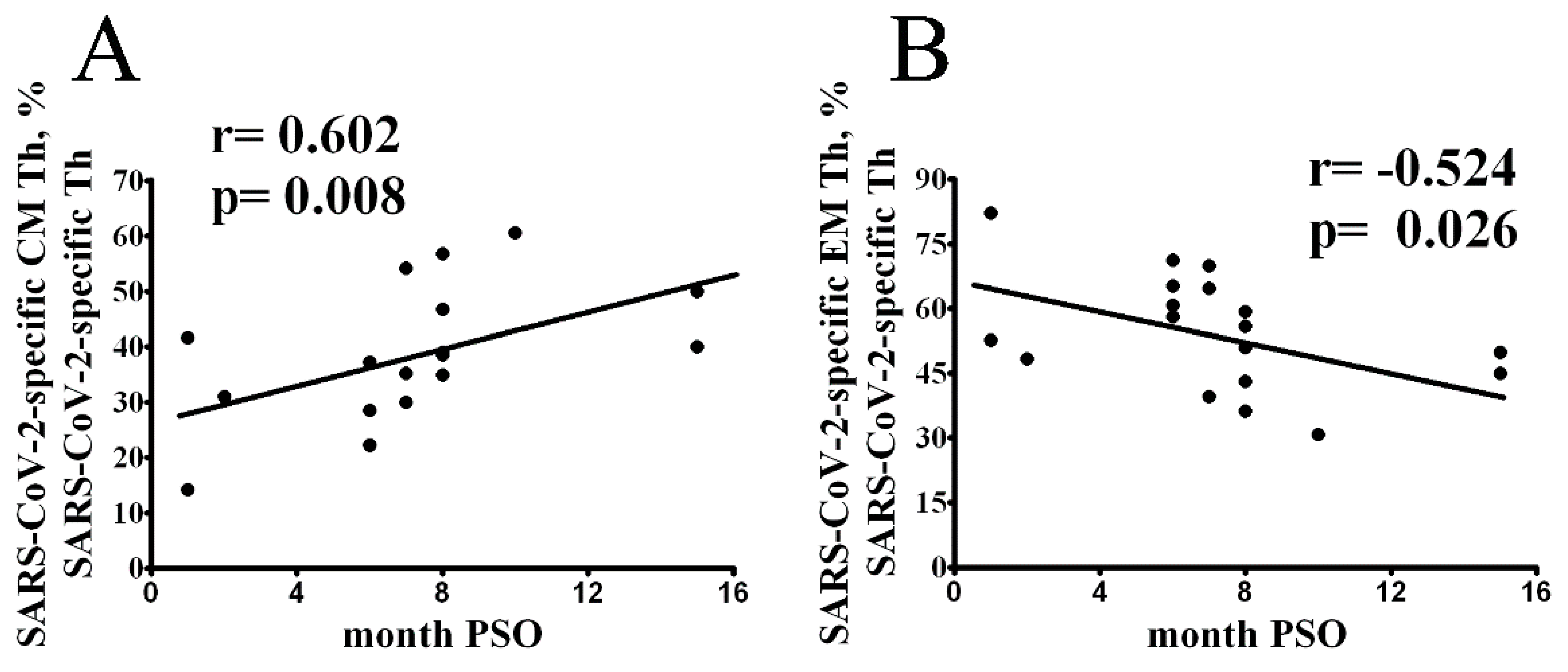
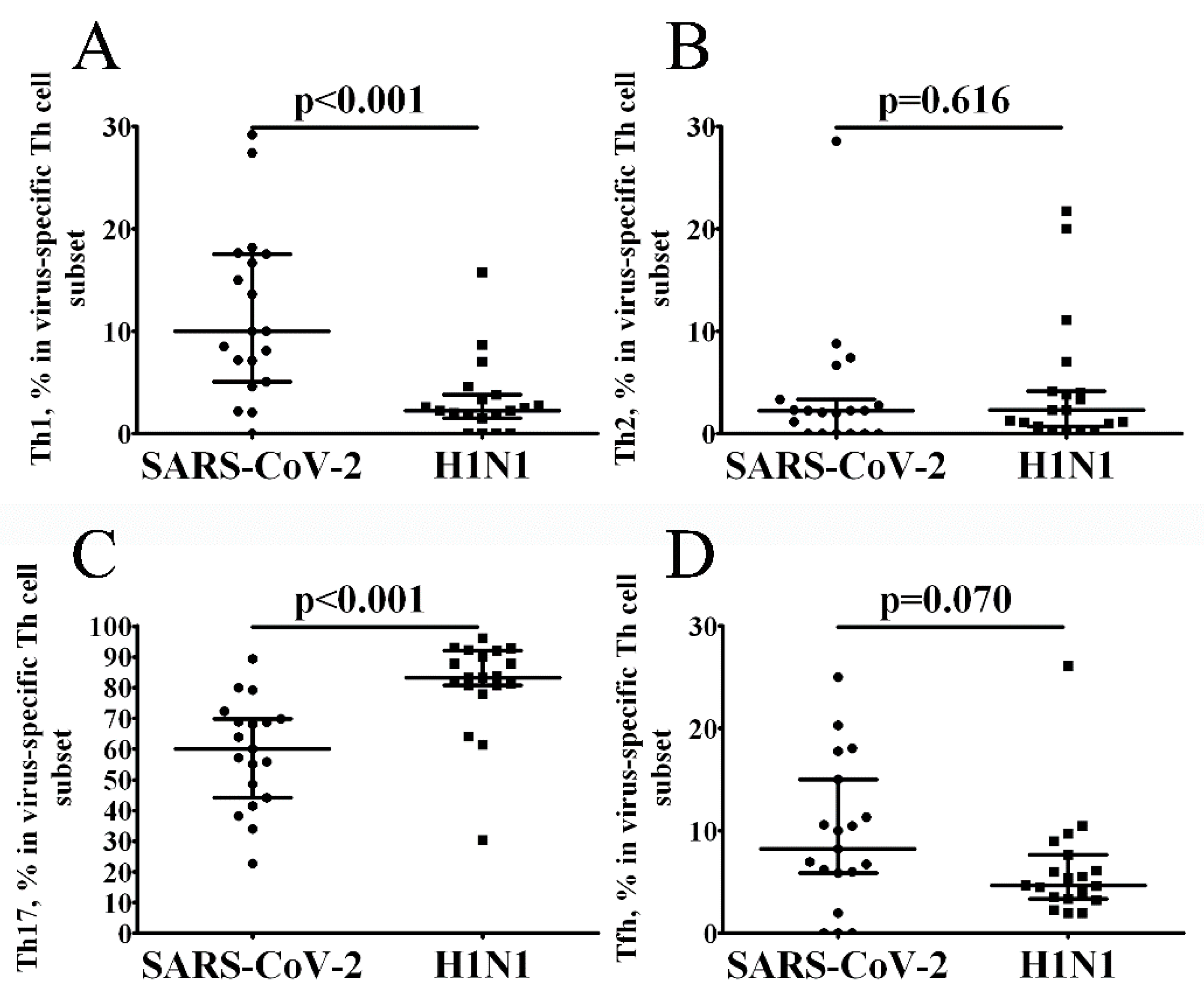
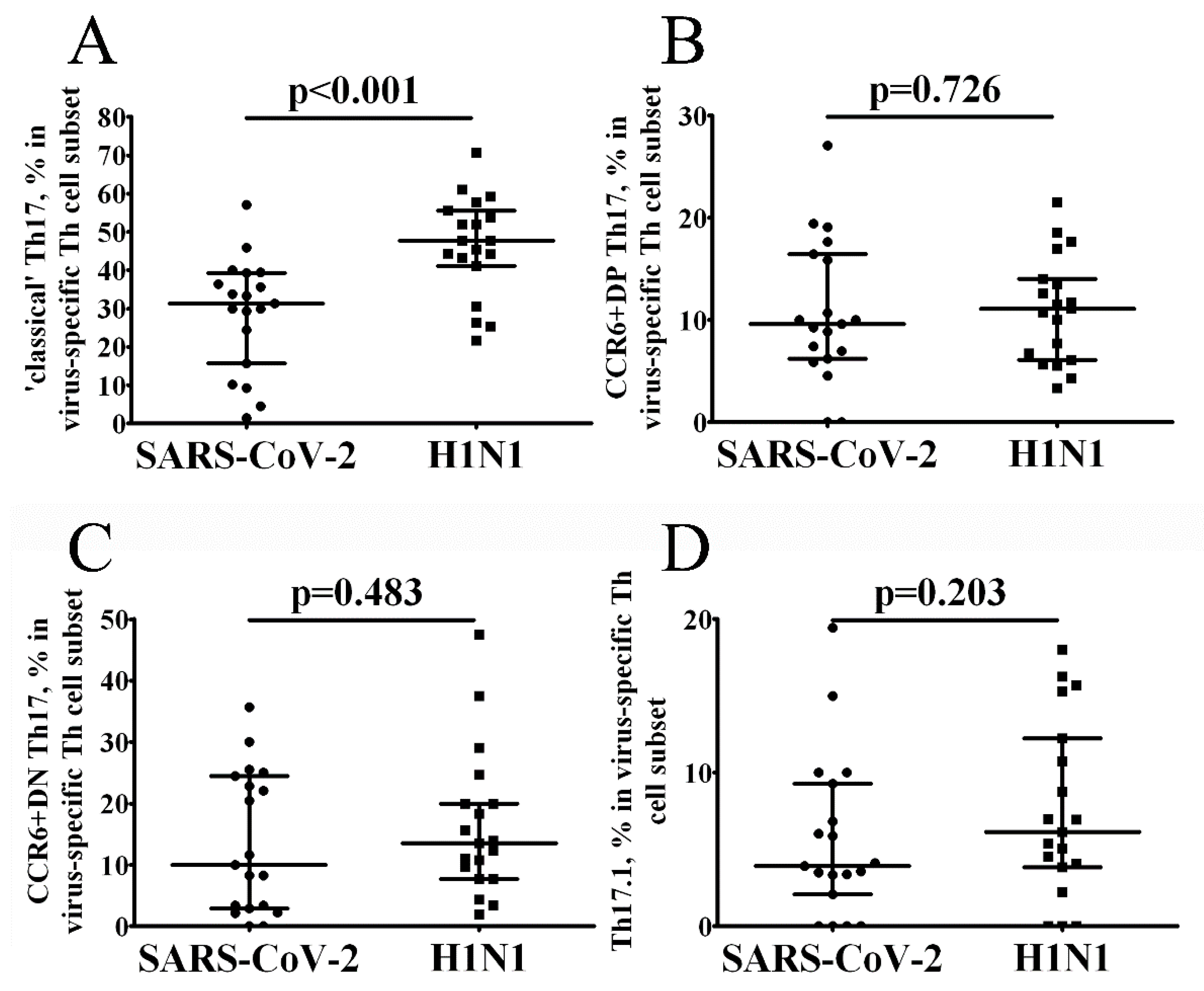
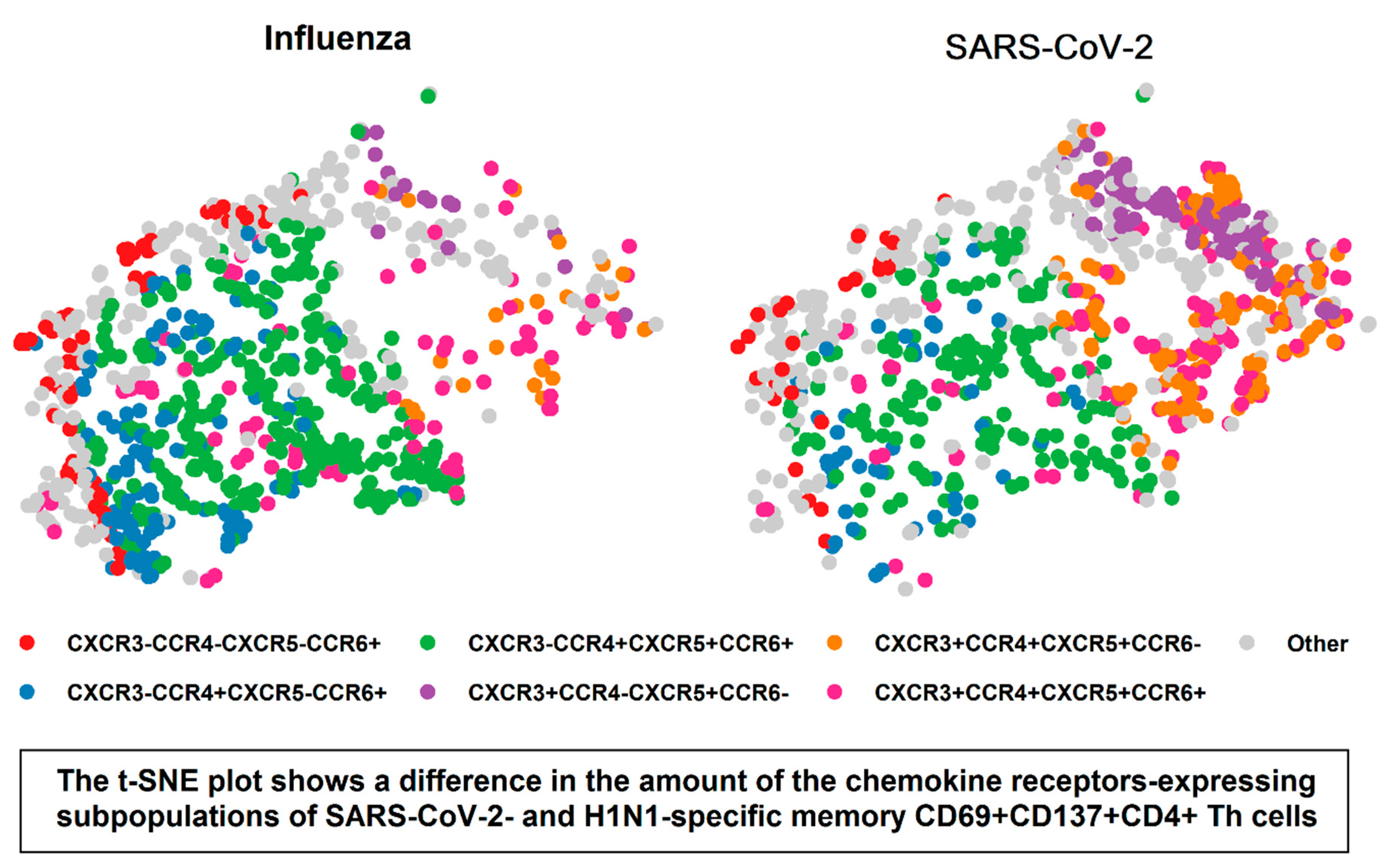
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). 2022. Available online: https://coronavirus.jhu.edu/map.html (accessed on 18 July 2022).
- Law, J.C.; Koh, W.H.; Budylowski, P.; Lin, J.; Yue, F.; Abe, K.T.; Rathod, B.; Girard, M.; Li, Z.; Rini, J.M.; et al. Systematic Examination of Antigen-Specific Recall T Cell Responses to SARS-CoV-2 versus Influenza Virus Reveals a Distinct Inflammatory Profile. J. Immunol. 2021, 206, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Aleebrahim-Dehkordi, E.; Molavi, B.; Mokhtari, M.; Deravi, N.; Fathi, M.; Fazel, T.; Mohebalizadeh, M.; Koochaki, P.; Shobeiri, P.; Hasanpour-Dehkordi, A. T helper type (Th1/Th2) responses to SARS-CoV-2 and influenza A (H1N1) virus: From cytokines produced to immune responses. Transpl. Immunol. 2021, 70, 101495. [Google Scholar] [CrossRef] [PubMed]
- Hirahara, K.; Nakayama, T. CD4+ T-cell subsets in inflammatory diseases: Beyond the Th1/Th2 paradigm. Int. Immunol. 2016, 28, 163–171. [Google Scholar] [CrossRef]
- Kudryavtsev, I.; Rubinstein, A.; Golovkin, A.; Kalinina, O.; Vasilyev, K.; Rudenko, L.; Isakova-Sivak, I. Dysregulated Immune Responses in SARS-CoV-2-Infected Patients: A Comprehensive Overview. Viruses 2022, 14, 1082. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; van den Akker, J.P.C.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e973. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef]
- Li, C.K.; Wu, H.; Yan, H.; Ma, S.; Wang, L.; Zhang, M.; Tang, X.; Temperton, N.J.; Weiss, R.A.; Brenchley, J.M.; et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008, 181, 5490–5500. [Google Scholar] [CrossRef]
- Bertoletti, A.; Tan, A.T.; Le Bert, N. The T-cell response to SARS-CoV-2: Kinetic and quantitative aspects and the case for their protective role. Oxford Open Immunol. 2021, 2, iqab006. [Google Scholar] [CrossRef]
- Matyushenko, V.; Isakova-Sivak, I.; Kudryavtsev, I.; Goshina, A.; Chistyakova, A.; Stepanova, E.; Prokopenko, P.; Sychev, I.; Rudenko, L. Detection of IFNgamma-Secreting CD4(+) and CD8(+) Memory T Cells in COVID-19 Convalescents after Stimulation of Peripheral Blood Mononuclear Cells with Live SARS-CoV-2. Viruses 2021, 13, 1490. [Google Scholar] [CrossRef]
- Amanat, F.; White, K.M.; Miorin, L.; Strohmeier, S.; McMahon, M.; Meade, P.; Liu, W.C.; Albrecht, R.A.; Simon, V.; Martinez-Sobrido, L.; et al. An In Vitro Microneutralization Assay for SARS-CoV-2 Serology and Drug Screening. Curr. Protoc. Microbiol. 2020, 58, e108. [Google Scholar] [CrossRef]
- Reed, L.J.; MUENCH, H. A Simple Method of Estimating Fifty Per Cent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- He, X.S.; Mahmood, K.; Maecker, H.T.; Holmes, T.H.; Kemble, G.W.; Arvin, A.M.; Greenberg, H.B. Analysis of the frequencies and of the memory T cell phenotypes of human CD8+ T cells specific for influenza A viruses. J. Infect. Dis. 2003, 187, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, L.; Isakova-Sivak, I.; Naykhin, A.; Kiseleva, I.; Stukova, M.; Erofeeva, M.; Korenkov, D.; Matyushenko, V.; Sparrow, E.; Kieny, M.P. H7N9 live attenuated influenza vaccine in healthy adults: A randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect. Dis. 2016, 16, 303–310. [Google Scholar] [CrossRef]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e1019. [Google Scholar] [CrossRef]
- Morou, A.; Brunet-Ratnasingham, E.; Dube, M.; Charlebois, R.; Mercier, E.; Darko, S.; Brassard, N.; Nganou-Makamdop, K.; Arumugam, S.; Gendron-Lepage, G.; et al. Altered differentiation is central to HIV-specific CD4(+) T cell dysfunction in progressive disease. Nat. Immunol. 2019, 20, 1059–1070. [Google Scholar] [CrossRef]
- Jung, J.H.; Rha, M.S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Golovkin, A.; Kalinina, O.; Bezrukikh, V.; Aquino, A.; Zaikova, E.; Karonova, T.; Melnik, O.; Vasilieva, E.; Kudryavtsev, I. Imbalanced Immune Response of T-Cell and B-Cell Subsets in Patients with Moderate and Severe COVID-19. Viruses 2021, 13, 1966. [Google Scholar] [CrossRef]
- Wacleche, V.S.; Goulet, J.P.; Gosselin, A.; Monteiro, P.; Soudeyns, H.; Fromentin, R.; Jenabian, M.A.; Vartanian, S.; Deeks, S.G.; Chomont, N.; et al. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology 2016, 13, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Hoechst, B.; Gamrekelashvili, J.; Ormandy, L.A.; Voigtlander, T.; Wedemeyer, H.; Ylaya, K.; Wang, X.W.; Hewitt, S.M.; Manns, M.P.; et al. Human CCR4+ CCR6+ Th17 cells suppress autologous CD8+ T cell responses. J. Immunol. 2012, 188, 6055–6062. [Google Scholar] [CrossRef] [PubMed]
- Juno, J.A.; Tan, H.X.; Lee, W.S.; Reynaldi, A.; Kelly, H.G.; Wragg, K.; Esterbauer, R.; Kent, H.E.; Batten, C.J.; Mordant, F.L.; et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020, 26, 1428–1434. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, A.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.R.; Li, Q.; Li, H.L.; Wang, X.; Wang, Q.; Zheng, X.S.; Geng, R.; Zhang, Y.L.; Li, B.; Jiang, R.D.; et al. Antibody-Dependent Enhancement of SARS-CoV-2 Infection of Human Immune Cells: In Vitro Assessment Provides Insight in COVID-19 Pathogenesis. Viruses 2021, 13, 2483. [Google Scholar] [CrossRef] [PubMed]
- Nelli, R.K.; Phadke, K.S.; Castillo, G.; Yen, L.; Saunders, A.; Rauh, R.; Nelson, W.; Bellaire, B.H.; Gimenez-Lirola, L.G. Enhanced apoptosis as a possible mechanism to self-limit SARS-CoV-2 replication in porcine primary respiratory epithelial cells in contrast to human cells. Cell Death Discov. 2021, 7, 383. [Google Scholar] [CrossRef]
- Pontelli, M.C.; Castro, I.A.; Martins, R.B.; Veras, F.P.; Serra, L.; Nascimento, D.C.; Cardoso, R.S.; Rosales, R.; Lima, T.M.; Souza, J.P.; et al. Infection of human lymphomononuclear cells by SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hou, H.; Zhang, Y.; Tang, G.; Luo, Y.; Liu, W.; Cheng, C.; Jiang, Y.; Xiong, Z.; Wu, S.; Sun, Z.; et al. Immunologic memory to SARS-CoV-2 in convalescent COVID-19 patients at 1 year postinfection. J. Allergy Clin. Immunol. 2021, 148, 1481–1492 e1482. [Google Scholar] [CrossRef]
- Cassaniti, I.; Percivalle, E.; Bergami, F.; Piralla, A.; Comolli, G.; Bruno, R.; Vecchia, M.; Sambo, M.; Colaneri, M.; Zuccaro, V.; et al. SARS-CoV-2 specific T-cell immunity in COVID-19 convalescent patients and unexposed controls measured by ex vivo ELISpot assay. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 27, 1029–1034. [Google Scholar] [CrossRef]
- Neidleman, J.; Luo, X.; George, A.F.; McGregor, M.; Yang, J.; Yun, C.; Murray, V.; Gill, G.; Greene, W.C.; Vasquez, J.; et al. Distinctive features of SARS-CoV-2-specific T cells predict recovery from severe COVID-19. Cell Rep. 2021, 36, 109414. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef] [PubMed]
- Neidleman, J.; Luo, X.; Frouard, J.; Xie, G.; Gill, G.; Stein, E.S.; McGregor, M.; Ma, T.; George, A.F.; Kosters, A.; et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep. Med. 2020, 1, 100081. [Google Scholar] [CrossRef] [PubMed]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e114. [Google Scholar] [CrossRef] [PubMed]
- Prussin, C.; Metcalfe, D.D. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J. Immunol. Methods 1995, 188, 117–128. [Google Scholar] [CrossRef]
- Wunsch, M.; Zhang, W.; Hanson, J.; Caspell, R.; Karulin, A.Y.; Recks, M.S.; Kuerten, S.; Sundararaman, S.; Lehmann, P.V. Characterization of the HCMV-Specific CD4 T Cell Responses that Are Associated with Protective Immunity. Viruses 2015, 7, 4414–4437. [Google Scholar] [CrossRef] [PubMed]
- Duechting, A.; Przybyla, A.; Kuerten, S.; Lehmann, P.V. Delayed Activation Kinetics of Th2- and Th17 Cells Compared to Th1 Cells. Cells 2017, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Kolls, J.K.; Zheng, Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 2008, 28, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Shahgolzari, M.; Yavari, A.; Arjeini, Y.; Miri, S.M.; Darabi, A.; Mozaffari Nejad, A.S.; Keshavarz, M. Immunopathology and Immunopathogenesis of COVID-19, what we know and what we should learn. Gene Rep. 2021, 25, 101417. [Google Scholar] [CrossRef] [PubMed]
- Muyayalo, K.P.; Huang, D.H.; Zhao, S.J.; Xie, T.; Mor, G.; Liao, A.H. COVID-19 and Treg/Th17 imbalance: Potential relationship to pregnancy outcomes. Am. J. Reprod. Immunol. 2020, 84, e13304. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Mahmoud Salehi Khesht, A.; Karpisheh, V.; Qubais Saeed, B.; Olegovna Zekiy, A.; Yapanto, L.M.; Nabi Afjadi, M.; Aksoun, M.; Nasr Esfahani, M.; Aghakhani, F.; Movahed, M.; et al. Different T cell related immunological profiles in COVID-19 patients compared to healthy controls. Int. Immunopharmacol. 2021, 97, 107828. [Google Scholar] [CrossRef] [PubMed]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef] [PubMed]
- Schultheiss, C.; Paschold, L.; Simnica, D.; Mohme, M.; Willscher, E.; von Wenserski, L.; Scholz, R.; Wieters, I.; Dahlke, C.; Tolosa, E.; et al. Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity 2020, 53, 442–455.e444. [Google Scholar] [CrossRef] [PubMed]
- Kalfaoglu, B.; Almeida-Santos, J.; Tye, C.A.; Satou, Y.; Ono, M. T-Cell Hyperactivation and Paralysis in Severe COVID-19 Infection Revealed by Single-Cell Analysis. Front. Immunol. 2020, 11, 589380. [Google Scholar] [CrossRef]
- Ghazavi, A.; Ganji, A.; Keshavarzian, N.; Rabiemajd, S.; Mosayebi, G. Cytokine profile and disease severity in patients with COVID-19. Cytokine 2021, 137, 155323. [Google Scholar] [CrossRef]
- Zhao, Y.; Kilian, C.; Turner, J.E.; Bosurgi, L.; Roedl, K.; Bartsch, P.; Gnirck, A.C.; Cortesi, F.; Schultheiss, C.; Hellmig, M.; et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci. Immunol. 2021, 6, 155323. [Google Scholar] [CrossRef]
- Mahallawi, W.H.; Khabour, O.F.; Zhang, Q.; Makhdoum, H.M.; Suliman, B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 2018, 104, 8–13. [Google Scholar] [CrossRef]
- Josset, L.; Menachery, V.D.; Gralinski, L.E.; Agnihothram, S.; Sova, P.; Carter, V.S.; Yount, B.L.; Graham, R.L.; Baric, R.S.; Katze, M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 2013, 4, e00165-00113. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.F.; Ortiz de Lejarazu, R.; Pumarola, T.; Rello, J.; Almansa, R.; Ramirez, P.; Martin-Loeches, I.; Varillas, D.; Gallegos, M.C.; Seron, C.; et al. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care 2009, 13, R201. [Google Scholar] [CrossRef]
- Frank, K.; Paust, S. Dynamic Natural Killer Cell and T Cell Responses to Influenza Infection. Front. Cell. Infect. Microbiol. 2020, 10, 425. [Google Scholar] [CrossRef]
- Lee, N.; Wong, C.K.; Chan, P.K.; Chan, M.C.; Wong, R.Y.; Lun, S.W.; Ngai, K.L.; Lui, G.C.; Wong, B.C.; Lee, S.K.; et al. Cytokine response patterns in severe pandemic 2009 H1N1 and seasonal influenza among hospitalized adults. PLoS ONE 2011, 6, e26050. [Google Scholar] [CrossRef] [PubMed]
- Almansa, R.; Socias, L.; Ramirez, P.; Martin-Loeches, I.; Valles, J.; Loza, A.; Rello, J.; Kelvin, D.J.; Leon, C.; Blanco, J.; et al. Imbalanced pro- and anti-Th17 responses (IL-17/granulocyte colony-stimulating factor) predict fatal outcome in 2009 pandemic influenza. Crit. Care 2011, 15, 448. [Google Scholar] [CrossRef] [PubMed]
- Sominina, A.; Danilenko, D.; Komissarov, A.; Karpova, L.; Pisareva, M.; Fadeev, A.; Konovalova, N.; Eropkin, M.; Stolyarov, K.; Shtro, A.; et al. Resurgence of Influenza Circulation in the Russian Federation during the Delta and Omicron COVID-19 Era. Viruses 2022, 14, 1909. [Google Scholar] [CrossRef]
- Reiss, S.; Baxter, A.E.; Cirelli, K.M.; Dan, J.M.; Morou, A.; Daigneault, A.; Brassard, N.; Silvestri, G.; Routy, J.P.; Havenar-Daughton, C.; et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE 2017, 12, e0186998. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryavtsev, I.; Matyushenko, V.; Stepanova, E.; Vasilyev, K.; Rudenko, L.; Isakova-Sivak, I. In Vitro Stimulation with Live SARS-CoV-2 Suggests Th17 Dominance In Virus-Specific CD4+ T Cell Response after COVID-19. Vaccines 2022, 10, 1544. https://doi.org/10.3390/vaccines10091544
Kudryavtsev I, Matyushenko V, Stepanova E, Vasilyev K, Rudenko L, Isakova-Sivak I. In Vitro Stimulation with Live SARS-CoV-2 Suggests Th17 Dominance In Virus-Specific CD4+ T Cell Response after COVID-19. Vaccines. 2022; 10(9):1544. https://doi.org/10.3390/vaccines10091544
Chicago/Turabian StyleKudryavtsev, Igor, Victoria Matyushenko, Ekaterina Stepanova, Kirill Vasilyev, Larisa Rudenko, and Irina Isakova-Sivak. 2022. "In Vitro Stimulation with Live SARS-CoV-2 Suggests Th17 Dominance In Virus-Specific CD4+ T Cell Response after COVID-19" Vaccines 10, no. 9: 1544. https://doi.org/10.3390/vaccines10091544
APA StyleKudryavtsev, I., Matyushenko, V., Stepanova, E., Vasilyev, K., Rudenko, L., & Isakova-Sivak, I. (2022). In Vitro Stimulation with Live SARS-CoV-2 Suggests Th17 Dominance In Virus-Specific CD4+ T Cell Response after COVID-19. Vaccines, 10(9), 1544. https://doi.org/10.3390/vaccines10091544







