The Role of Cellular Immunity in the Protective Efficacy of the SARS-CoV-2 Vaccines
Abstract
:1. Introduction
1.1. Humoral and Cellular Immune Responses Induced by SARS-CoV-2 Vaccines
| Name of Vaccine | Platform | Spike- or RBD-Specific Antibody Geometric Mean Titer (GMT) | CD4+ | CD8+ | Clinical Efficacy |
|---|---|---|---|---|---|
| mRNA-1273 | RNA | 1,192,154 compared to 142,140 of HCS | Th1 and Tfh | Potent response | 63.0% efficacy for infections (95% CI, from 56.6 to 68.5) and 98.2% (95% CI, from 92.8 to 99.6) efficacy for severe COVID-19 [13,38,39] |
| BNT162b2 | RNA | 25,006 compared to 602 of HCS | Th1 and Tfh | Potent response | 95% efficacy for prevention of COVID-19 (95% CI, from 90.3 to 97.6) [7,13,40] |
| BBIBP-CorV | Inactivated virus | Neutralizing antibody, 228.7; no comparison to that of HCS | CD4+ or CD8+ response non-distinguishable in humans | CD4+ or CD8+ response non-distinguishable in humans | 83.5% efficacy for infections (95% CI 65.4–92.1) and 100% efficacy for moderate hospitalization [18,19,41] |
| INO-4800 | DNA | From 655.5 to 994.2; no comparison to that of HCS | Not significant | Potent response | Not mentioned [42] |
| ZyCoV-D | DNA | 884.04; slightly lower than that of HCS | CD4+ or CD8+ response non-distinguishable in humans | CD4+ or CD8+ response non-distinguishable in humans | 66.6% efficacy for infections (95% CI 47.6–80.7) and 100% efficacy for moderate–severe COVID-19 [36,43] |
| Ad5-nCoV (Adenovirus type 5 vector) | Ad5 (nonreplicating adeno virus) | 615.8–1445.8; no comparison to that of HCS | CD4+ or CD8+ response non-distinguishable in humans | CD4+ or CD8+ response non-distinguishable in humans | 57.5% efficacy (95% CI, from 39.7 to 70) for infections and 91.7% efficacy for severe COVID-19 [44,45] |
| Ad26.COV2.S | Ad26 (nonreplicating adenovirus) | 1677–2292 compared to 899 of HCS | Th1 | Moderate response | 66.1–76% efficacy (95% CI, 75–77%) for infections and 81% efficacy (95% CI, 78–82%) for COVID-19-related hospitalizations [13,46,47,48,49] |
| NVX-CoV2373 | Protein subunit | 63160 compared to 8344 of HCS | Th1 | CD8+ response not detected in humans | 89.7% efficacy for infections (95% CI, 80.2 to 94.6) 100% efficacy for severe COVID-19 [13,50,51] |
| ZF2001 | Protein subunit | 2777; no comparison to that of HCS | Th1 and Th2 | CD8+ response not detected in humans | Not mentioned [52] |
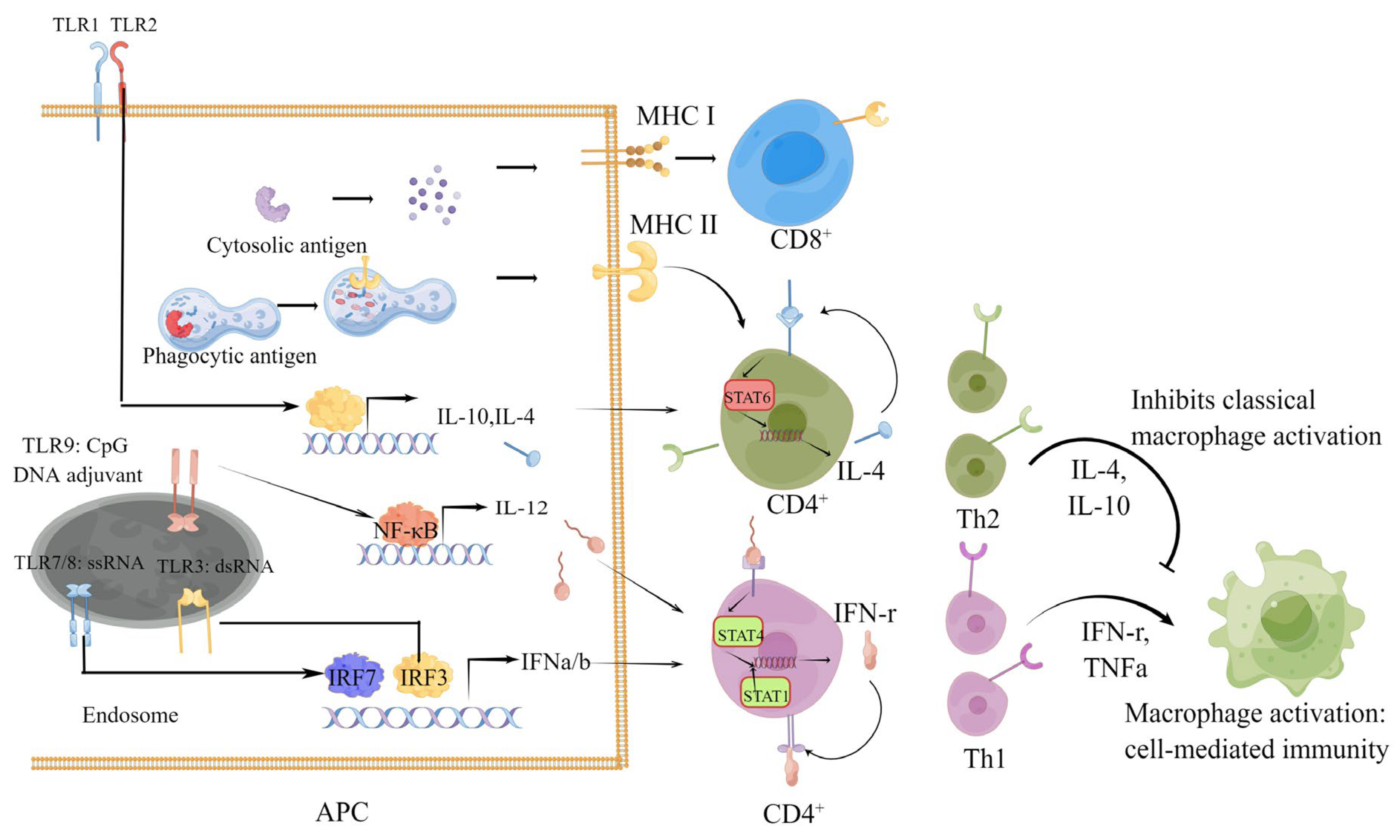
1.2. Role of Cellular Immunity in the Process of SARS-CoV-2 Infection and COVID-19
1.3. Types and Protective Effect of Cellular Immunity Induced by SARS-CoV-2 Infection and Vaccines
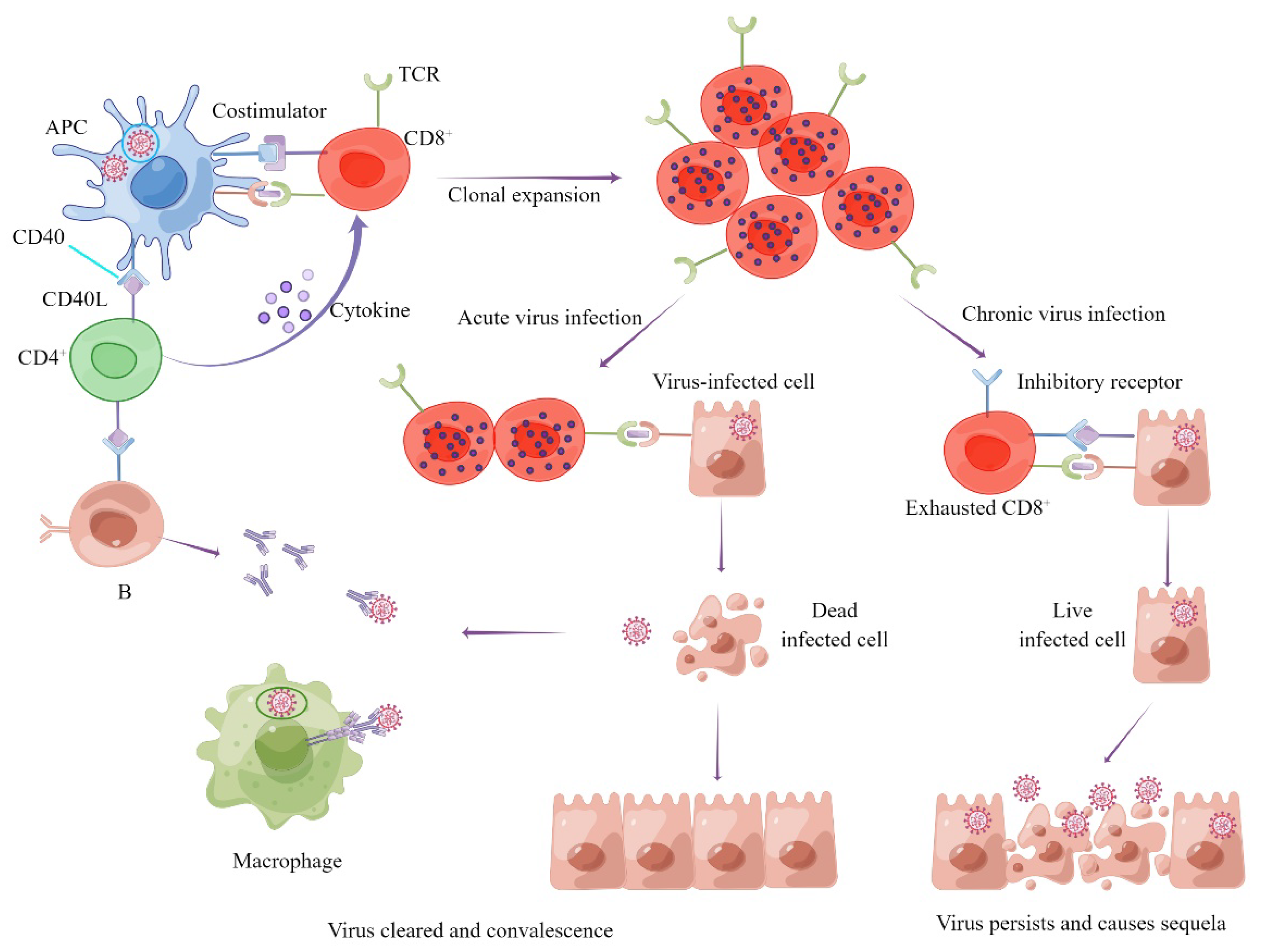
1.4. SARS-CoV-2 Variants Can Escape Vaccine-Induced Humoral Rather Than Cellular Immunity
1.5. Role of Cellular Immunity in the Prevention of Severe Illness Caused by SARS-CoV-2 Variants
1.6. Cellular Immunity May Have the Potential to Ease the Sequelae of COVID-19
1.7. Strategy for Cellular Immunity Vaccine Design and Methods for Cellular Immunity Evaluation
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. Remdesivir in COVID-19: A critical review of pharmacology, pre-clinical and clinical studies. Diabetol. Metab. Syndr. 2020, 14, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Tuccori, M.; Ferraro, S.; Convertino, I.; Cappello, E.; Valdiserra, G.; Blandizzi, C.; Maggi, F.; Focosi, D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: Clinical pipeline. MAbs 2020, 12, 1854149. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. Concurrent human antibody and TH1 type T-cell responses elicited by a COVID-19 RNA vaccine. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Dong, Y.; Dai, T.; Wang, B.; Zhang, L.; Zeng, L.H.; Huang, J.; Yan, H.; Zhang, L.; Zhou, F. The way of SARS-CoV-2 vaccine development: Success and challenges. Signal Transduct. Target. Ther. 2021, 6, 387. [Google Scholar] [CrossRef]
- Hodgson, S.H.; Mansatta, K.; Mallett, G.; Harris, V.; Emary, K.R.W.; Pollard, A.J. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 2021, 21, e26–e35. [Google Scholar] [CrossRef]
- Tesfaye, D.Y.; Gudjonsson, A.; Bogen, B.; Fossum, E. Targeting Conventional Dendritic Cells to Fine-Tune Antibody Responses. Front. Immunol. 2019, 10, 1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Mallajosyula, V.; Tato, C.M.; Tan, G.S.; Wang, T.T. SARS-CoV-2 vaccines in advanced clinical trials: Where do we stand? Adv. Drug Deliv. Rev. 2021, 172, 314–338. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef]
- Fadlyana, E.; Rusmil, K.; Tarigan, R.; Rahmadi, A.R.; Prodjosoewojo, S.; Sofiatin, Y.; Khrisna, C.V.; Sari, R.M.; Setyaningsih, L.; Surachman, F.; et al. A phase III, observer-blind, randomized, placebo-controlled study of the efficacy, safety, and immunogenicity of SARS-CoV-2 inactivated vaccine in healthy adults aged 18-59 years: An interim analysis in Indonesia. Vaccine 2021, 39, 6520–6528. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Vályi-Nagy, I.; Matula, Z.; Gönczi, M.; Tasnády, S.; Bekő, G.; Réti, M.; Ajzner, É.; Uher, F. Comparison of antibody and T cell responses elicited by BBIBP-CorV (Sinopharm) and BNT162b2 (Pfizer-BioNTech) vaccines against SARS-CoV-2 in healthy adult humans. Geroscience 2021, 43, 2321–2331. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Mateus, J.; Dan, J.M.; Zhang, Z.; Rydyznski Moderbacher, C.; Lammers, M.; Goodwin, B.; Sette, A.; Crotty, S.; Weiskopf, D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 2021, 374, eabj9853. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.P. Review of COVID-19 mRNA Vaccines: BNT162b2 and mRNA-1273. J. Pharm. Pract. 2021. [Google Scholar] [CrossRef] [PubMed]
- Collier, A.Y.; Yu, J.; McMahan, K.; Liu, J.; Chandrashekar, A.; Maron, J.S.; Atyeo, C.; Martinez, D.R.; Ansel, J.L.; Aguayo, R.; et al. Differential Kinetics of Immune Responses Elicited by Covid-19 Vaccines. N. Engl. J. Med. 2021, 385, 2010–2012. [Google Scholar] [CrossRef]
- Dickerman, B.A.; Gerlovin, H.; Madenci, A.L.; Kurgansky, K.E.; Ferolito, B.R.; Figueroa Muñiz, M.J.; Gagnon, D.R.; Gaziano, J.M.; Cho, K.; Casas, J.P.; et al. Comparative Effectiveness of BNT162b2 and mRNA-1273 Vaccines in U.S. Veterans. N. Engl. J. Med. 2022, 386, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Self, W.H.; Tenforde, M.W.; Rhoads, J.P.; Gaglani, M.; Ginde, A.A.; Douin, D.J.; Olson, S.M.; Talbot, H.K.; Casey, J.D.; Mohr, N.M.; et al. Comparative Effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) Vaccines in Preventing COVID-19 Hospitalizations Among Adults Without Immunocompromising Conditions—United States, March-August 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1337–1343. [Google Scholar] [PubMed]
- Lim, W.W.; Mak, L.; Leung, G.M.; Cowling, B.J.; Peiris, M. Comparative immunogenicity of mRNA and inactivated vaccines against COVID-19. Lancet Microbe 2021, 2, e423. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.; Mellinghoff, S.C.; Shamsrizi, P.; Addo, M.M.; Dahlke, C. Correlates of Vaccine-Induced Protection against SARS-CoV-2. Vaccines 2021, 9, 238. [Google Scholar] [CrossRef]
- Earle, K.A.; Ambrosino, D.M.; Fiore-Gartland, A.; Goldblatt, D.; Gilbert, P.B.; Siber, G.R.; Dull, P.; Plotkin, S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 2021, 39, 4423–4428. [Google Scholar] [CrossRef]
- Soresina, A.; Moratto, D.; Chiarini, M.; Paolillo, C.; Baresi, G.; Focà, E.; Bezzi, M.; Baronio, B.; Giacomelli, M.; Badolato, R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020, 31, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Montero-Escribano, P.; Matías-Guiu, J.; Gómez-Iglesias, P.; Porta-Etessam, J.; Pytel, V.; Matias-Guiu, J.A. Anti-CD20 and COVID-19 in multiple sclerosis and related disorders: A case series of 60 patients from Madrid, Spain. Mult. Scler. Relat. Disord. 2020, 42, 102185. [Google Scholar] [CrossRef] [PubMed]
- Novi, G.; Mikulska, M.; Briano, F.; Toscanini, F.; Tazza, F.; Uccelli, A.; Inglese, M. COVID-19 in a MS patient treated with ocrelizumab: Does immunosuppression have a protective role? Mult. Scler. Relat. Disord. 2020, 42, 102120. [Google Scholar] [CrossRef] [PubMed]
- Safavi, F.; Nourbakhsh, B.; Azimi, A.R. B-cell depleting therapies may affect susceptibility to acute respiratory illness among patients with multiple sclerosis during the early COVID-19 epidemic in Iran. Mult. Scler. Relat. Disord. 2020, 43, 102195. [Google Scholar] [CrossRef]
- Tang, P.; Hasan, M.R.; Chemaitelly, H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; AlMukdad, S.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021, 27, 2136–2143. [Google Scholar] [CrossRef]
- Dey, A.; Chozhavel Rajanathan, T.M.; Chandra, H.; Pericherla, H.P.R.; Kumar, S.; Choonia, H.S.; Bajpai, M.; Singh, A.K.; Sinha, A.; Saini, G.; et al. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models. Vaccine 2021, 39, 4108–4116. [Google Scholar] [CrossRef]
- Momin, T.; Kansagra, K.; Patel, H.; Sharma, S.; Sharma, B.; Patel, J.; Mittal, R.; Sanmukhani, J.; Maithal, K.; Dey, A.; et al. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): Results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine 2021, 38, 101020. [Google Scholar] [CrossRef]
- Mousa, M.; Albreiki, M.; Alshehhi, F.; AlShamsi, S.; Marzouqi, N.A.; Alawadi, T.; Alrand, H.; Alsafar, H.; Fikri, A. Similar effectiveness of the inactivated vaccine BBIBP-CorV (Sinopharm) and the mRNA vaccine BNT162b2 (Pfizer-BioNTech) against COVID-19 related hospitalizations during the Delta outbreak in the United Arab Emirates. J. Travel Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- El Sahly, H.M.; Baden, L.R.; Essink, B.; Doblecki-Lewis, S.; Martin, J.M.; Anderson, E.J.; Campbell, T.B.; Clark, J.; Jackson, L.A.; Fichtenbaum, C.J.; et al. Efficacy of the mRNA-1273 SARS-CoV-2 Vaccine at Completion of Blinded Phase. N. Engl. J. Med. 2021, 385, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Doğanay, H.L.; Akova, M.; Güner, H.R.; Azap, A.; Akhan, S.; Köse, Ş.; Erdinç, F.; Akalın, E.H.; Tabak, Ö.F.; et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): Interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 2021, 398, 213–222. [Google Scholar] [CrossRef]
- Tebas, P.; Yang, S.; Boyer, J.D.; Reuschel, E.L.; Patel, A.; Christensen-Quick, A.; Andrade, V.M.; Morrow, M.P.; Kraynyak, K.; Agnes, J.; et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine 2021, 31, 100689. [Google Scholar] [CrossRef]
- Khobragade, A.; Bhate, S.; Ramaiah, V.; Deshpande, S.; Giri, K.; Phophle, H.; Supe, P.; Godara, I.; Revanna, R.; Nagarkar, R.; et al. Efficacy, safety, and immunogenicity of the DNA SARS-CoV-2 vaccine (ZyCoV-D): The interim efficacy results of a phase 3, randomised, double-blind, placebo-controlled study in India. Lancet 2022, 399, 1313–1321. [Google Scholar] [CrossRef]
- Zhu, F.C.; Li, Y.H.; Guan, X.H.; Hou, L.H.; Wang, W.J.; Li, J.X.; Wu, S.P.; Wang, B.S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Halperin, S.A.; Ye, L.; MacKinnon-Cameron, D.; Smith, B.; Cahn, P.E.; Ruiz-Palacios, G.M.; Ikram, A.; Lanas, F.; Lourdes Guerrero, M.; Muñoz Navarro, S.R.; et al. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: An international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet 2022, 399, 237–248. [Google Scholar]
- Corchado-Garcia, J.; Zemmour, D.; Hughes, T.; Bandi, H.; Cristea-Platon, T.; Lenehan, P.; Pawlowski, C.; Bade, S.; O’Horo, J.C.; Gores, G.J.; et al. Analysis of the Effectiveness of the Ad26.COV2.S Adenoviral Vector Vaccine for Preventing COVID-19. JAMA Netw. Open 2021, 4, e2132540. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Polinski, J.M.; Weckstein, A.R.; Batech, M.; Kabelac, C.; Kamath, T.; Harvey, R.; Jain, S.; Rassen, J.A.; Khan, N.; Schneeweiss, S. Durability of the Single-Dose Ad26.COV2.S Vaccine in the Prevention of COVID-19 Infections and Hospitalizations in the US Before and During the Delta Variant Surge. JAMA Netw. Open 2022, 5, e222959. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, Y.; Dai, L.; Wang, J.; He, P.; Li, C.; Fang, X.; Wang, C.; Zhao, X.; Huang, E.; et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: Two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021, 21, 1107–1119. [Google Scholar] [CrossRef]
- Liu, Z.; Long, W.; Tu, M.; Chen, S.; Huang, Y.; Wang, S.; Zhou, W.; Chen, D.; Zhou, L.; Wang, M.; et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020, 81, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, T. CD4+T, CD8+T counts and severe COVID-19: A meta-analysis. J. Infect. 2020, 81, e82–e84. [Google Scholar] [CrossRef] [PubMed]
- Swadling, L.; Maini, M.K. T cells in COVID-19—united in diversity. Nat. Immunol. 2020, 21, 1307–1308. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Kroemer, M.; Spehner, L.; Vettoretti, L.; Bouard, A.; Eberst, G.; Pili Floury, S.; Capellier, G.; Lepiller, Q.; Orillard, E.; Mansi, L.; et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J. Infect. 2021, 82, 282–327. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2021, 601, 110–117. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, J.; Mangalam, A.K.; Channappanavar, R.; Fett, C.; Meyerholz, D.K.; Agnihothram, S.; Baric, R.S.; David, C.S.; Perlman, S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 2016, 44, 1379–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Lu, Z.; Zhang, L.; Fan, T.; Xiong, R.; Shen, X.; Feng, H.; Meng, H.; Lin, W.; Jiang, W.; et al. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020, 127, 104361. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e19. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Sagar; Daul, F.; Salvat Lago, M.; Decker, A.; et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021, 27, 78–85. [Google Scholar] [CrossRef]
- Zuo, J.; Dowell, A.C.; Pearce, H.; Verma, K.; Long, H.M.; Begum, J.; Aiano, F.; Amin-Chowdhury, Z.; Hoschler, K.; Brooks, T.; et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021, 22, 620–626. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Stephenson, K.E.; Le Gars, M.; Sadoff, J.; de Groot, A.M.; Heerwegh, D.; Truyers, C.; Atyeo, C.; Loos, C.; Chandrashekar, A.; McMahan, K.; et al. Immunogenicity of the Ad26.COV2.S Vaccine for COVID-19. JAMA 2021, 325, 1535–1544. [Google Scholar] [CrossRef]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell 2020, 183, 1340–1353.e16. [Google Scholar] [CrossRef] [PubMed]
- Juno, J.A.; Tan, H.X.; Lee, W.S.; Reynaldi, A.; Kelly, H.G.; Wragg, K.; Esterbauer, R.; Kent, H.E.; Batten, C.J.; Mordant, F.L.; et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020, 26, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Sattler, A.; Angermair, S.; Stockmann, H.; Heim, K.M.; Khadzhynov, D.; Treskatsch, S.; Halleck, F.; Kreis, M.E.; Kotsch, K. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J. Clin. Investig. 2020, 130, 6477–6489. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Arunachalam, P.S.; O’Hagan, D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021, 20, 454–475. [Google Scholar] [CrossRef]
- Zhuang, Z.; Lai, X.; Sun, J.; Chen, Z.; Zhang, Z.; Dai, J.; Liu, D.; Li, Y.; Li, F.; Wang, Y.; et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 2021, 218, e20202187. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, S.; Haque, A.; Vandierendonck, A.; Weidensee, B.; Plovyt, M.; Stuchlíková, M.; François, N.; Valembois, S.; Heyndrickx, L.; Michiels, J.; et al. A dual-antigen self-amplifying RNA SARS-CoV-2 vaccine induces potent humoral and cellular immune responses and protects against SARS-CoV-2 variants through T cell-mediated immunity. Mol. Ther. 2022. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. Nature 2021, 602, 294–299. [Google Scholar]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Cheng, L.; Song, S.; Zhou, B.; Ge, X.; Yu, J.; Zhang, M.; Ju, B.; Zhang, Z. Impact of the N501Y substitution of SARS-CoV-2 Spike on neutralizing monoclonal antibodies targeting diverse epitopes. Virol. J. 2021, 18, 87. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Krammer, F.; Simon, V.; Martinez-Sobrido, L.; Garcia-Sastre, A.; Schotsaert, M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. MedRxiv 2021, 2, e283–e284. [Google Scholar]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Methot, N.; Yu, E.D.; Zhang, Y.; Dan, J.M.; Goodwin, B.; Rubiro, P.; Sutherland, A.; Wang, E.; et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021, 2, 100355. [Google Scholar] [CrossRef]
- Alter, G.; Yu, J.; Liu, J.; Chandrashekar, A.; Borducchi, E.N.; Tostanoski, L.H.; McMahan, K.; Jacob-Dolan, C.; Martinez, D.R.; Chang, A.; et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 2021, 596, 268–272. [Google Scholar] [CrossRef]
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e11. [Google Scholar] [CrossRef]
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jäger, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2021, 601, 617–622. [Google Scholar] [CrossRef]
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 229–237. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef]
- Lundgren, J.D.; Grund, B.; Barkauskas, C.E.; Holland, T.L.; Gottlieb, R.L.; Sandkovsky, U.; Brown, S.M.; Knowlton, K.U.; Self, W.H.; Files, D.C.; et al. A Neutralizing Monoclonal Antibody for Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 905–914. [Google Scholar]
- Mehandru, S.; Merad, M. Pathological sequelae of long-haul COVID. Nat. Immunol. 2022, 23, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; Tabernero, J.; Bower, M.; Scotti, L.; Patel, M.; Colomba, E.; Dolly, S.; Loizidou, A.; Chester, J.; Mukherjee, U.; et al. Prevalence and impact of COVID-19 sequelae on treatment and survival of patients with cancer who recovered from SARS-CoV-2 infection: Evidence from the OnCovid retrospective, multicentre registry study. Lancet Oncol. 2021, 22, 1669–1680. [Google Scholar] [CrossRef]
- Carod-Artal, F.J. Post-COVID-19 syndrome: Epidemiology, diagnostic criteria and pathogenic mechanisms involved. Rev. Neurol. 2021, 72, 384–396. [Google Scholar] [PubMed]
- Douaud, G.; Lee, S.; Alfaro-Almagro, F.; Arthofer, C.; Wang, C.; McCarthy, P.; Lange, F.; Andersson, J.L.R.; Griffanti, L.; Duff, E.; et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature 2022, 604, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Chen, Y.; Wang, Q.H.; Wang, L.R.; Jiang, L.; Yang, Y.; Chen, X.; Li, Y.; Cen, Y.; Xu, C.; et al. One-Year Trajectory of Cognitive Changes in Older Survivors of COVID-19 in Wuhan, China: A Longitudinal Cohort Study. JAMA Neurol. 2022, 79, 509–517. [Google Scholar] [CrossRef]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 16144. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Bowe, B.; Xie, Y. Long Covid after Breakthrough COVID-19: The post-acute sequelae of breakthrough COVID-19. Nat. Med. 2021. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Yong, S.J. Long COVID or post-COVID-19 syndrome: Putative pathophysiology, risk factors, and treatments. Infect. Dis. 2021, 53, 737–754. [Google Scholar] [CrossRef]
- Huang, L.; Yao, Q.; Gu, X.; Wang, Q.; Ren, L.; Wang, Y.; Hu, P.; Guo, L.; Liu, M.; Xu, J.; et al. 1-year outcomes in hospital survivors with COVID-19: A longitudinal cohort study. Lancet 2021, 398, 747–758. [Google Scholar] [CrossRef]
- Ramakrishnan, R.K.; Kashour, T.; Hamid, Q.; Halwani, R.; Tleyjeh, I.M. Unraveling the Mystery Surrounding Post-Acute Sequelae of COVID-19. Front. Immunol. 2021, 12, 686029. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.A.; Leavy, O.C.; Richardson, M.; Elneima, O.; McCauley, H.J.; Shikotra, A.; Singapuri, A.; Sereno, M.; Saunders, R.M.; Harris, V.C.; et al. Clinical characteristics with inflammation profiling of long COVID and association with 1-year recovery following hospitalisation in the UK: A prospective observational study. Lancet Respir. Med. 2022. [Google Scholar] [CrossRef]
- Su, Y.; Yuan, D.; Chen, D.G.; Ng, R.H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, 881–895.e20. [Google Scholar] [CrossRef] [PubMed]
- Gold, J.E.; Okyay, R.A.; Licht, W.E.; Hurley, D.J. Investigation of Long COVID Prevalence and Its Relationship to Epstein-Barr Virus Reactivation. Pathogens 2021, 10, 763. [Google Scholar] [CrossRef] [PubMed]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Crook, H.; Raza, S.; Nowell, J.; Young, M.; Edison, P. Long covid-mechanisms, risk factors, and management. BMJ 2021, 374, n1648. [Google Scholar] [CrossRef]
- Siripanthong, B.; Asatryan, B.; Hanff, T.C.; Chatha, S.R.; Khanji, M.Y.; Ricci, F.; Muser, D.; Ferrari, V.A.; Nazarian, S.; Santangeli, P.; et al. The Pathogenesis and Long-Term Consequences of COVID-19 Cardiac Injury. JACC Basic Trans. Sci. 2022, 7, 294–308. [Google Scholar] [CrossRef]
- Young, A.R.; Locke, M.C.; Cook, L.E.; Hiller, B.E.; Zhang, R.; Hedberg, M.L.; Monte, K.J.; Veis, D.J.; Diamond, M.S.; Lenschow, D.J. Dermal and muscle fibroblasts and skeletal myofibers survive chikungunya virus infection and harbor persistent RNA. PLoS Pathog. 2019, 15, e1007993. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Saeidi, A.; Zandi, K.; Cheok, Y.Y.; Saeidi, H.; Wong, W.F.; Lee, C.Y.Q.; Cheong, H.C.; Yong, Y.K.; Larsson, M.; Shankar, E.M. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Front. Immunol. 2018, 9, 2569. [Google Scholar] [CrossRef] [PubMed]
- Peluso, M.J.; Deitchman, A.N.; Torres, L.; Iyer, N.S.; Munter, S.E.; Nixon, C.C.; Donatelli, J.; Thanh, C.; Takahashi, S.; Hakim, J.; et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021, 36, 109518. [Google Scholar] [CrossRef]
- Nehme, M.; Braillard, O.; Salamun, J.; Jacquerioz, F.; Courvoisier, D.S.; Spechbach, H.; Guessous, I. Symptoms After COVID-19 Vaccination in Patients with Post-Acute Sequelae of SARS-CoV-2. J. Gen. Intern. Med. 2022, 37, 1585–1588. [Google Scholar] [CrossRef]
- Wisnivesky, J.P.; Govindarajulu, U.; Bagiella, E.; Goswami, R.; Kale, M.; Campbell, K.N.; Meliambro, K.; Chen, Z.; Aberg, J.A.; Lin, J.J. Association of Vaccination with the Persistence of Post-COVID Symptoms. J. Gen. Intern. Med. 2022, 37, 1748–1753. [Google Scholar] [CrossRef] [PubMed]
- Visvabharathy, L.; Hanson, B.; Orban, Z.; Lim, P.H.; Palacio, N.M.; Jain, R.; Clark, J.R.; Graham, E.L.; Liotta, E.M.; Penaloza-MacMaster, P.; et al. Neuro-COVID long-haulers exhibit broad dysfunction in T cell memory generation and responses to vaccination. MedRxiv 2021. [Google Scholar] [CrossRef]
- Ferretti, A.P.; Kula, T.; Wang, Y.; Nguyen, D.M.V.; Weinheimer, A.; Dunlap, G.S.; Xu, Q.; Nabilsi, N.; Perullo, C.R.; Cristofaro, A.W.; et al. Unbiased Screens Show CD8(+) T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity 2020, 53, 1095–1107.e3. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680.e2. [Google Scholar] [CrossRef]
- Hartung, E.; Becker, M.; Bachem, A.; Reeg, N.; Jäkel, A.; Hutloff, A.; Weber, H.; Weise, C.; Giesecke, C.; Henn, V.; et al. Induction of potent CD8 T cell cytotoxicity by specific targeting of antigen to cross-presenting dendritic cells in vivo via murine or human XCR1. J. Immunol. 2015, 194, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Grunwitz, C.; Salomon, N.; Vascotto, F.; Selmi, A.; Bukur, T.; Diken, M.; Kreiter, S.; Türeci, Ö.; Sahin, U. HPV16 RNA-LPX vaccine mediates complete regression of aggressively growing HPV-positive mouse tumors and establishes protective T cell memory. Oncoimmunology 2019, 8, e1629259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahase, E. Covid-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Li, Y.H.; Wang, L.L.; Liu, H.Q.; Lu, S.Y.; Liu, Y.; Li, K.; Liu, B.; Li, S.Y.; Shao, F.M.; et al. Azvudine is a thymus-homing anti-SARS-CoV-2 drug effective in treating COVID-19 patients. Signal Transduct. Target. Ther. 2021, 6, 414. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Wu, T.; Xie, H.; Li, Y.; Zhang, J.; Su, X.; Qi, H. The Role of Cellular Immunity in the Protective Efficacy of the SARS-CoV-2 Vaccines. Vaccines 2022, 10, 1103. https://doi.org/10.3390/vaccines10071103
Sun Z, Wu T, Xie H, Li Y, Zhang J, Su X, Qi H. The Role of Cellular Immunity in the Protective Efficacy of the SARS-CoV-2 Vaccines. Vaccines. 2022; 10(7):1103. https://doi.org/10.3390/vaccines10071103
Chicago/Turabian StyleSun, Zhongjie, Tingxin Wu, Huangfan Xie, Yuhuan Li, Jinlan Zhang, Xuncheng Su, and Hailong Qi. 2022. "The Role of Cellular Immunity in the Protective Efficacy of the SARS-CoV-2 Vaccines" Vaccines 10, no. 7: 1103. https://doi.org/10.3390/vaccines10071103
APA StyleSun, Z., Wu, T., Xie, H., Li, Y., Zhang, J., Su, X., & Qi, H. (2022). The Role of Cellular Immunity in the Protective Efficacy of the SARS-CoV-2 Vaccines. Vaccines, 10(7), 1103. https://doi.org/10.3390/vaccines10071103







