Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies
Abstract
1. Introduction
2. Materials and Methods
2.1. PCR Amplification
2.2. Construction of pRSET B-M2e-NP Expression Plasmid
2.3. Site-Directed Mutagenesis PCR
2.4. Optimization of Protein Expression (M2e-NP and mM2e-NP Proteins)
2.5. Purification of Recombinant Proteins Using Affinity Column
2.6. Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blotting
2.7. Enzyme-Linked Immunosorbent Assay (ELISA)
3. Results
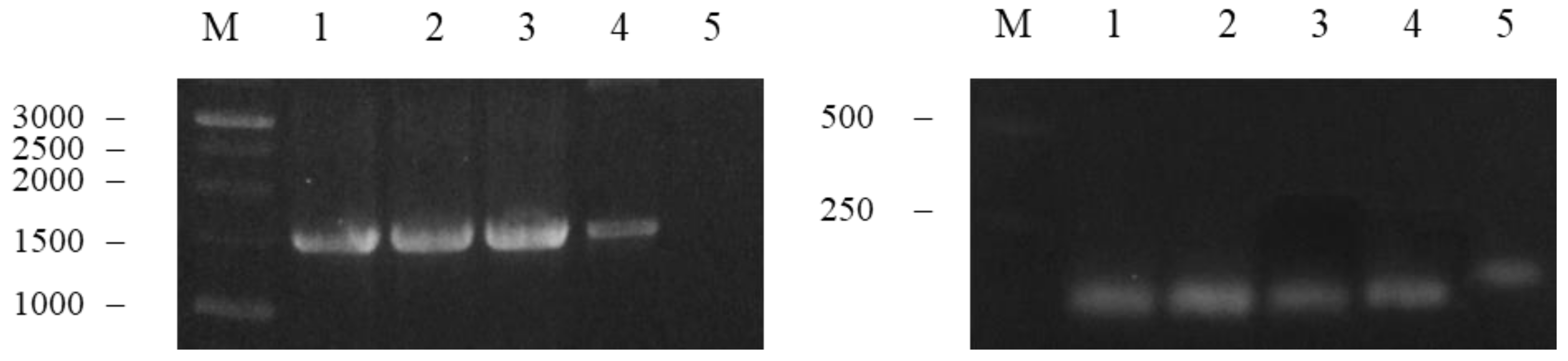
3.1. Amplification and Fusion of M2e and NP Genes
3.2. Synthesis of M2e-NP Fusion Gene
3.3. Selection of Positive Clones Carrying pRSET B-M2e-NP and DNA Sequencing
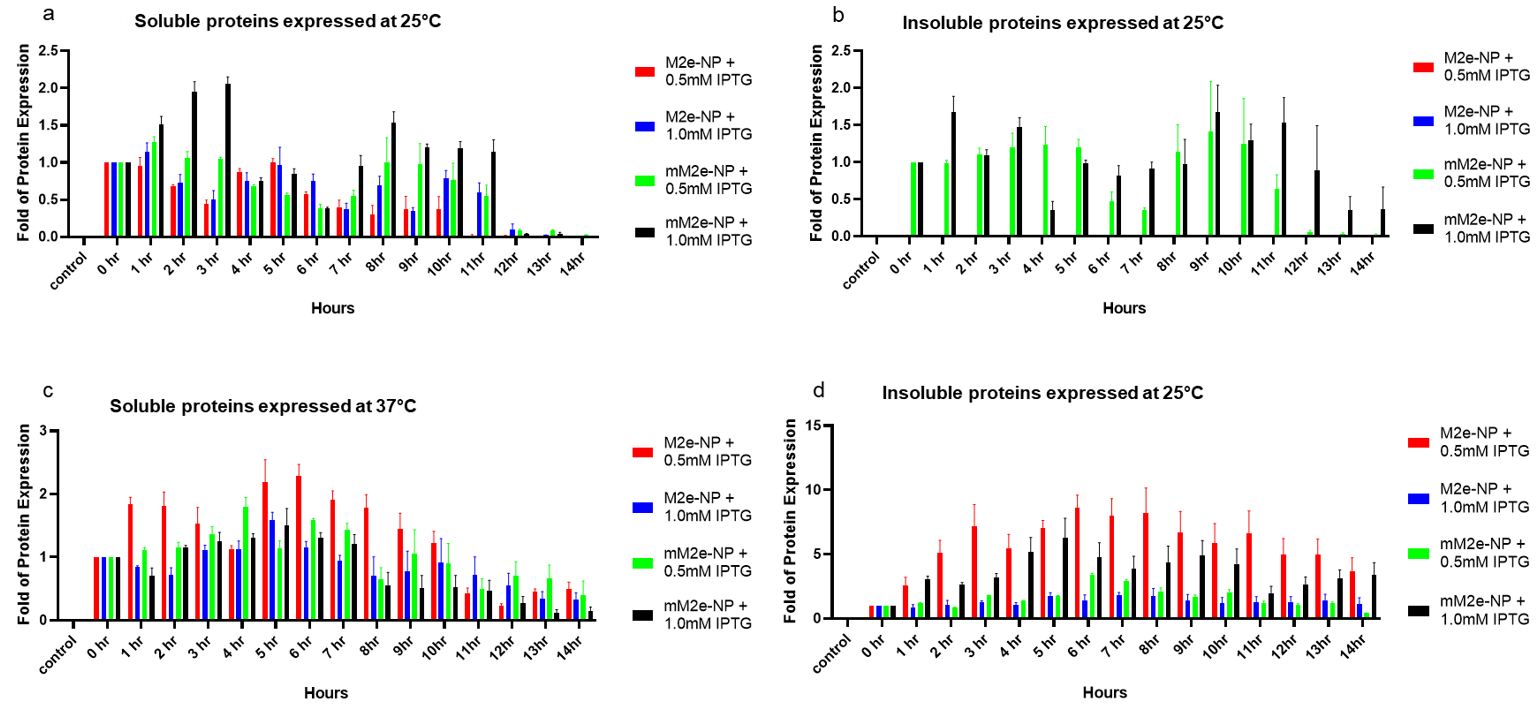
3.4. Optimization of M2e-NP Protein Expression
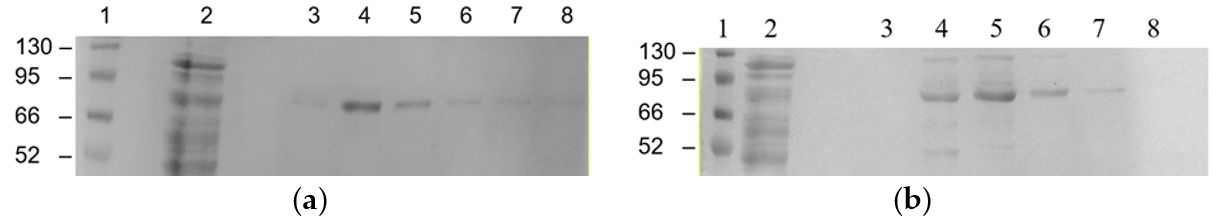
3.5. Affinity Purification of M2e-NP and mM2e-NP Proteins
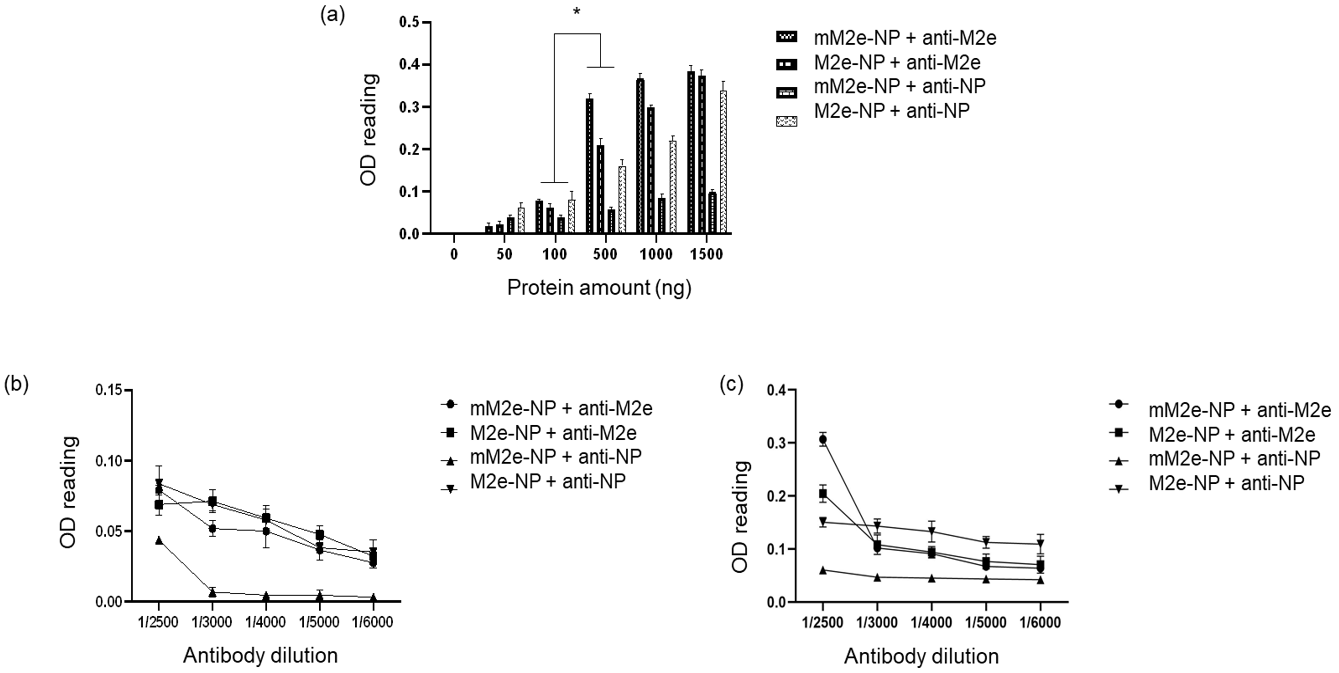
3.6. Antigenicity of M2e-NP Protein
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. Influenza A virus isolation, culture and identification. Nat. Protoc. 2014, 9, 2663. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, S.; Kawaoka, Y. The pathogenesis of influenza virus infections: The contributions of virus and host factors. Curr. Opin. Immunol. 2011, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef]
- Stouffer, A.L.; Acharya, R.; Salom, D.; Levine, A.S.; Di Costanzo, L.; Soto, C.S.; Tereshko, V.; Nanda, V.; Stayrook, S.; DeGrado, W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451, 596–599. [Google Scholar] [CrossRef]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef]
- Gao, X.; Wang, W.; Li, Y.; Zhang, S.; Duan, Y.; Xing, L.; Zhao, Z.; Zhang, P.; Li, Z.; Li, R. Enhanced Influenza VLP vaccines comprising matrix-2 ectodomain and nucleoprotein epitopes protects mice from lethal challenge. Antivir. Res. 2013, 98, 4–11. [Google Scholar] [CrossRef]
- Wu, F.; Huang, J.-H.; Yuan, X.-Y.; Huang, W.-S.; Chen, Y.-H. Characterization of immunity induced by M2e of influenza virus. Vaccine 2007, 25, 8868–8873. [Google Scholar] [CrossRef]
- Ciampor, F.; Thompson, C.; Grambas, S.; Hay, A. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992, 22, 247–258. [Google Scholar] [CrossRef]
- Tan, M.P.; Tan, W.S.; Mohamed Alitheen, N.B.; Yap, W.B. M2e-Based Influenza Vaccines with Nucleoprotein: A Review. Vaccines 2021, 9, 739. [Google Scholar] [CrossRef]
- Ng, A.K.L.; Zhang, H.; Tan, K.; Li, Z.; Liu, J.h.; Chan, P.K.S.; Li, S.M.; Chan, W.Y.; Au, S.W.N.; Joachimiak, A. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008, 22, 3638–3647. [Google Scholar] [CrossRef]
- Chenavas, S.; Estrozi, L.F.; Slama-Schwok, A.; Delmas, B.; Di Primo, C.; Baudin, F.; Li, X.; Crépin, T.; Ruigrok, R.W. Monomeric nucleoprotein of influenza A virus. PLoS Pathog. 2013, 9, e1003275. [Google Scholar] [CrossRef]
- Yap, W.B.; Tey, B.T.; Ng, M.Y.; Ong, S.T.; Tan, W.S. N-terminally His-tagged hepatitis B core antigens: Construction, expression, purification and antigenicity. J. Virol. Methods 2009, 160, 125–131. [Google Scholar] [CrossRef]
- Tan, T.; Syed Hassan, S.; Yap, W. Expression of surface-bound nonstructural 1 (NS 1) protein of influenza virus A H5N1 on Lactobacillus casei strain C1. Lett. Appl. Microbiol. 2017, 64, 446–451. [Google Scholar] [CrossRef]
- Treanor, J.J. Influenza vaccination. N. Engl. J. Med. 2016, 375, 1261–1268. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, M.; Mozdzanowska, K.; Zharikova, D.; Hoff, H.; Wunner, W.; Couch, R.B.; Gerhard, W. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006, 3, 1–13. [Google Scholar] [CrossRef]
- Helenius, A. Unpacking the incoming influenza virus. Cell 1992, 69, 577–578. [Google Scholar] [CrossRef]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef]
- Kang, S.-M.; Song, J.-M.; Quan, F.-S.; Compans, R.W. Influenza vaccines based on virus-like particles. Virus Res. 2009, 143, 140–146. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ngunjiri, J.M.; Xia, M.; Elaish, M.; Jang, H.; Mahesh, K.; Abundo, M.C.; Jiang, X.; Lee, C.-W. Heterosubtypic protection against avian influenza virus by live attenuated and chimeric norovirus P-particle-M2e vaccines in chickens. Vaccine 2019, 37, 1356–1364. [Google Scholar] [CrossRef]
- Yap, W.B.; Tey, B.T.; Alitheen, N.B.M.; Tan, W.S. Display of the antigenic region of Nipah virus nucleocapsid protein on hepatitis B virus capsid. J. Biosci. Bioeng. 2012, 113, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Santiago, F.; Lambert, K.; Takimoto, T.; Topham, D.J. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 2011, 85, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Laidlaw, B.J.; Decman, V.; Ali, M.-A.A.; Abt, M.C.; Wolf, A.I.; Monticelli, L.A.; Mozdzanowska, K.; Angelosanto, J.M.; Artis, D.; Erikson, J. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 2013, 9, e1003207. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, Y.; Lee, Y.T.; Bouchard, K.R.; Benechet, A.; Khanna, K.; Cauley, L.S. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 2014, 95, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Rodriguez, J.; Meijerhof, T.; Niesters, H.G.; Stjernholm, G.; Hovden, A.-O.; Sørensen, B.; Ökvist, M.; Sommerfelt, M.A.; Huckriede, A. A novel peptide-based vaccine candidate with protective efficacy against influenza A in a mouse model. Virology 2018, 515, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, W.; Spencer, W.; Rhoads, R. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990, 18, 6409–6412. [Google Scholar] [CrossRef]
- Dutton, C.M.; Christine, P.; Sommer, S.S. General method for amplifying regions of very high G+ C content. Nucleic Acids Res. 1993, 21, 2953–2954. [Google Scholar] [CrossRef]
- Ravin, N.V.; Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Shaldjan, A.A.; Kovaleva, A.A.; Tsybalova, L.M.; Skryabin, K.G. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant loop region of hepatitis B core antigen: Insertion of multiple copies of M2e increases immunogenicity and protective efficiency. Vaccine 2015, 33, 3392–3397. [Google Scholar]
- Robinson, C.R.; Sauer, R.T. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 5929–5934. [Google Scholar] [CrossRef]
- Chowdhury, M.Y.; Kim, T.-H.; Uddin, M.B.; Kim, J.-H.; Hewawaduge, C.; Ferdowshi, Z.; Sung, M.-H.; Kim, C.-J.; Lee, J.-S. Mucosal vaccination of conserved sM2, HA2 and cholera toxin subunit A1 (CTA1) fusion protein with poly gamma-glutamate/chitosan nanoparticles (PC NPs) induces protection against divergent influenza subtypes. Vet. Microbiol. 2017, 201, 240–251. [Google Scholar] [CrossRef]
- El Aliani, A.; El Abid, H.; Kassal, Y.; Khyatti, M.; Attaleb, M.; Ennaji, M.M.; El Mzibri, M. HPV16 L1 diversity and its potential impact on the vaccination-induced immunity. Gene 2020, 747, 144682. [Google Scholar] [CrossRef]
- Tagmyer, T.L.; Craigo, J.K.; Cook, S.J.; Even, D.L.; Issel, C.J.; Montelaro, R.C. Envelope determinants of equine infectious anemia virus vaccine protection and the effects of sequence variation on immune recognition. J. Virol. 2008, 82, 4052–4063. [Google Scholar] [CrossRef]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Ahmad, I.; Nawaz, N.; Darwesh, N.M.; ur Rahman, S.; Mustafa, M.Z.; Khan, S.B.; Patching, S.G. Overcoming challenges for amplified expression of recombinant proteins using Escherichia coli. Protein Expr. Purif. 2018, 144, 12–18. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; d’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Feng, Y.-L.; Le, X.-H.; Wu, S.-P. Pathogenesis and Pathological Changes of Avian Influenza in Human. In Avian Influenza in Human; Springer: Berlin/Heidelberg, Germany, 2021; pp. 29–40. [Google Scholar]
- Zykova, A.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Stepanova, L.A.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Highly Immunogenic Nanoparticles Based on a Fusion Protein Comprising the M2e of Influenza A Virus and a Lipopeptide. Viruses 2020, 12, 1133. [Google Scholar] [CrossRef]
- Chen, L.; Zanker, D.; Xiao, K.; Wu, C.; Zou, Q.; Chen, W. Immunodominant CD4+ T-cell responses to influenza A virus in healthy individuals focus on matrix 1 and nucleoprotein. J. Virol. 2014, 88, 11760–11773. [Google Scholar] [CrossRef]
- Li, J.; Mahajan, A.; Tsai, M.-D. Ankyrin repeat: A unique motif mediating protein− protein interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef]
- Zhou, D.; Wu, T.-L.; Lasaro, M.O.; Latimer, B.P.; Parzych, E.M.; Bian, A.; Li, Y.; Li, H.; Erikson, J.; Xiang, Z. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol. Ther. 2010, 18, 2182–2188. [Google Scholar] [CrossRef]
- Jennings, R.; Smith, T.; Potter, C. Use of the enzyme-linked immunosorbent assay (ELISA) for the estimation of serum antibodies in an influenza virus vaccine study. Med. Microbiol. Immunol. 1981, 169, 247–258. [Google Scholar] [CrossRef] [PubMed]



| Primer | Sequencing |
|---|---|
| M2e forward primer | 5′-ATA AAG GAT CCG AGT CTT CTA ACC GAG GTC GAA ACG CCT ACC AGA AAC GAA-3′ |
| M2e reverse primer | 5′-GGT GCC TTG AGA CGC ACC ACC ACT ACC ACC ATT TGC GGC AAC AGC-3′ |
| NP forward primer | 5′-GCT GTT GCC GCA AAT GGT GGT AGT GGT GGT GCG TCT CAA GGC ACC-3′ |
| NP reverse primer | 5′-TCA AGC TTC ATT GTC ATA CTC CTC TGC-3′ |
| Primer | Sequence |
|---|---|
| Forward 1F(M2e forward primer) | 5′-ATA AAG GAT CCG AGT CTT CTA ACC GAG GTC GAA ACG CCT ACC AGA AAC GAA-3′ |
| Reverse 1R | 5′-GCC TTG AGA CGC ACC ACC ACT ACC ACC ATT TGC GGC AAC AGC AAT AGG-3 |
| Forward 2F | 5′-GCT GTT GCC GCA AAT GGT GGT AGT GGT GGT GCG TCT CAA GGC ACC-3′ |
| Reverse 2R | 5′-ACT CTT ATG TGC TGG ATT CTC ATT TGG TCT AAT-3′ |
| Forward 3F | 5′-GAG AAT CCA GCA CAT AAG AGT CAA TTA GTG TGG-3′ |
| Reverse 3R | 5′-GGA GTC CAT TGC CTC CAT GTT CTC-3′ |
| Forward 4F | 5′-GAG AAC ATG GAG GCA ATG GAC TCC-3′ |
| Reverse 4R | 5′-TAC CGA GAA AGT GGG CTG AAC GCT-3′ |
| Forward 5F | 5′-AGC GTT CAG CCC ACT TTC TCG GTA-3′ |
| Reverse 5R | 5′-ACT TTC CAT CAT TCT TAT GAT TTC-3′ |
| Forward 6F | 5′-GAA ATC ATA AGA ATG ATG GAA AGT-3′ |
| Reverse 6R(NP reverse primer) | 5′-TCA AGC TTC ATT GTC ATA CTC CTC TGC-3′ |
| Gene | Nucleotide Change(5′-3′) | Amino Acid Change |
|---|---|---|
| M2e | …CTT… …ATT… | Leucine (L25) → Isoluecine (I25) |
| NP | …GAT… …AAT… | Aspartic acid (D353) → Asparagine (N353) |
| …CGA… …CAA… | Arginine (R361) → Glutamine (Q361) | |
| …ACA… …GCA… | Threonine (T407) → Alanine (A407) | |
| …GCT… …ACT… | Alanine (A445) → Threonine (T445) | |
| …AAA… …AGA… | Lysine (K480) → Arginine (R480) |
| Expression Temperature ( °C) | IPTG Concentration (mM) | Protein | Type of Lysate | Western Blotting Results |
|---|---|---|---|---|
| 25 | 0.5 | mM2e-NP | Supernatant/Soluble |  |
| Pellet/Insoluble | 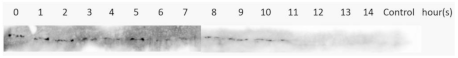 | |||
| M2e-NP | Supernatant/Soluble |  | ||
| Pellet/Insoluble |  | |||
| 1.0 | mM2e-NP | Supernatant/Soluble |  | |
| Pellet/Insoluble |  | |||
| M2e-NP | Supernatant/Soluble |  | ||
| Pellet/Insoluble |  | |||
| 37 | 0.5 | mM2e-NP | Supernatant/Soluble | 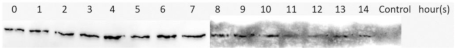 |
| Pellet/Insoluble |  | |||
| M2e-NP | Supernatant/Soluble | 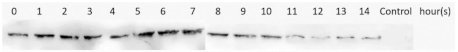 | ||
| Pellet/Insoluble |  | |||
| 1.0 | mM2e-NP | Supernatant/Soluble |  | |
| Pellet/Insoluble |  | |||
| M2e-NP | Supernatant/Soluble | 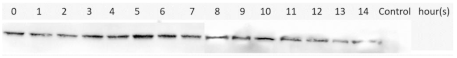 | ||
| Pellet/Insoluble | 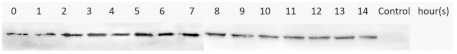 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.P.; Mohamed Alitheen, N.B.; Tan, W.S.; Yap, W.B. Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies. Vaccines 2022, 10, 2066. https://doi.org/10.3390/vaccines10122066
Tan MP, Mohamed Alitheen NB, Tan WS, Yap WB. Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies. Vaccines. 2022; 10(12):2066. https://doi.org/10.3390/vaccines10122066
Chicago/Turabian StyleTan, Mei Peng, Noorjahan Banu Mohamed Alitheen, Wen Siang Tan, and Wei Boon Yap. 2022. "Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies" Vaccines 10, no. 12: 2066. https://doi.org/10.3390/vaccines10122066
APA StyleTan, M. P., Mohamed Alitheen, N. B., Tan, W. S., & Yap, W. B. (2022). Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies. Vaccines, 10(12), 2066. https://doi.org/10.3390/vaccines10122066





