Low Genetic Polymorphism in the Immunogenic Sequences of Rhipicephalus microplus Clade C
Abstract
1. Introduction
2. Materials and Methods
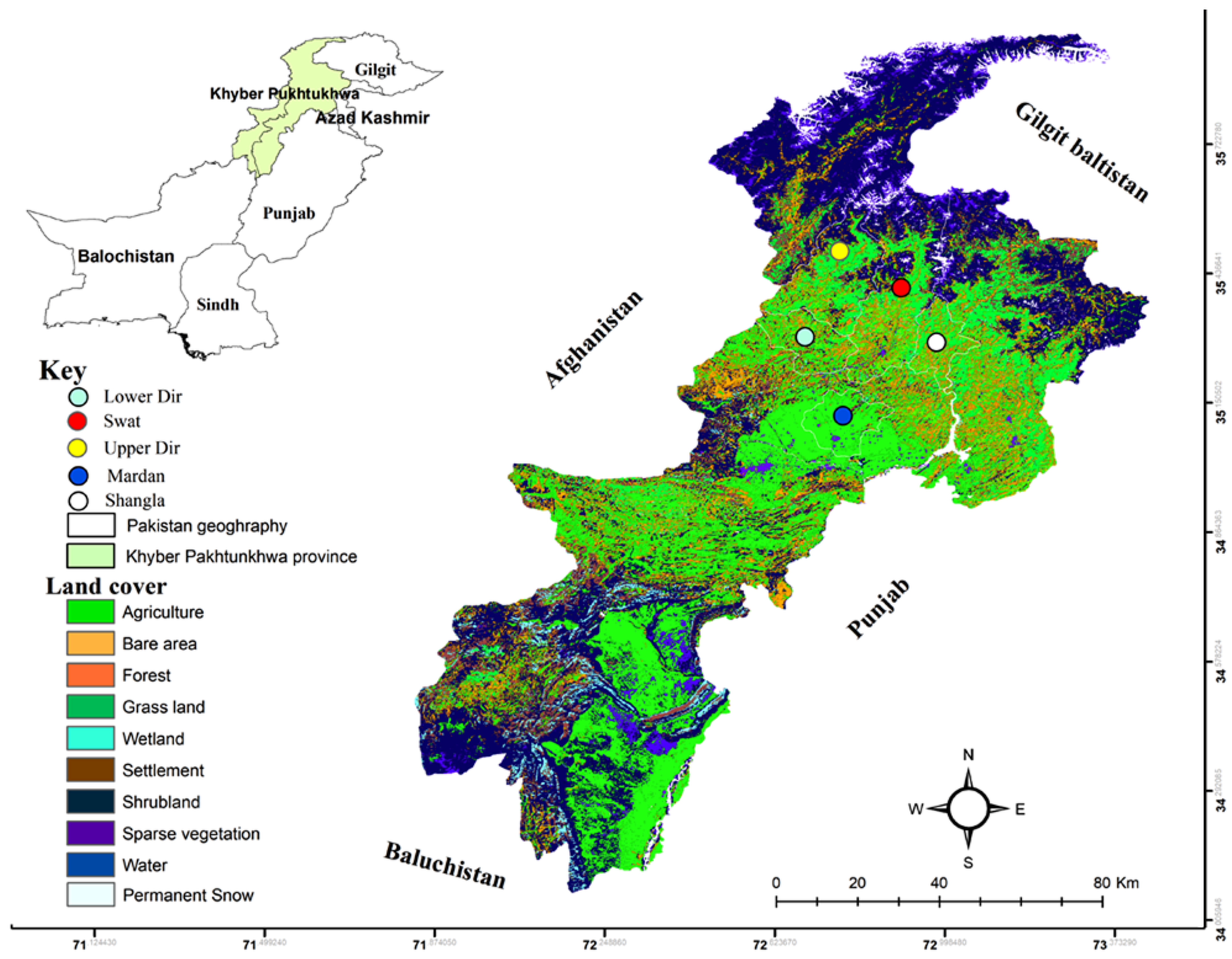
2.1. Study Area
2.2. Ethical Approval
2.3. Sample Collection and Morphological Identification
2.4. Nucleic Acid Extraction and cDNA Synthesis
2.5. Primer Synthesis
2.6. PCR Amplification
| Organism/Gene | Primer Sequence | Tm °C, s | Amplicon Size | References |
|---|---|---|---|---|
| Ticks/cox | F: GGA ACAA TATA TTT AAT TTT TGG R: ATC TAT CCC TAC TGT AAA TAT ATG | 55 °C, 60 s | 801 | [38] |
| Piroplasms (Theileria/Babesia spp.)/18S rRNA | F: ACC GTGCTAA TTGT AGGGCTA ATAC R: GAACCCAAAGACTTTGATTTCTCTC | 55 °C, 30 s | 897 | This study |
| Rickettsia spp./gltA | F: GCAAGTATCGGTGAGGATGTAAT R: GCTTCCTTAAAATTCAATAAATCAGG | 50 °C, 30 s | 401 | [39] |
| Anaplasma spp./16S rRNA | F: GGTACCYACAGAAGAAGTCC R: TGCA CTCA TCGT TTACAG | 55 °C, 30 s | 345 | [40] |
| Tick’s full-length ORF coding genes | ||||
| Cysteine protease inhibitor (cystatin 2b) | F: ATGGCTTCTTTGAGAATCACCCCG R: TTAGGTAGATGTGCTGCTTCCTTCG | 60 °C, 30 s | 423 | This study |
| Cathepsin L-like cysteine proteinase (cathepsin-L) | F: ATGCTTAGATTAAGCGTACTTTGCG R: TTAGACGAGBGGGTAGCTGGCCTG | 57 °C, 30 s | 999 | This study |
| Glutathione S-transferase (GST) | F: ATGGCTCCTGTGCTCGGCTAC R: GCTTGTTTCATGGCTTCTTCTGC | 54 °C, 30 s | 672 | This study |
| Ferritin 1 | F: ATGTTTTGGTCGATGTTATGC R: CTAGTCTGACAGGGTCTCCTTGTCA | 60 °C, 30 s | 654 | This study |
| 60S acidic ribosomal protein (P0) | F: ATGGTCAGGGAGGAYAAGAC R: CTAGTCGAAGAGTCCGAAGCCCAT | 50 °C, 30 s | 957 | This study |
| Aquaporin 2 | F: AAT TCAGCAGC AGGAG AAGC R: CTGA TGCATA AAAAA CTCAG CAT | 50 °C, 30 s | 1043 | This study |
| ATAQ | F: ATG GGAA GAATG AACA ACG AACGC R: TCAG GCCTC TTCCTC CGTTG GAAGC | 60 °C, 30 s | 1818 | This study |
| R. microplus 05 antigen (Rm05Uy) | F: ATGGT GGCTT TCAAG GCAG CCC R: TTAA CCATGG GCCGG CGC ACCA | 60 °C, 30 s | 516 | This study |
| Actin | F: GCATCCACGAGACCACG R: GGGGTGTAGAAGGAAGG | 54 °C, 30 s | 339 | [41] |
2.7. Purification, Cloning and Sequencing
2.8. Sequence and Phylogenetic Analysis
3. Results
3.1. Sequences Analysis
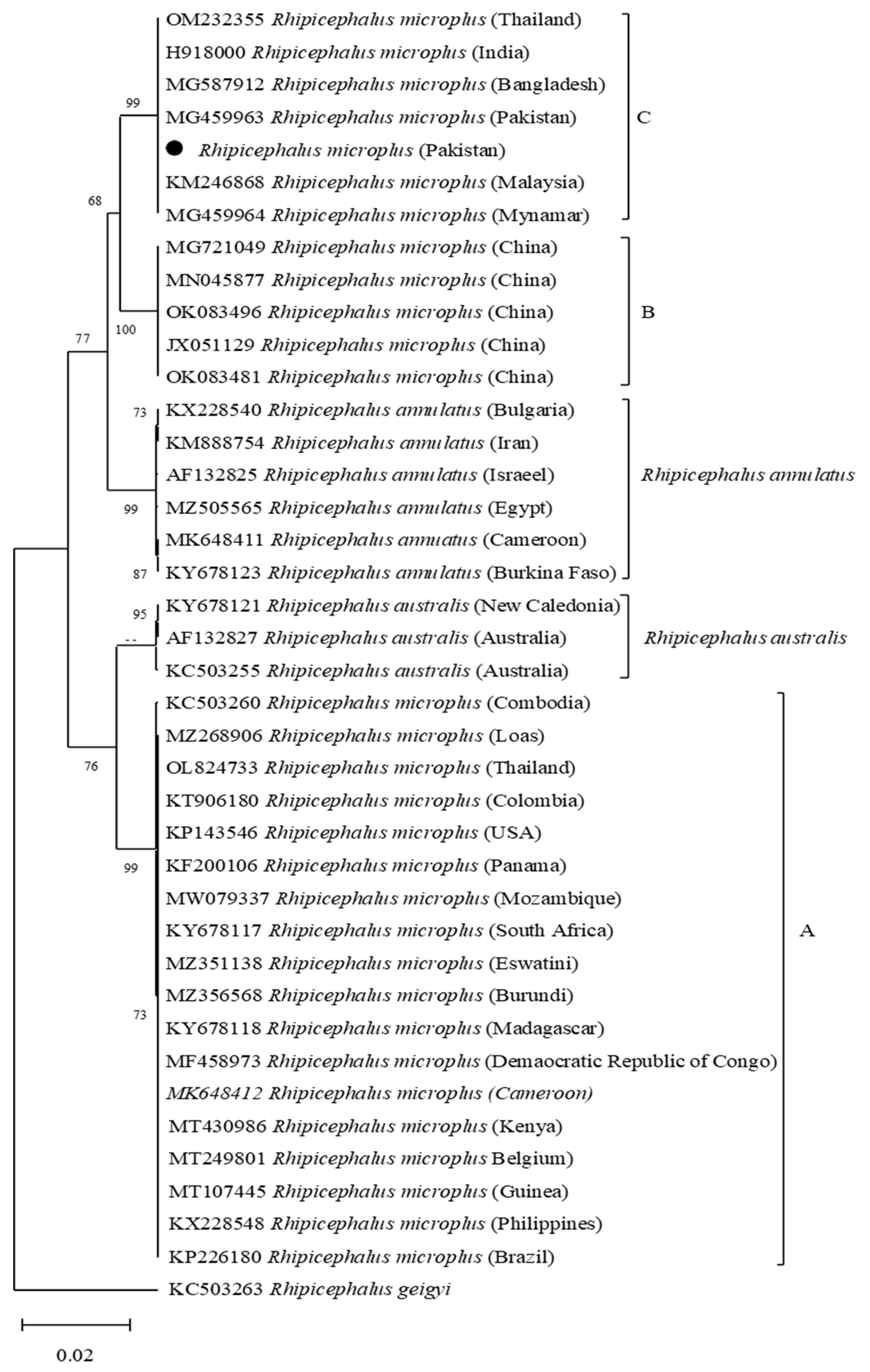
3.2. Phylogenetic Analysis of cox
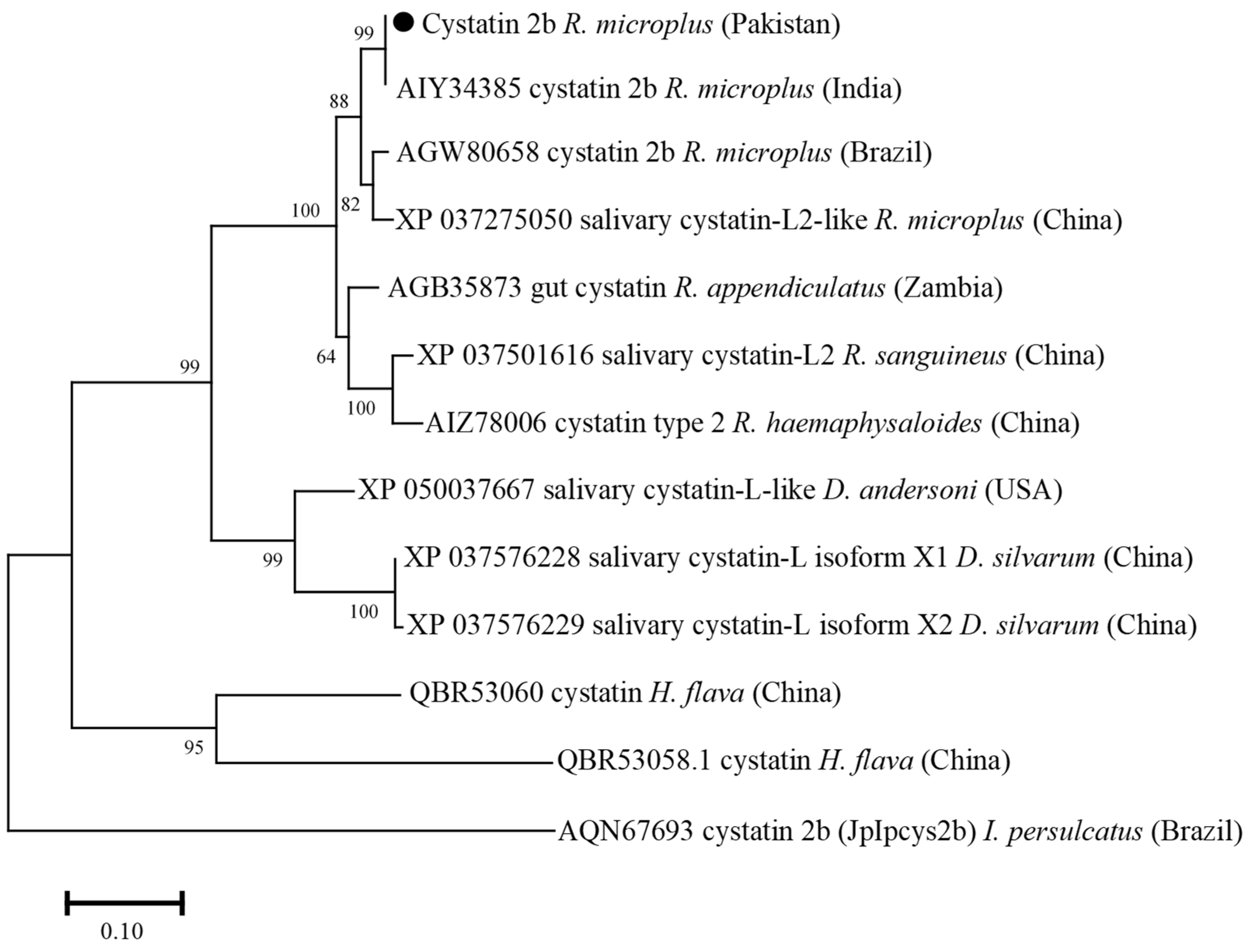
3.3. Phylogenetic Analysis of Cystatin 2b
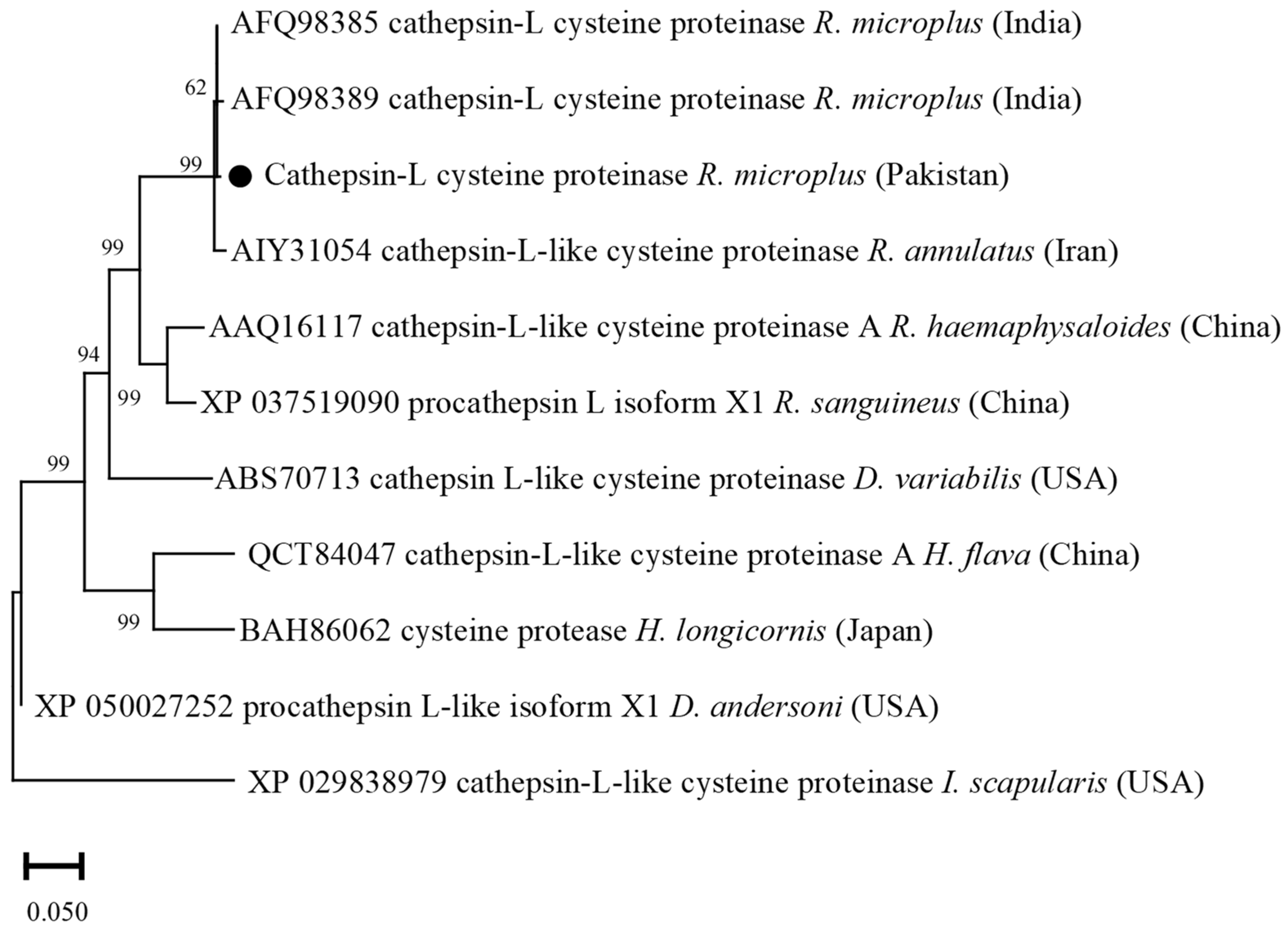
3.4. Phylogenetic Analysis of Cathepsin L-like Cysteine Proteinase
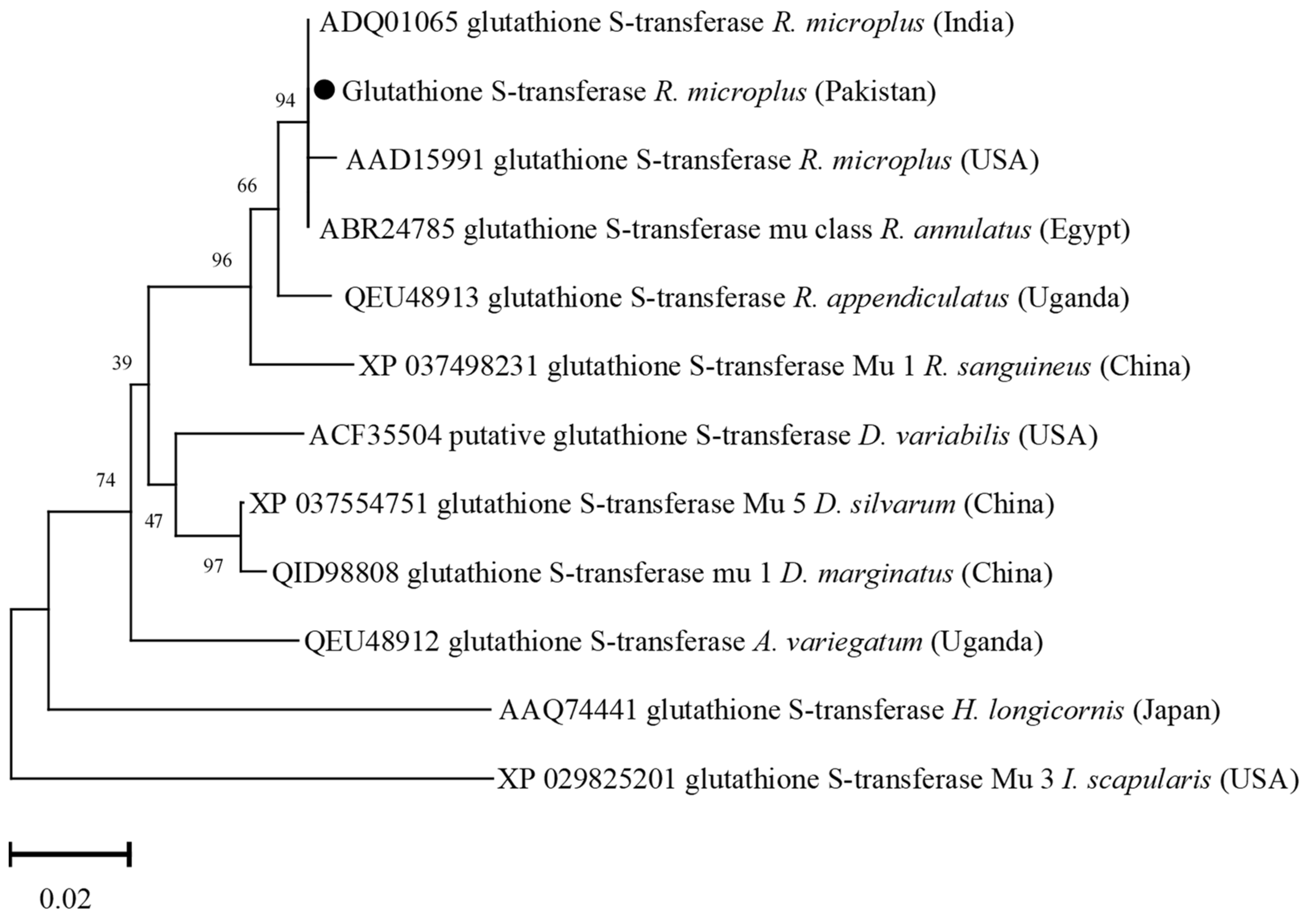
3.5. Phylogenetic Analysis of Glutathione S-Transferase
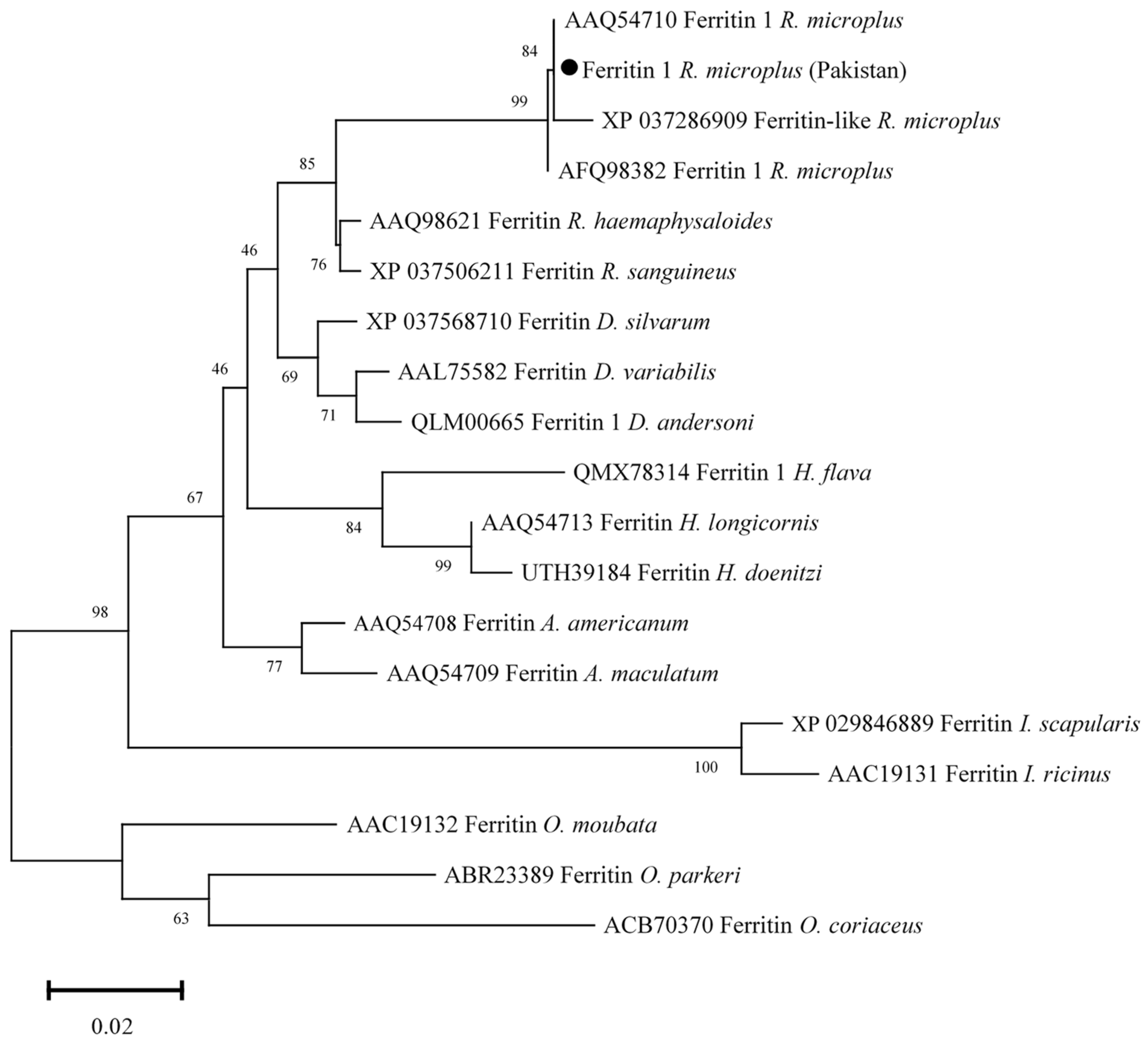
3.6. Phylogenetic Analysis of Ferritin 1
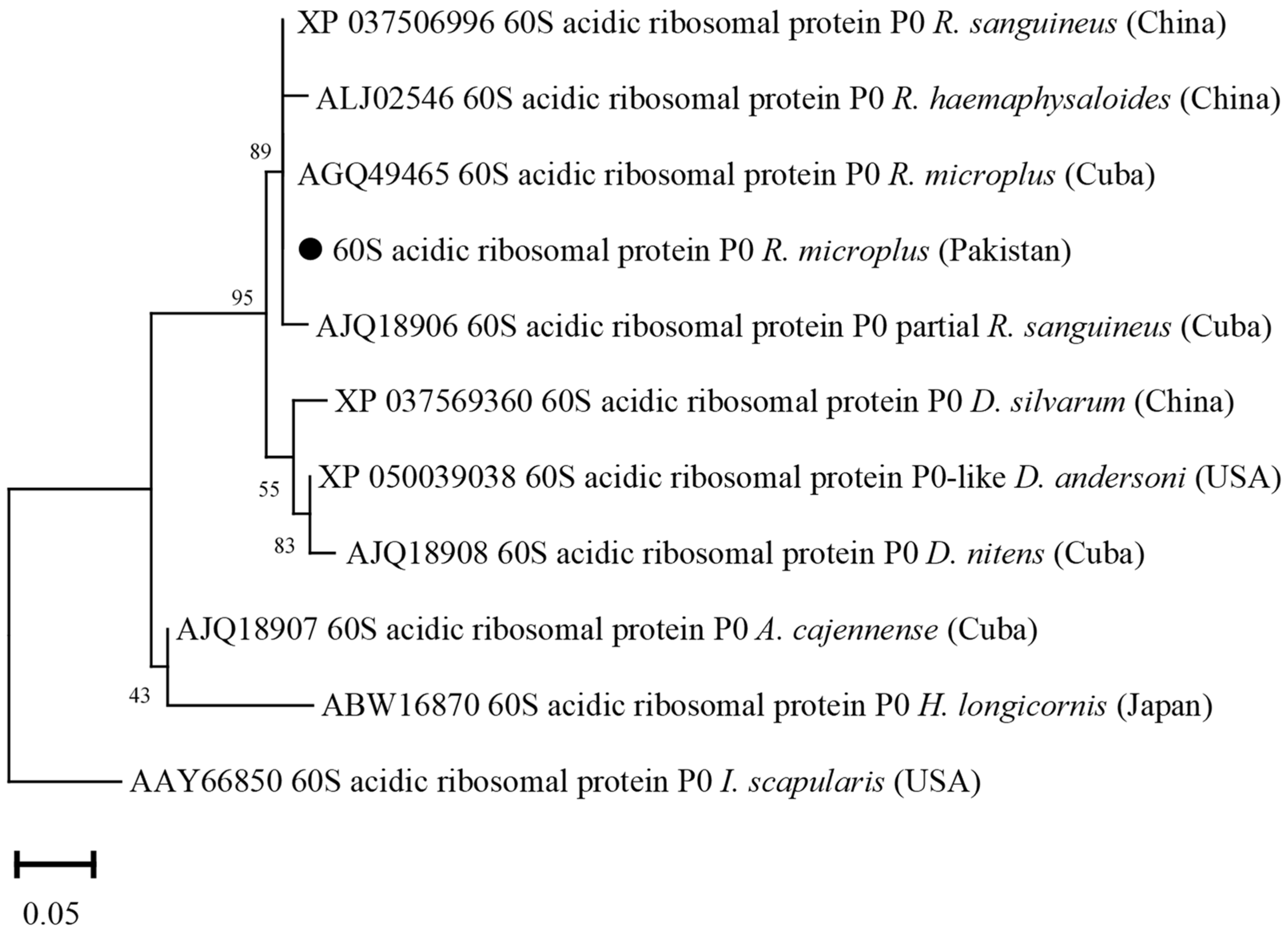
3.7. Phylogenetic Analysis of 60S Acidic Ribosomal Protein (P0)
3.8. Phylogenetic Analysis of Aquaporin 2
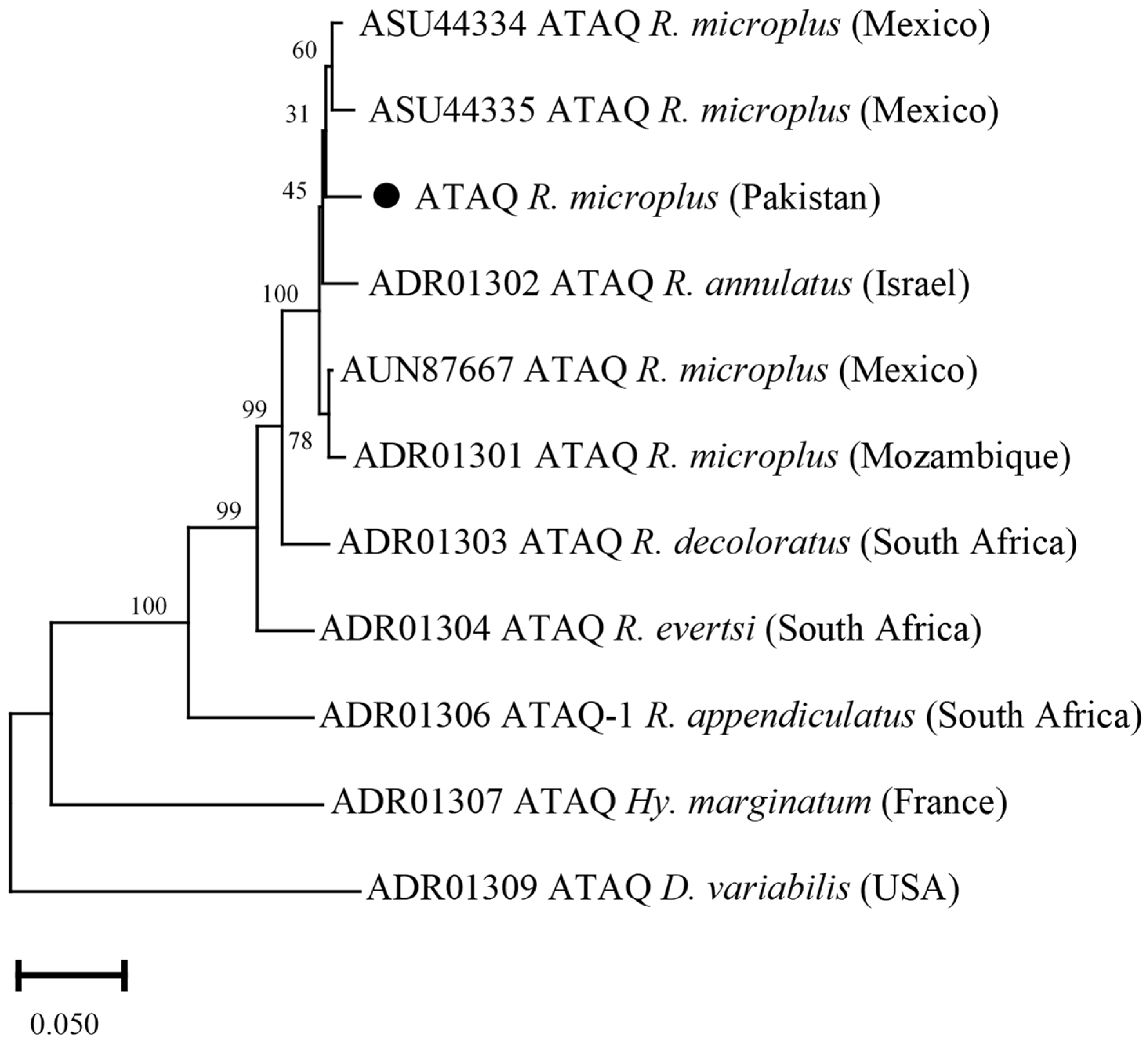
3.9. Phylogenetic Analysis of ATAQ
3.10. Phylogenetic Analysis of Rm05Uy
3.11. Nucleotide Polymorphism
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parola, P.; Raoult, D. Ticks and tick-borne bacterial diseases in humans: An emerging infectious threat. Clin. Infect. Dis. 2001, 32, 897–928. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Zeb, I.; Alouffi, A.; Zahid, H.; Almutairi, M.M.; Ayed Alshammari, F.; Mohammed Alrouji, M.; Termignoni, C.; da Silva Vaz, I., Jr.; Tanaka, T. Host Immune Responses to Salivary Components-A Critical Facet of Tick-Host Interactions. Front. Cell. Infect. Microbiol. 2022, 12, 809052. [Google Scholar] [CrossRef] [PubMed]
- Agwunobi, D.O.; Yu, Z.; Liu, J. A retrospective review on ixodid tick resistance against synthetic acaricides: Implications and perspectives for future resistance prevention and mitigation. Pestic. Biochem. Physiol. 2021, 173, 104776. [Google Scholar] [CrossRef] [PubMed]
- Obaid, M.K.; Islam, N.; Alouffi, A.; Khan, A.Z.; da Silva Vaz, I., Jr.; Tanaka, T.; Ali, A. Acaricides Resistance in Ticks: Selection, Diagnosis, Mechanisms, and Mitigation. Front. Cell. Infect. Microbial. 2022, 885, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Willadsen, P.; Riding, G.A.; McKenna, R.V.; Kemp, D.H.; Tellam, R.L.; Nielsen, J.N.; Gough, J.M. Immunologic control of a parasitic arthropod. Identification of a protective antigen from Boophilus microplus. J. Immunol. 1989, 143, 1346–1351. [Google Scholar]
- De la Fuente, J.; Contreras, M. Additional considerations for anti-tick vaccine research. Expert Rev. Vaccines 2022, 21, 1019–1021. [Google Scholar] [CrossRef]
- Parizi, L.F.; Githaka, N.W.; Acevedo, C.; Benavides, U.; Seixas, A.; Logullo, C.; da Silva Vaz, I., Jr. Sequence characterization and immunogenicity of cystatins from the cattle tick Rhipicephalus (Boophilus) microplus. Ticks Tick-Borne Dis. 2013, 4, 492–499. [Google Scholar] [CrossRef]
- Kumar, B.; Manjunathachar, H.V.; Nagar, G.; Ravikumar, G.; de la Fuente, J.; Saravanan, B.C.; Ghosh, S. Functional characterization of candidate antigens of Hyalomma anatolicum and evaluation of its cross-protective efficacy against Rhipicephalus microplus. Vaccine 2017, 35, 5682–5692. [Google Scholar] [CrossRef]
- De Lima, M.R.; Ferreira, C.S.; De Freitas, D.J.; Valenzuela, J.G.; Masuda, A. Cloning and partial characterization of a Boophilus microplus (Acari: Ixodidae) glutathione S-transferase. Insect Biochem. Mol. Biol. 2002, 32, 747–754. [Google Scholar] [CrossRef]
- Xu, G.; Fang, Q.Q.; Keirans, J.E.; Durden, L.A. Ferritin gene coding sequences are conserved among eight hard tick species (Ixodida: Ixodidae). Ann. Entomol. Soc. Am. 2004, 97, 567–573. [Google Scholar] [CrossRef]
- Kaewmongkol, S.; Kaewmongkol, G.; Jittapalapong, S. Molecular cloning and expression of the ferritin gene from midgut and salivary glands of brown dog ticks (Rhipicephalus sanguineus) in Thailand. J. Trop. Med. Parasitol. 2015, 38, 2. [Google Scholar]
- Rodríguez-Mallón, A.; Fernández, E.; Encinosa, P.E.; Bello, Y.; Méndez-Pérez, L.; Ruiz, L.C.; Estrada, M.P. A novel tick antigen shows high vaccine efficacy against the dog tick, Rhipicephalus sanguineus. Vaccine 2012, 30, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Burdin, M.; Hoppler, S.; Bowmana, A.S. Role of an aquaporin in the sheep tick Ixodes ricinus, Assessment as a potential control target. Int. J. Parasitol. 2018, 40, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Nijhof, A.M.; Balk, J.A.; Postigo, M.; Rhebergen, A.M.; Taoufik, A.; Jongejan, F. Bm86 homologues and novel ATAQ proteins with multiple epidermal growth factor (EGF)-like domains from hard and soft ticks. Int. J. Parasitol. 2010, 40, 1587–1597. [Google Scholar] [CrossRef][Green Version]
- Alzugaray, M.F.; Parizi, L.F.; Seixas, A.; Benavides, U.; da Silva Vaz, I., Jr. Molecular and functional characterization of Bm05br antigen from Rhipicephalus microplus. Ticks Tick-Borne Dis. 2017, 8, 320–329. [Google Scholar] [CrossRef]
- Mans, B.J.; Featherston, J.; De Castro, M.H.; Pienaar, R. Gene duplication and protein evolution in tick-host interactions. Front. Cell. Infect. Microbiol. 2017, 7, 413. [Google Scholar] [CrossRef]
- García-García, J.C.; Gonzalez, I.L.; González, D.M.; Valdés, M.; Méndez, L.; Lamberti, J.; D’Agostino, B.; Citroni, D.; Fragoso, H.; Ortiz, M.; et al. Sequence variations in the Boophilus microplus Bm86 locus and implications for immunoprotection in cattle vaccinated with this antigen. Exp. Appl. Acarol. 1999, 23, 883–895. [Google Scholar] [CrossRef]
- Freeman, J.M.; Davey, R.B.; Kappmeyer, L.S.; Kammlah, D.M.; Olafson, P.U. Bm86 midgut protein sequence variation in South Texas cattle fever ticks. Parasit Vectors 2010, 3, 101. [Google Scholar] [CrossRef]
- Blecha, I.M.Z.; Csordas, B.G.; Aguirre, A.D.A.R.; Cunha, R.C.; Garcia, M.V.; Andreotti, R. Analysis of Bm86 conserved epitopes: Is a global vaccine against Cattle Tick Rhipicephalus microplus possible? Rev. Bras. DE Parasitol. 2018, 27, 267–279. [Google Scholar] [CrossRef]
- Martínez-Arzate, S.G.; Sánchez-Bermúdez, J.C.; Sotelo-Gómez, S.; Diaz-Albiter, H.M.; Hegazy-Hassan, W.; Tenorio-Borroto, E.; Barbabosa-Pliego, A.; Vázquez-Chagoyán, J.C. Genetic diversity of Bm86 sequences in Rhipicephalus (Boophilus) microplus ticks from Mexico: Analysis of haplotype distribution patterns. BMC Genet. 2019, 20, 56. [Google Scholar] [CrossRef]
- Del-Castillo, S.M.L.C.; Hernández-Ortiz, R.; Gómez-Romero, N.; Martínez-Velázquez, M.; Castro-Saines, E.; Lagunes-Quintanilla, R. Genetic diversity of the ATAQ gene in Rhipicephalus microplus collected in Mexico and implications as anti-tick vaccine. Parasitol. Res. 2020, 119, 3523–3529. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Almazán, C.; Canales, M.; Pérez de la Lastra, J.M.; Kocan, K.M.; Willadsen, P. A ten-year review of commercial vaccine performance for control of tick infestations on cattle. Anim. Health Res. Rev. 2007, 8, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Sossai, S.; Peconick, A.P.; Sales-Junior, P.A.; Marcelino, F.C.; Vargas, M.I.; Neves, E.S.; Patarroyo, J.H. Polymorphism of the Bm86 gene in South American strains of the cattle tick Boophilus microplus. Exp. Appl. Acarol. 2005, 37, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Nijhof, A.M.; Balk, J.A.; Postigo, M.; Jongejan, F. Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol. Cell Biol. 2009, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Peña, A.; de la Fuente, J. The ecology of ticks and epidemiology of tick-borne viral diseases. Antivir. Res. 2014, 108, 104–128. [Google Scholar] [CrossRef]
- Low, V.L.; Tay, S.T.; Kho, K.L.; Koh, F.X.; Tan, T.K.; Lim, Y.A.L.; Sofian-Azirun, M. Molecular characterisation of the tick Rhipicephalus microplus in Malaysia: New insights into the cryptic diversity and distinct genetic assemblages throughout the world. Parasites Vectors 2015, 8, 341. [Google Scholar] [CrossRef]
- Burger, T.D.; Shao, R.; Barker, S.C. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol. Phylogenetics Evol. 2014, 76, 241–253. [Google Scholar] [CrossRef]
- Ali, A.; Khan, M.A.; Zahid, H.; Yaseen, P.M.; Qayash, K.M.; Nawab, J.; Ur Rehman, Z.; Ateeq, M.; Khan, S.; Ibrahim, M. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 2019, 10, 793. [Google Scholar] [CrossRef]
- Karim, S.; Budachetri, K.; Mukherjee, N.; Williams, J.; Kausar, A.; Hassan, M.J.; Adamson, S.; Dowd, S.E.; Apanskevich, D.; Arijo, A.; et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl. Trop. Dis. 2017, 11, e0005681. [Google Scholar] [CrossRef]
- Economic Survey of Pakistan. Government of Pakistan Finance Division, 2020–2021. Available online: https://www.finance.gov.pk/survey_2021.html (accessed on 21 January 2022).
- Aiman, O.; Ullah, S.; Chitimia-Dobler, L.; Nijhof, A.M.; Ali, A. First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick-Borne Dis. 2022, 13, 101899. [Google Scholar] [CrossRef]
- Khan, Z.; Shehla, S.; Alouffi, A.; Kashif Obaid, M.; Khan, A.; Almutairi, M.M.; Numan, M.; Aiman, O.; Alam, S.; Ullah, S.; et al. Molecular survey and genetic characterization of Anaplasma marginale in ticks collected from livestock hosts in Pakistan. Animals 2022, 12, 1708. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Khan, M.; Alouffi, A.; Almutairi, M.M.; Ullah, S.; Numan, M.; Ali, A. Spatio-Temporal Patterns of Ticks and Molecular Survey of Anaplasma marginale, with Notes on Their Phylogeny. Microorganisms 2022, 10, 1663. [Google Scholar] [CrossRef] [PubMed]
- Ullah, K.; Numan, M.; Alouffi, A.; Almutairi, M.M.; Zahid, H.; Khan, M.; Ul Islam, Z.; Kamil, A.; Safi, Z.S.; Ahmed, H.; et al. Molecular Characterization and assessment of risk factors associated with Theileria annulata infection. Microorganisms 2022, 10, 1614. [Google Scholar] [CrossRef] [PubMed]
- Parizi, L.F.; Sabadin, G.A.; Alzugaray, M.F.; Seixas, A.; Logullo, C.; Konnai, S.; da Silva Vaz, I., Jr. Rhipicephalus microplus and Ixodes ovatus cystatins in tick blood digestion and evasion of host immune response. Parasites Vectors 2015, 8, 122. [Google Scholar] [CrossRef]
- Roy, B.C.; Estrada-Peña, A.; Krücken, J.; Rehman, A.; Nijhof, A.M. Morphological and phylogenetic analyses of Rhipicephalus microplus ticks from Bangladesh, Pakistan and Myanmar. Ticks Tick-Borne Dis. 2018, 9, 1069–1079. [Google Scholar] [CrossRef]
- ESRI. ArcGIS Desktop, version 10.3.1, Windows; Environmental Systems Research Institute Inc.: Redlands, CA, USA, 2015. [Google Scholar]
- Chitimia, L.; Lin, R.Q.; Cosoroaba, I.; Wu, X.Y.; Song, H.Q.; Yuan, Z.G.; Zhu, X.Q. Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox1 and nad5 genes. Exp. Appl. Acarol. 2010, 52, 305–311. [Google Scholar] [CrossRef]
- Labruna, M.B.; Whitworth, T.; Bouyer, D.H.; McBride, J.; Camargo, L.M.A.; Camargo, E.P.; Walker, D.H. Rickettsia bellii and Rickettsia amblyommii in Amblyomma ticks from the state of Rondônia, Western Amazon, Brazil. J. Med. Entomol. 2004, 41, 1073–1081. [Google Scholar] [CrossRef]
- Inokuma, H.; Raoult, D.; Brouqui, P. Detection of Ehrlichia platys DNA in Brown Dog Ticks (Rhipicephalus sanguineus) in Okinawa Island, Japan. J. Clin. Microbiol. 2000, 38, 4219–4221. [Google Scholar] [CrossRef]
- Vaz Junior, I.D.; Imamura, S.; Nakajima, C.; de Cardoso, F.C.; Ferreira, C.A.S.; Renard, G.; Masuda, A.; Ohashi, K.; Onuma, N. Molecular cloning and sequence analysis of cDNAs encoding for Boophilus microplus, Haemaphysalis longicornis and Rhipicephalus appendiculatus actins. Vet. Parasitol. 2005, 127, 147–155. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DNASP V5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinfromatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Vivas, R.I.; Jonsson, N.N.; Bhushan, C. Strategies for the control of Rhipicephalus microplus ticks in a world of conventional acaricide and macrocyclic lactone resistance. Parasitol. Res. 2018, 117, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Wu, S.; Zhang, Y.; Chen, Y.; Feng, C.; Yuan, X.; Lin, X. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasites Vectors 2014, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Rachinsky, A.; Guerrero, F.D.; Scoles, G.A. Differential protein expression in ovaries of uninfected and Babesia-infected southern cattle ticks, Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2007, 37, 1291–1308. [Google Scholar] [CrossRef]
- Antunes, S.; Couto, J.; Ferrolho, J.; Rodrigues, F.; Nobre, J.; Santos, A.S.; Santos-Silva, M.M.; de la Fuente, J.; Domingos, A. Rhipicephalus bursa sialotranscriptomic response to blood feeding and Babesia ovis infection: Identification of candidate protective antigens. Front. Cell. Infect. Microbiol. 2018, 8, 116. [Google Scholar] [CrossRef]
- Scoles, G.A.; Hussein, H.E.; Olds, C.L.; Mason, K.L.; Davis, S.K. Vaccination of cattle with synthetic peptides corresponding to predicted extracellular domains of Rhipicephalus (Boophilus) microplus aquaporin 2 reduced the number of ticks feeding to repletion. Parasites Vectors 2022, 15, 49. [Google Scholar] [CrossRef]
- Rodríguez-Mallón, A.; Encinosa, P.E.; Méndez-Pérez, L.; Bello, Y.; Fernández, R.R.; Garay, H.; Estrada, M.P. High efficacy of a 20 amino acid peptide of the acidic ribosomal protein P0 against the cattle tick, Rhipicephalus microplus. Ticks Tick-Borne Dis. 2015, 6, 530–537. [Google Scholar] [CrossRef]
- Parizi, L.F.; Rangel, C.K.; Sabadin, G.A.; Saggin, B.F.; Kiio, I.; Xavier, M.A.; da Silva Vaz, I., Jr. Rhipicephalus microplus cystatin as a potential cross-protective tick vaccine against Rhipicephalus appendiculatus. Ticks Tick-Borne Dis. 2020, 11, 101378. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; da Rocha, L.A.; Torquato, R.J.; Junior, I.D.S.V.; Florin-Christensen, M.; Tanaka, A.S. A novel type 1 cystatin involved in the regulation of Rhipicephalus microplus midgut cysteine proteases. Ticks Tick-Borne Dis. 2020, 11, 101374. [Google Scholar] [CrossRef] [PubMed]
- Shahein, Y.E.; Amr El Sayed, E.H.; Abouelella, A.M.K.; Hamed, R.R.; Allam, S.A.M.; Farid, N.M. Molecular cloning, expression and characterization of a functional GSTmu class from the cattle tick Boophilus annulatus. Vet. Parasitol. 2008, 152, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Duscher, G.G.; Galindo, R.C.; Tichy, A.; Hummel, K.; Kocan, K.M.; de la Fuente, J. Glutathione S-transferase affects permethrin detoxification in the brown dog tick, Rhipicephalus sanguineus. Ticks Tick-Borne Dis. 2014, 5, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Saidi, S.; Nabian, S.; Ebrahimzade, E.; Najafi, A.; Moghaddam, M.M.; Sazmand, A.; Tabrizi, S.S. Identification and characterization of a cathepsin L-like cysteine protease from Rhipicephalus (Boophilus) annulatus. Exp. Appl. Acarol. 2016, 68, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, E.P.; Kusakisako, K.; Talactac, M.R.; Galay, R.L.; Hatta, T.; Matsuo, T.; Tanaka, T. Characterization and expression analysis of a newly identified glutathione S-transferase of the hard tick Haemaphysalis longicornis during blood-feeding. Parasites Vectors 2018, 11, 91. [Google Scholar] [CrossRef]
- Kopáček, P.; Ždychová, J.; Yoshiga, T.; Weise, C.; Rudenko, N.; Law, J.H. Molecular cloning, expression and isolation of ferritins from two tick species—Ornithodoros moubata and Ixodes ricinus. Insect Biochem. Mol. Biol. 2003, 33, 103–113. [Google Scholar] [CrossRef]
- García-García, J.C.; Montero, C.; Redondo, M.; Vargas, M.; Canales, M.; Boue, O.; de la Fuente, J. Control of ticks resistant to immunization with Bm86 in cattle vaccinated with the recombinant antigen Bm95 isolated from the cattle tick, Boophilus microplus. Vaccine 2000, 18, 2275–2287. [Google Scholar] [CrossRef]
- Medlock, J.M.; Hansford, K.M.; Bormane, A.; Derdakova, M.; Estrada-Peña, A.; George, J.C.; Van Bortel, W. Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasites Vectors 2013, 6, 1. [Google Scholar] [CrossRef]
- Artigas-Jerónimo, S.; Villar, M.; Cabezas-Cruz, A.; Valdés, J.J.; Estrada-Peña, A.; Alberdi, P.; De la Fuente, J. Functional evolution of subolesin/akirin. Front. Physiol. 2018, 9, 1612. [Google Scholar] [CrossRef]
- Baron, S.; van der Merwe, N.A.; Maritz-Olivier, C. The genetic relationship between Rhipicephalus microplus and Rhipicephalus decoloratus ticks in South Africa and their population structure. Mol. Phylogenet. Evol. 2018, 129, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Cid, J.A.; Pérez de la Lastra, J.M.; Villar, M.; Jiménez, M.; Pinal, R.; Estrada-Peña, A.; Molina, R.; Lucientes, J.; Gortázar, C.; de la Fuente, J. Control of multiple arthropod vector infestations with subolesin/akirin vaccines. Vaccine 2013, 31, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Popara, M.; Villar, M.; Mateos-Hernández, L.; Fernández de Mera, I.G.; Marina, A.; del Valle, M.; Almazán, C.; Domingos, A.; de la Fuente, J. Lesser protein degradation machinery correlates with higher BM86 tick vaccine efficacy in Rhipicephalus annulatus when compared to Rhipicephalus microplus. Vaccine 2013, 31, 4728–4735. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Mallón, A. Developing anti-tick vaccines. In Vaccine Design; Humana Press: New York, NY, USA, 2016; pp. 243–259. [Google Scholar]
- Sabadin, G.A.; Parizi, L.F.; Kiio, I.; Xavier, M.A.; da Silva Matos, R.; Camargo-Mathias, M.I.; da Silva Vaz, I., Jr. Effect of recombinant glutathione S-transferase as vaccine antigen against Rhipicephalus appendiculatus and Rhipicephalus sanguineus infestation. Vaccine 2017, 35, 6649–6656. [Google Scholar] [CrossRef] [PubMed]


| Gene | Country | Accession Number | Identity (%) | Polymorphic Nucleotides | Nonsynonymous Polymorphism |
|---|---|---|---|---|---|
| Cysteine protease inhibitor (cystatin 2b) | India | KM588294 | 100 | - | - |
| Cathepsin L-like cysteine proteinase (cathepsin-L) | India | JX502822 | 99.40 | T189C, A528C, A684G, A732G, T837C, T940A | Ser246Gly, Trp281Arg, Val315Asp |
| Glutathione S-transferase (GST) | India | HQ337620 | 100 | - | - |
| Ferritin 1 | USA | AY277902 | 99.39 | G330A, A396G, T507C, T514C | - |
| 60S acidic ribosomal protein (P0) | Cuba | KC845304 | 99.48 | C75A, T151C, C195T, (T354C, A762G | - |
| Aquaporin 2 | USA | KP406519 | 99.62 | C781A, C873A, C961A, T963A | Leu254Ile, |
| ATAQ | Mexico | MG437296 | 98.46 | G90A, A177G, G405A, C794A, C855G, A858G, A1001G, A1131G, G1137A, G1152A, C1170T, T1275C, A1284G, C1296A, C1318T, G1327A, G1415T, C1416T, T1483C, A1991C, A1562C, A1411C, G1640A, G1671A, C1695A, A1701G, T1710A, A1711C | Glu265Ala, Gln285His, Ile320Val, Gln334Arg, Pro440Ser, Val443Ile, Leu473Met, Cys495byArg, Glu497Asp, Asp521Ala, Gln537His, Ile567Met, Ile571Leu, |
| R. microplus 05 antigen (Rm05Uy) | Uruguay | KX611484 | 99.61 | T207C, T319C | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeb, I.; Almutairi, M.M.; Alouffi, A.; Islam, N.; Parizi, L.F.; Safi, S.Z.; Tanaka, T.; da Silva Vaz, I., Jr.; Ali, A. Low Genetic Polymorphism in the Immunogenic Sequences of Rhipicephalus microplus Clade C. Vaccines 2022, 10, 1909. https://doi.org/10.3390/vaccines10111909
Zeb I, Almutairi MM, Alouffi A, Islam N, Parizi LF, Safi SZ, Tanaka T, da Silva Vaz I Jr., Ali A. Low Genetic Polymorphism in the Immunogenic Sequences of Rhipicephalus microplus Clade C. Vaccines. 2022; 10(11):1909. https://doi.org/10.3390/vaccines10111909
Chicago/Turabian StyleZeb, Ismail, Mashal M. Almutairi, Abdulaziz Alouffi, Nabila Islam, Luís Fernando Parizi, Sher Zaman Safi, Tetsuya Tanaka, Itabajara da Silva Vaz, Jr., and Abid Ali. 2022. "Low Genetic Polymorphism in the Immunogenic Sequences of Rhipicephalus microplus Clade C" Vaccines 10, no. 11: 1909. https://doi.org/10.3390/vaccines10111909
APA StyleZeb, I., Almutairi, M. M., Alouffi, A., Islam, N., Parizi, L. F., Safi, S. Z., Tanaka, T., da Silva Vaz, I., Jr., & Ali, A. (2022). Low Genetic Polymorphism in the Immunogenic Sequences of Rhipicephalus microplus Clade C. Vaccines, 10(11), 1909. https://doi.org/10.3390/vaccines10111909










