Updating the National Antigen Bank in Korea: Protective Efficacy of Synthetic Vaccine Candidates against H5Nx Highly Pathogenic Avian Influenza Viruses Belonging to Clades 2.3.2.1 and 2.3.4.4
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Synthesis of the HA Gene and Vaccine Development
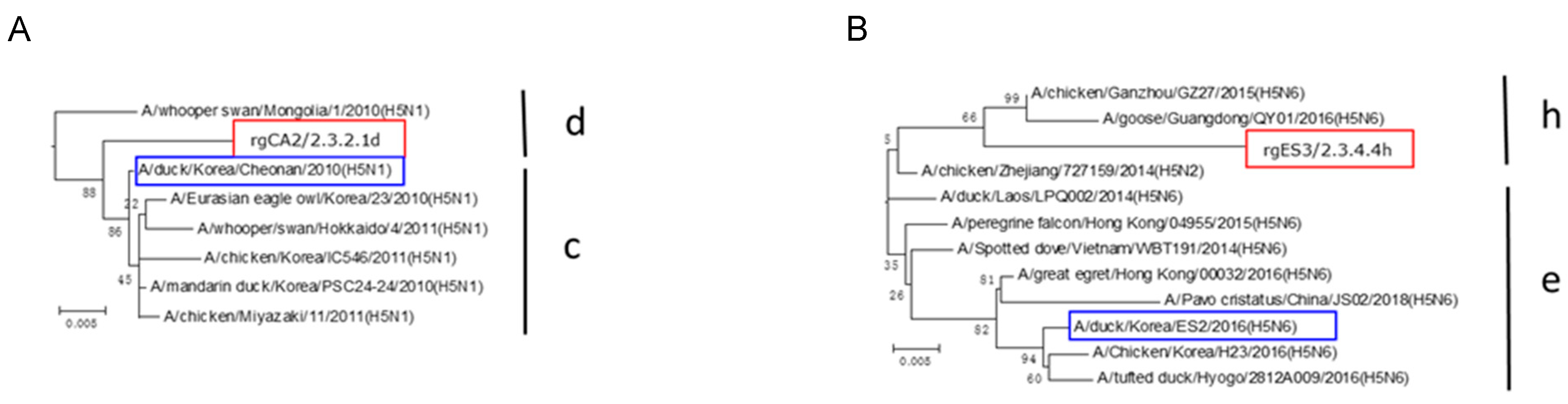
2.3. Phylogenetic Analysis
2.4. Vaccination and Challenge of Chickens to Determine Vaccine Potency and Efficacy
2.4.1. Serology and Antibody Assays
2.4.2. Post-Challenge Virus Shedding
2.5. Antibody Persistence
2.6. Statistical Analysis
3. Results
3.1. Vaccine Development
3.2. Vaccine Potency
3.2.1. Clinical Protection
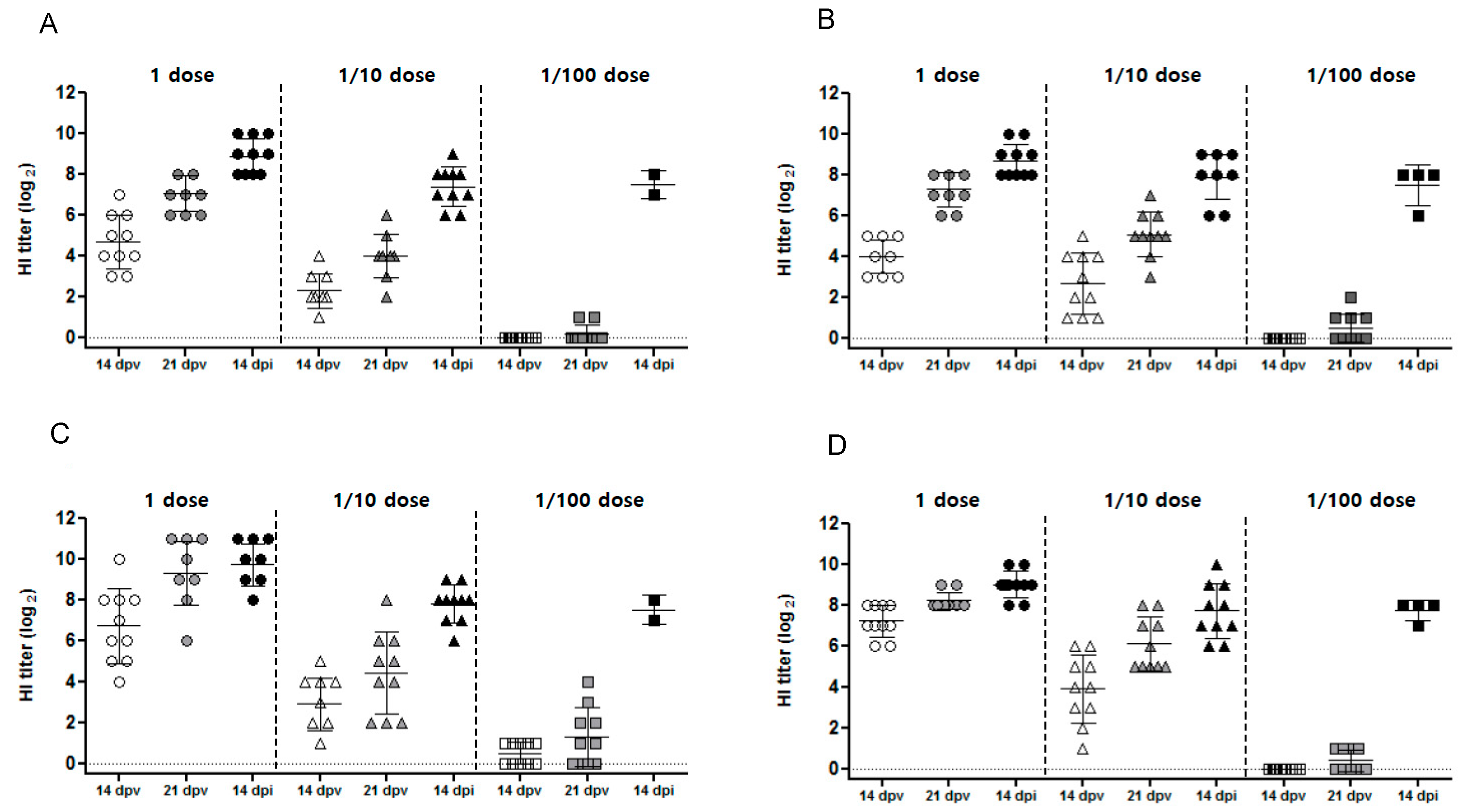
3.2.2. Serology
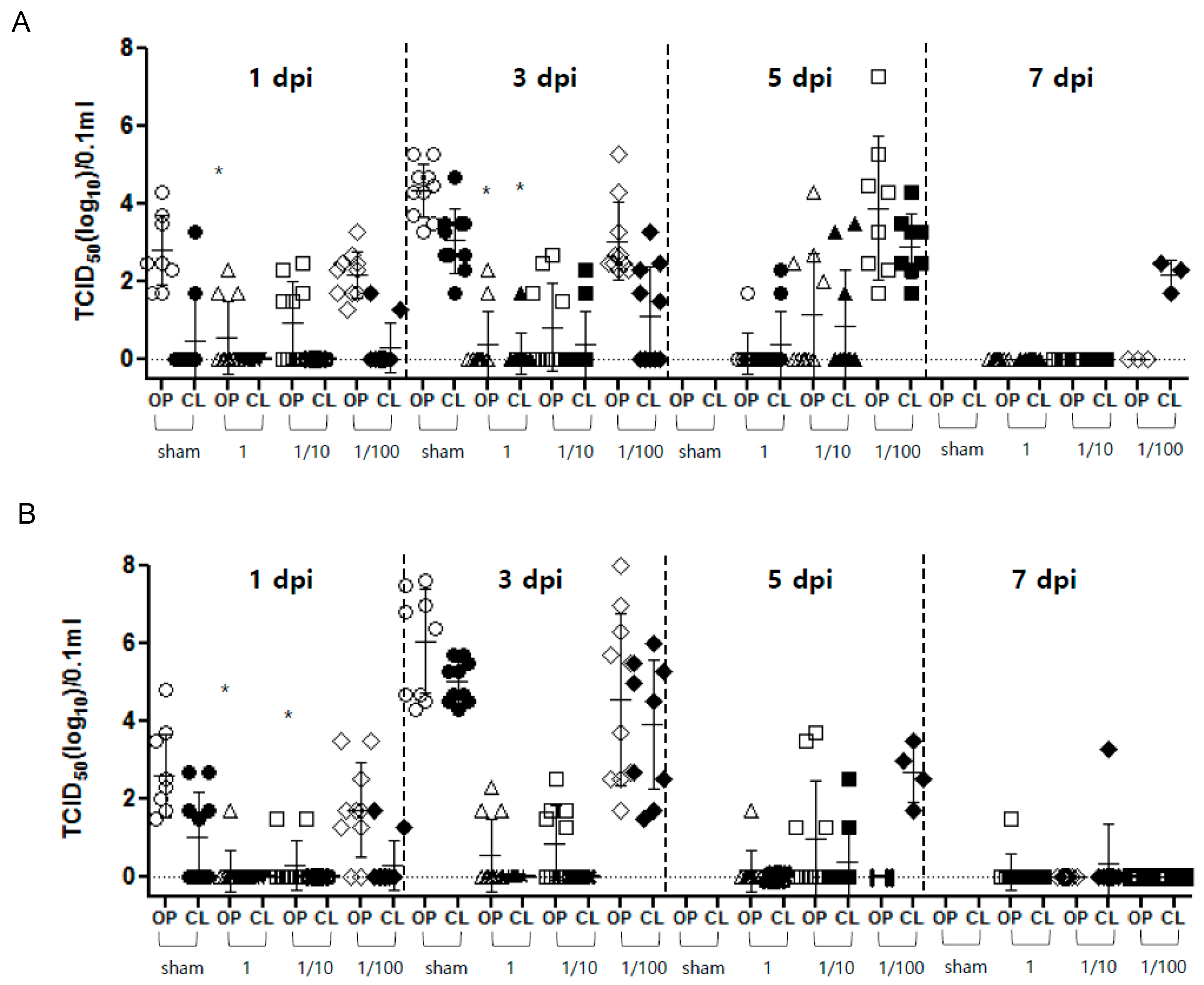
3.2.3. Virus Shedding
3.3. Antibody Persistence
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Li, M.; Li, Y.; Tian, J.; Bai, X.; Yang, C.; Shi, J.; Ai, R.; Chen, W.; Zhang, W.; et al. Outbreaks of Highly Pathogenic Avian Influenza (H5N6) Virus Subclade 2.3.4.4h in Swans, Xinjiang, Western China, 2020. Emerg. Infect. Dis. 2020, 26, 2956–2960. [Google Scholar] [CrossRef]
- Park, Y.; Lee, Y.; Lee, D.; Baek, Y.; Si, Y.; Meeduangchanh, P.; Theppangna, W.; Douangngeun, B.; Kye, S.; Lee, M.; et al. Genetic and pathogenic characteristics of clade 2.3.2.1c H5N1 highly pathogenic. Transbounding Emerg. Dis. Ease 2019, 67, 947–955. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; Li, B.; Guo, Y.; Xing, J.; Xu, Q.; Liu, L.; Zhang, J.; Qi, W.; Jia, W.; et al. Continuous Reassortment of Clade 2.3.4.4 H5N6 Highly Pathogenetic Avian Influenza Viruses Demonstrating High Risk to Public Health. Pathogens 2020, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Yuyun, I.; Wibawa, H.; Setiaji, G.; Kusumastuti, T.A.; Nugroho, W.S. Determining highly pathogenic H5 avian influenza clade 2.3.2.1c seroprevalence in ducks, Purbalingga, Central Java, Indonesia. Vet. World 2020, 13, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Suarez, D.L.; Tumpey, T.M.; Sung, H.-W.; Kwon, Y.-K.; Lee, Y.-J.; Choi, J.-G.; Joh, S.-J.; Kim, M.-C.; Lee, E.-K.; et al. Characterization of highly pathogenic H5N1 avian influenza A viruses isolated from South Korea. J. Virol. 2005, 79, 3692–3702. [Google Scholar] [CrossRef]
- Yoon, H.; Park, C.-K.; Nam, H.-M.; Wee, S.-H. Virus spread pattern within infected chicken farms using regression model: The 2003-2004 HPAI epidemic in the Republic of Korea. J. Vet. Med. B Infect. Dis. Vet. Public Health 2005, 52, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Song, B.-M.; Lee, Y.-N.; Lee, E.-K.; Heo, G.-B.; Kye, S.-J.; Lee, K.-H.; Bae, Y.-C.; Lee, Y.-J.; Kim, B. Pathogenicity of clade 2.3.4.4 H5N6 highly pathogenic avian influenza virus in three chicken breeds from South Korea in 2016/2017. J. Vet. Sci. 2019, 20, e27. [Google Scholar] [CrossRef]
- Jeong, J.; Kang, H.-M.; Lee, E.-K.; Song, B.-M.; Kwon, Y.-K.; Kim, H.-R.; Choi, K.-S.; Kim, J.-Y.; Lee, H.-J.; Moon, O.-K.; et al. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet. Microbiol. 2014, 173, 249–257. [Google Scholar] [CrossRef]
- Ooka, T.; Al, E.; Kim, H.-R.; Lee, Y.-J.; Park, C.-K.; Oem, J.-K.; Lee, O.-S.; Kang, H.-M.; Choi, J.-G.; Bae, Y.-C. Highly pathogenic avian influenza (H5N1) outbreaks in wild birds and poultry, South Korea. Emerg. Infect. Dis. 2012, 18, 480–483. [Google Scholar] [CrossRef]
- Lee, E.-K.; Lee, Y.-N.; Kye, S.-J.; Lewis, N.S.; Brown, I.H.; Sagong, M.; Heo, G.-B.; Kang, Y.-M.; Cho, H.-K.; Kang, H.-M.; et al. Characterization of a novel reassortant H5N6 highly pathogenic avian influenza virus clade 2.3.4.4 in Korea, 2017. Emerg. Microbes Infect. 2018, 7, 103. [Google Scholar] [CrossRef]
- Cui, P.; Shi, J.; Wang, C.; Zhang, Y.; Xing, X.; Kong, H.; Yan, C.; Zeng, X.; Liu, L.; Tian, G.; et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3.4.4b HA gene and biologic analysis of the ones detected in China. Emerg. Microbes Infect. 2022, 11, 1693–1704. [Google Scholar] [CrossRef]
- Gu, W.; Shi, J.; Cui, P.; Yan, C.; Zhang, Y.; Wang, C.; Zhang, Y.; Xing, X.; Zeng, X.; Liu, L.; et al. Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 2022, 11, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Sagong, M.; Lee, Y.; Song, S.; Cha, R.M.; Lee, E.; Kang, Y.; Cho, H.; Kang, H.; Lee, Y.; Lee, K. Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Transbound. Emerg. Dis. 2022, 69, e3255–e3260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, K.; Su, Q.; Chen, X.; Wang, X.; Li, Q.; Wang, W.; Mao, X.; Xu, J.; Zhou, X.; et al. Clinical features of the first critical case of acute encephalitis caused by the avian influenza A (H5N6) virus. Emerg. Microbes Infect. 2022, 11, 2437–2446. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-M.; Cho, H.-K.; Kim, H.-M.; Lee, M.-H.; To, T.L.; Kang, H.-M. Protective efficacy of vaccines of the Korea national antigen bank against the homologous H5Nx clade 2.3.2.1 and clade 2.3.4.4 highly pathogenic avian influenza viruses. Vaccine 2020, 38, 663–672. [Google Scholar] [CrossRef]
- David, E.; Swayne, D.R.K. Vaccines and vaccination for avian influenza in poultry. In Animal Influenza, 2nd ed.; Swayne, D.E., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2022; pp. 378–416. [Google Scholar]
- Monne, I.; Fusaro, A.; Nelson, M.I.; Bonfanti, L.; Mulatti, P.; Hughes, J.; Murcia, P.R.; Schivo, A.; Valastro, V.; Moreno, A.; et al. Emergence of a Highly Pathogenic Avian Influenza Virus from a Low-Pathogenic Progenitor. J. Virol. 2014, 88, 4375–4388. [Google Scholar] [CrossRef]
- An, S.-H.; Lee, C.-Y.; Hong, S.-M.; Choi, J.-G.; Lee, Y.-J.; Jeong, J.-H.; Kim, J.-B.; Song, C.-S.; Kim, J.-H.; Kwon, H.-J. Bioengineering a highly productive vaccine strain in embryonated chicken eggs and mammals from a non-pathogenic clade 2·3·4·4 H5N8 strain. Vaccine 2019, 37, 6154–6616. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Suphaphiphat, P.; Gibson, D.G.; Wentworth, D.E.; Stockwell, T.B.; Algire, M.A.; Alperovich, N.; Barro, M.; Brown, D.M.; Craig, S.; et al. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci. Transl. Med. 2013, 5, 185. [Google Scholar] [CrossRef]
- Yan, J.; Villarreal, D.O.; Racine, T.; Chu, J.S.; Walters, J.N.; Morrow, M.P.; Khan, A.S.; Sardesai, N.Y.; Kim, J.J.; Kobinger, G.P.; et al. Protective immunity to H7N9 influenza viruses elicited by synthetic DNA vaccine. Vaccine 2014, 32, 2833–2842. [Google Scholar] [CrossRef]
- Song, M.-S.; Baek, Y.H.; Pascua, P.N.Q.; Kwon, H.-I.; Park, S.-J.; Kim, E.-H.; Lim, G.-J.; Choi, Y.-K. Establishment of Vero cell RNA polymerase I-driven reverse genetics for Influenza A virus and its application for pandemic (H1N1) 2009 influenza virus vaccine production. J. Gen. Virol. 2013, 94 Pt 6, 1230–1235. [Google Scholar] [CrossRef]
- Hoffmann, E.; Krauss, S.; Perez, D.; Webby, R.; Webster, R.G. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 2002, 20, 3165–3170. [Google Scholar] [CrossRef]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Liu, M.; Liu, F.; Liu, D.F.; Zhang, Y.; Pan, W.Q.; Chen, H.; Wan, C.H.; Sun, E.C.; Li, H.T.; et al. Evaluation of several adjuvants in avian influenza vaccine to chickens and ducks. Virol. J. 2011, 8, 321. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-Y.; Kang, Y.-M.; Cho, H.-K.; Park, S.-J.; Lee, M.-H.; Lee, Y.-J.; Kang, H.-M. Development of a recombinant H9N2 influenza vaccine candidate against the Y280 lineage field virus and its protective efficacy. Vaccine 2021, 39, 6201–6205. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002. Available online: https://apps.who.int/iris/bitstream/handle/10665/68026/WHO_CDS_CSR_NCS_2002.5.pdf?sequence=1&isAllowed=y (accessed on 4 February 2022).
- Kapczynski, D.R.; Pantin-Jackwood, M.J.; Spackman, E.; Chrzastek, K.; Suarez, D.; Swayne, D. Homologous and heterologous antigenic matched vaccines containing different H5 hemagglutinins provide variable protection of chickens from the 2014 U.S. H5N8 and H5N2 clade 2.3.4.4 highly pathogenic avian influenza viruses. Vaccine 2017, 35, 6345–6353. [Google Scholar] [CrossRef]
- Zeng, X.; Tian, G.; Shi, J.; Deng, G.; Li, C.; Chen, H. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci. China Life Sci. 2018, 61, 1465–1473. [Google Scholar] [CrossRef]
- Avian influenza (Including Infection with High Pathogenicity Avian Influenza Viruses). In OIE Terrestrial Manual 2021; World organization for Animal Health (WOAH): Paris, France, 2021.
- Zeng, X.; Chen, P.; Liu, L.; Deng, G.; Li, Y.; Shi, J.; Kong, K.; Feng, H.; Bai, J.; Li, X.; et al. Protective Efficacy of an H5N1 Inactivated Vaccine Against Challenge with Lethal H5N1, H5N2, H5N6, and H5N8 Influenza Viruses in Chickens. Avian Dis. 2016, 60 (Suppl. S1), 253–255. [Google Scholar] [CrossRef]
- Guo, C.-Y.; Zhang, H.-X.; Zhang, J.-J.; Sun, L.-J.; Li, H.-J.; Liang, D.-Y.; Feng, Q.; Li, Y.; Feng, Y.-M.; Xie, X.; et al. Localization Analysis of Heterophilic Antigen Epitopes of H1N1 Influenza Virus Hemagglutinin. Virol. Sin. 2019, 34, 3536–3542. [Google Scholar] [CrossRef]
- Leung, Y.C.; Luk, G.; Sia, S.-F.; Wu, Y.-O.; Ho, C.-K.; Chow, K.-C.; Tang, S.-C.; Guan, Y.; Peiris, J.M. Experimental challenge of chicken vaccinated with commercially available H5 vaccines reveals loss of protection to some highly pathogenic avian influenza H5N1 strains circulating in Hong Kong/China. Vaccine 2013, 31, 3536–3542. [Google Scholar] [CrossRef]
- Kang, Y.-M.; Cho, H.-K.; Kim, H.-M.; Lee, C.-H.; Kim, D.-Y.; Choi, S.-H.; Lee, M.-H.; Kang, H.-M. Protection of layers and breeders against homologous or heterologous HPAIv by vaccines from Korean national antigen bank. Sci. Rep. 2020, 10, 9436. [Google Scholar] [CrossRef]

| Vaccine | Growth Properties in Eggs | Challenge Virus | Vaccine Efficacy | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EID50 (log10/0.1 mL) | HA Titer (log2) | Homology with the Vaccine (%) | Antigen Dose 1 | HI Titer (log2) 4 Against | Clinical Signs | Peak Shedding (3 dpi) 3 | Survival (%) (MDT) 2 | PD50 | Antibody Persistence (24 wpv) 5 | |||
| Challenge Strain Antigen | Vaccine Strain Antigen | OP | CL | |||||||||
| rgCA2/2.3.2.1d | 8.2 | 8 | CA/2.3.2.1 97.0 | 1 | 10/10 (7.1) | 10/10 (9.3) | Diarrhea | 2/10 * (2.0) | 1/10 * (1.7) | 100 | 42 | 8.3 |
| 1/10 | 10/10 (4.0) | 10/10 (4.4) | Diarrhea | 4/10 (2.1) | 2/10 (2.0) | 100 | ||||||
| 1/100 | 2/10 (0.5) | 5/10 (0.5) | Lethargy, green feces, anorexia, Death | 10/10 (3.1) | 5/10 (2.3) | 20 (4.5) | ||||||
| Sham | 0/10 | 0/10 | Death | 10/10 (4.4) | 10/10 (3.1) | 0 (2.2) | ||||||
| rgES3/2.3.4.4h | 8.4 | 8 | ES2/2.3.4.4 95.4 | 1 | 10/10 (7.3) | 10/10 (8.2) | Diarrhea | 3/10 (1.9) | 0/10 | 100 | 68 | 7.8 |
| 1/10 | 10/10 (5.1) | 10/10 (5.9) | Diarrhea | 5/10 (1.7) | 0/10 | 100 | ||||||
| 1/100 | 4/10 (0.5) | 4/10 (0.4) | Lethargy, green feces, Death | 10/10 (4.6) | 10/10 (3.9) | 40 (2.7) | ||||||
| Sham | 0/10 | 0/10 | Death | 10/10 (6.1) | 10/10 (5.0) | 0 (2.6) | ||||||
| Variable Residue 1 | 2.3.2.1 | 2.3.4.4 | ||
|---|---|---|---|---|
| Vaccine Strain 2 | Challenge Strain | Vaccine Strain 2 | Challenge Strain | |
| rgCA2/2.3.2.1d | CA/2.3.2.1 | rgES3/2.3.4.4h | ES2/2.3.4.4 | |
| 69 | E | E | E | E |
| 71 | T | I | I | I |
| 83 | A | A | L | L |
| 95 | F | F | A | A |
| 133 | S | S | A | S |
| 140 | N | N | V | V |
| 162 | I | I | I | I |
| 183 | D | D | A | A |
| 189 | R | R | L | L |
| 194 | P | P | T | T |
| 270 | E | E | E | E |
| Homology 3 (%) | 97.0 | 95 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Y.-M.; Cho, H.-K.; An, S.-J.; Kim, H.-J.; Lee, Y.-J.; Kang, H.-M. Updating the National Antigen Bank in Korea: Protective Efficacy of Synthetic Vaccine Candidates against H5Nx Highly Pathogenic Avian Influenza Viruses Belonging to Clades 2.3.2.1 and 2.3.4.4. Vaccines 2022, 10, 1860. https://doi.org/10.3390/vaccines10111860
Kang Y-M, Cho H-K, An S-J, Kim H-J, Lee Y-J, Kang H-M. Updating the National Antigen Bank in Korea: Protective Efficacy of Synthetic Vaccine Candidates against H5Nx Highly Pathogenic Avian Influenza Viruses Belonging to Clades 2.3.2.1 and 2.3.4.4. Vaccines. 2022; 10(11):1860. https://doi.org/10.3390/vaccines10111860
Chicago/Turabian StyleKang, Yong-Myung, Hyun-Kyu Cho, Sung-Jun An, Hyun-Jun Kim, Youn-Jeong Lee, and Hyun-Mi Kang. 2022. "Updating the National Antigen Bank in Korea: Protective Efficacy of Synthetic Vaccine Candidates against H5Nx Highly Pathogenic Avian Influenza Viruses Belonging to Clades 2.3.2.1 and 2.3.4.4" Vaccines 10, no. 11: 1860. https://doi.org/10.3390/vaccines10111860
APA StyleKang, Y.-M., Cho, H.-K., An, S.-J., Kim, H.-J., Lee, Y.-J., & Kang, H.-M. (2022). Updating the National Antigen Bank in Korea: Protective Efficacy of Synthetic Vaccine Candidates against H5Nx Highly Pathogenic Avian Influenza Viruses Belonging to Clades 2.3.2.1 and 2.3.4.4. Vaccines, 10(11), 1860. https://doi.org/10.3390/vaccines10111860





