A Novel Dry-Stabilized Whole Blood Microsampling and Protein Extraction Method for Testing of SARS-CoV-2 Antibody Titers
Abstract
:1. Introduction
2. Materials and Methods
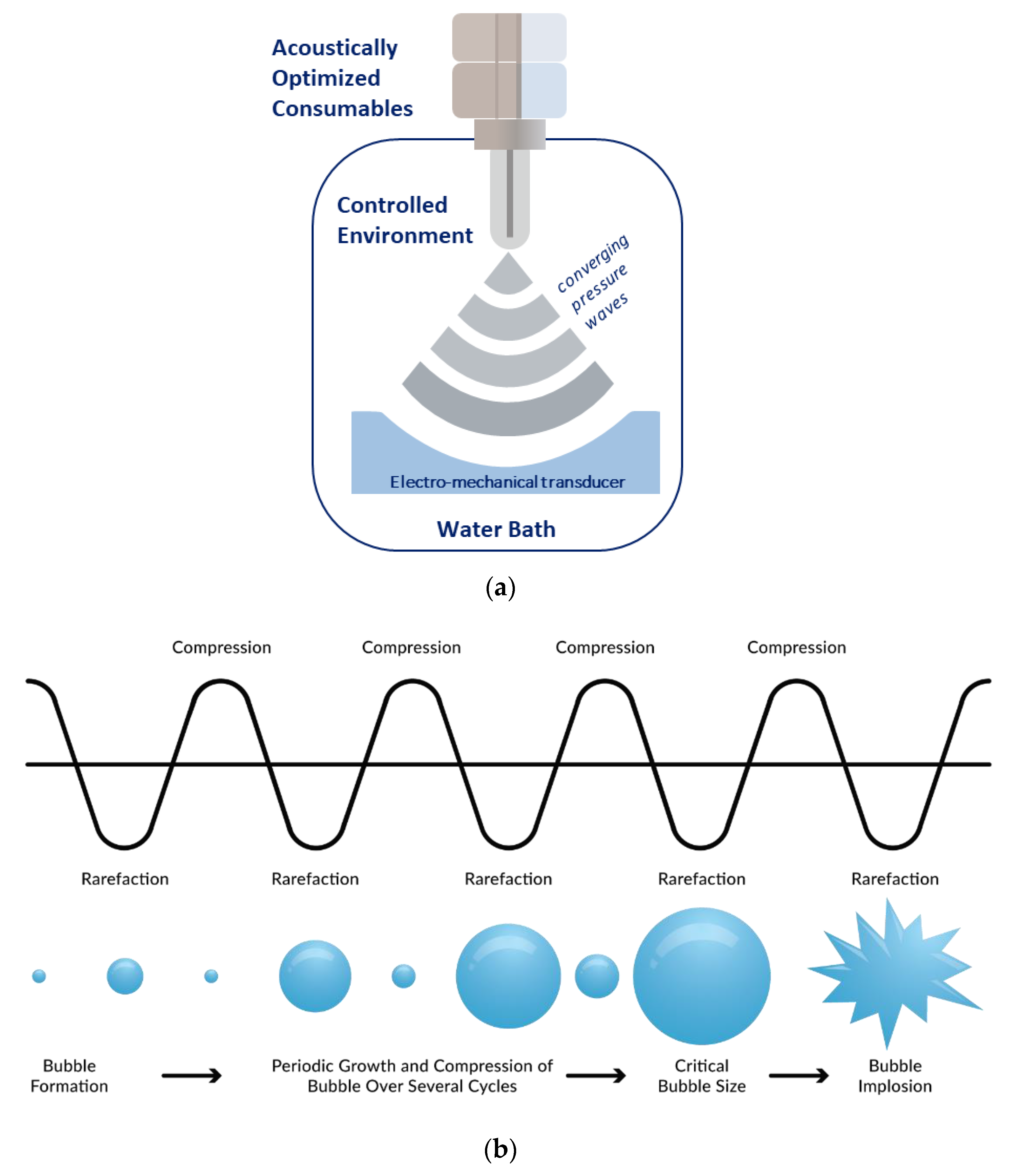
2.1. Adaptive Focused Acoustics
2.2. Blood Donor Demographics and Ethics
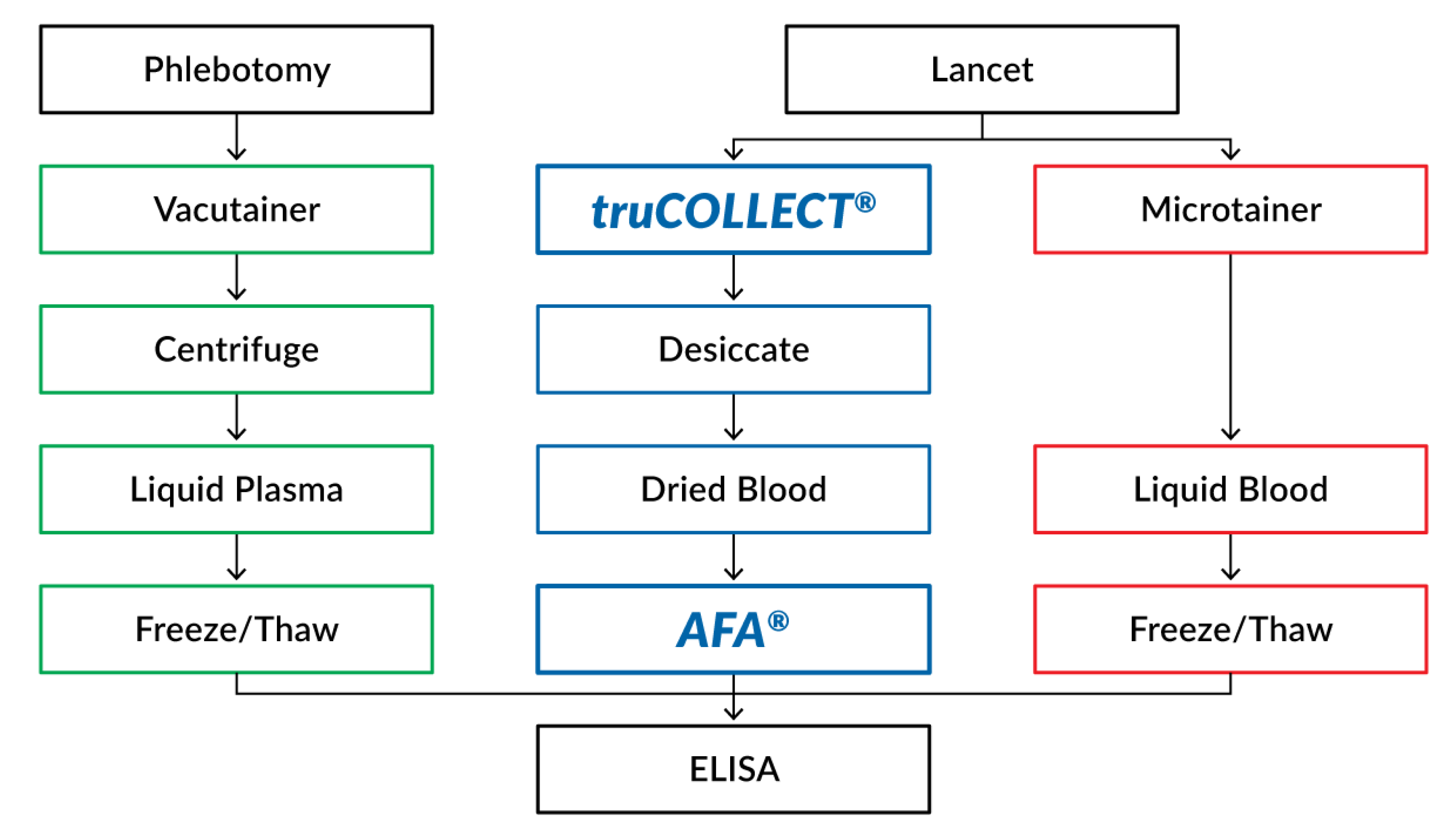
2.3. Venipuncture Blood Sample Collection and Processing
2.4. Capillary Blood Sample Collection
2.5. truCOLLECT Sample Processing
2.6. Serology
2.7. Analytical Analysis
3. Results
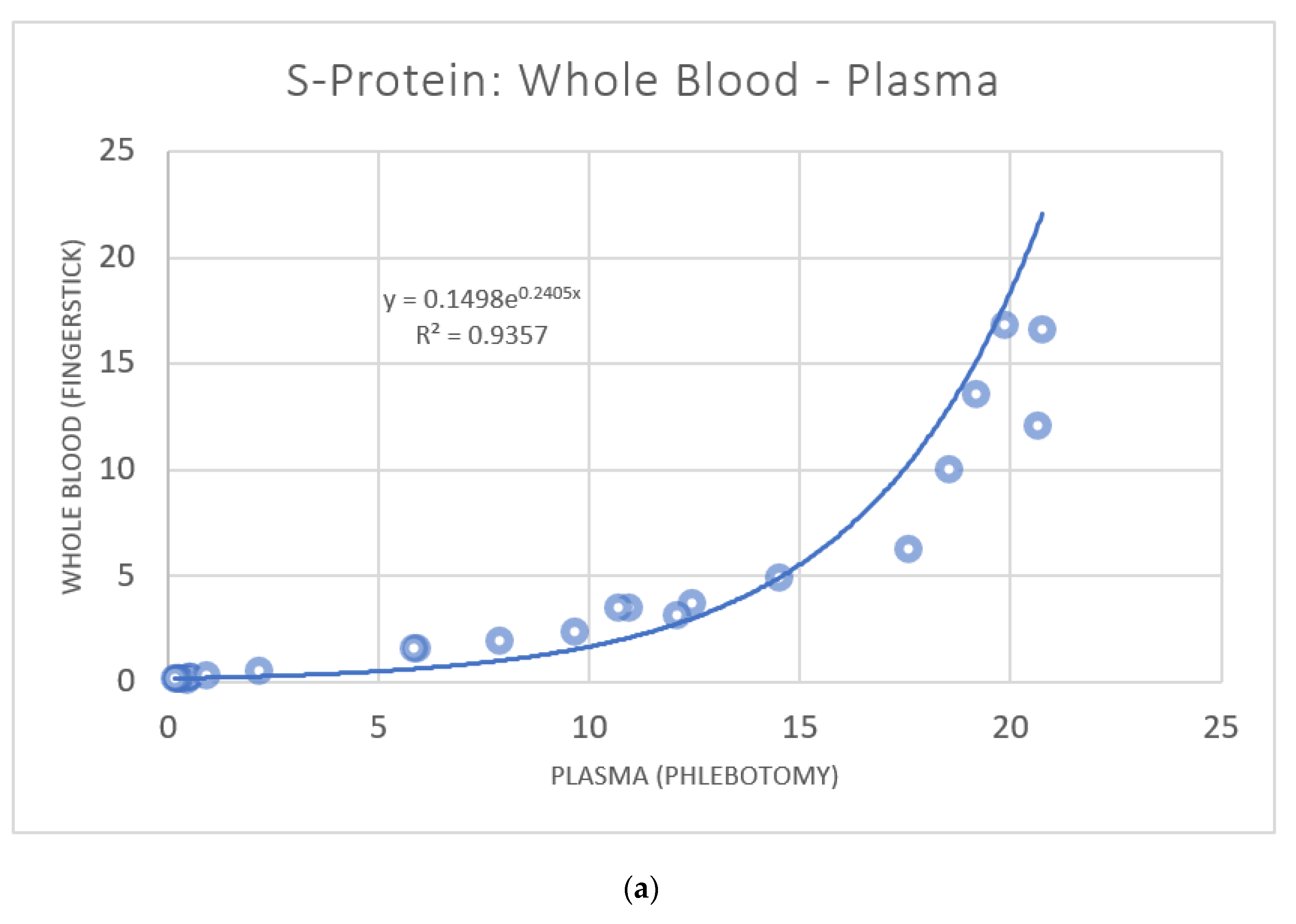
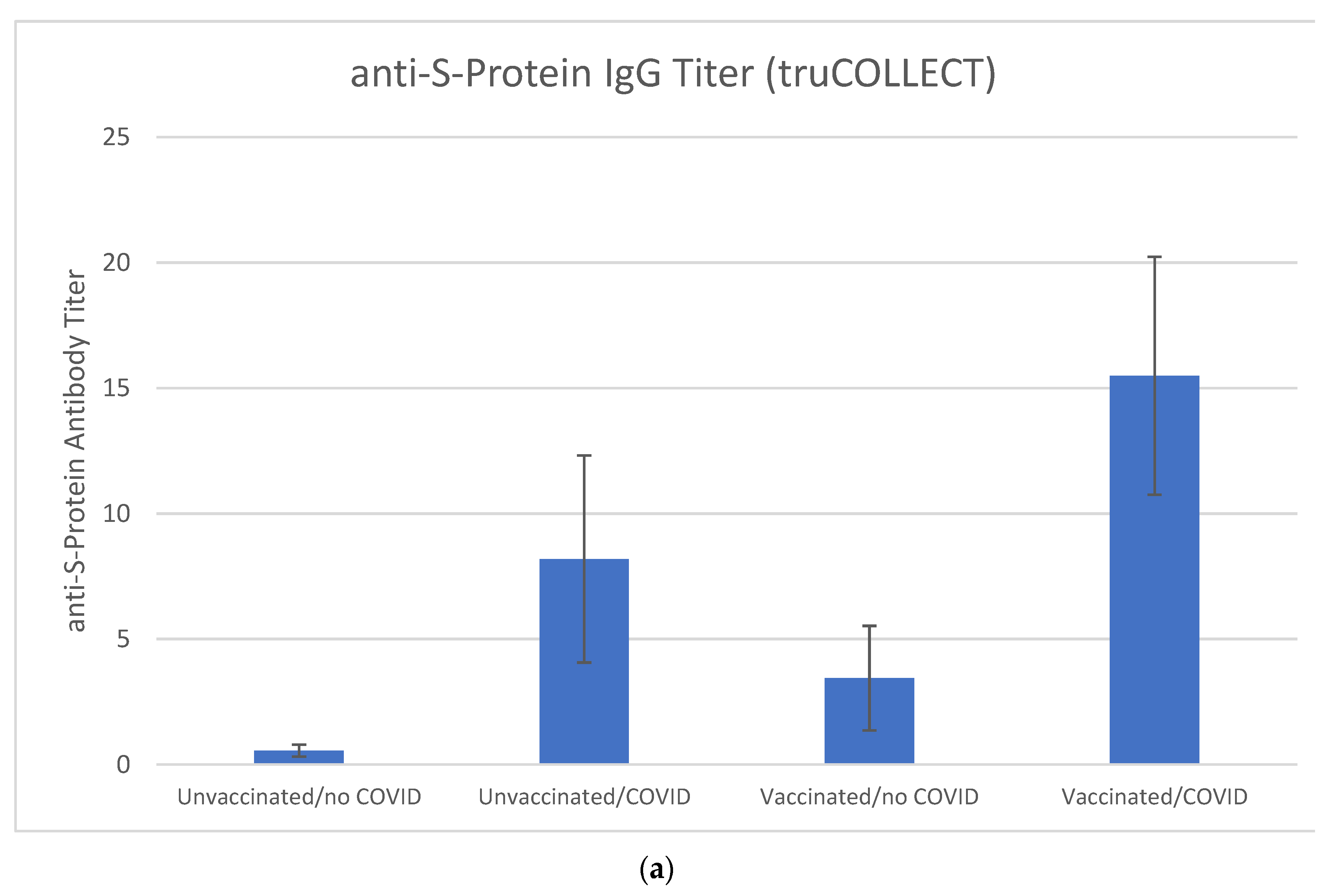
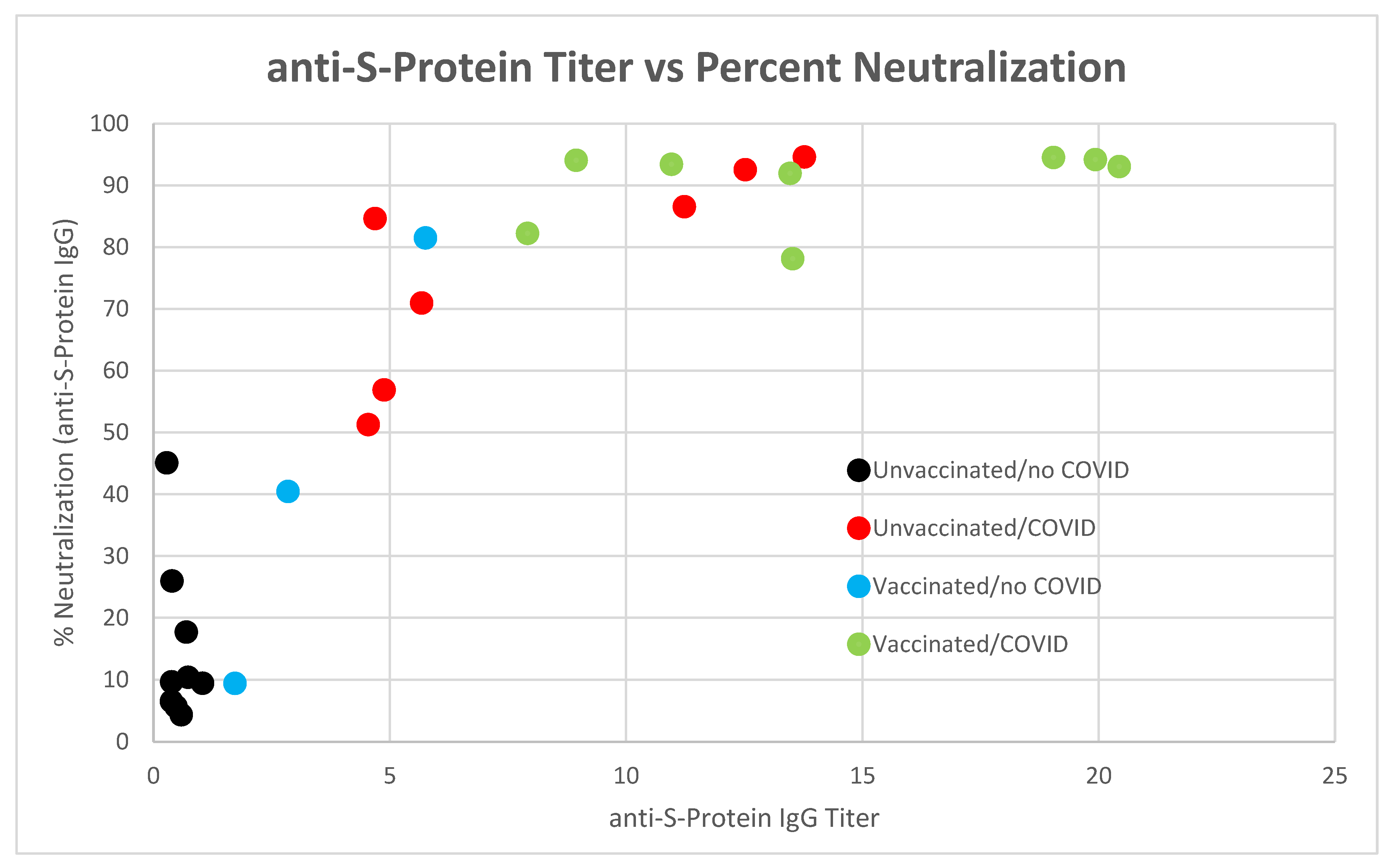
3.1. anti-SARS-CoV-2 Spike Protein Titer Analysis
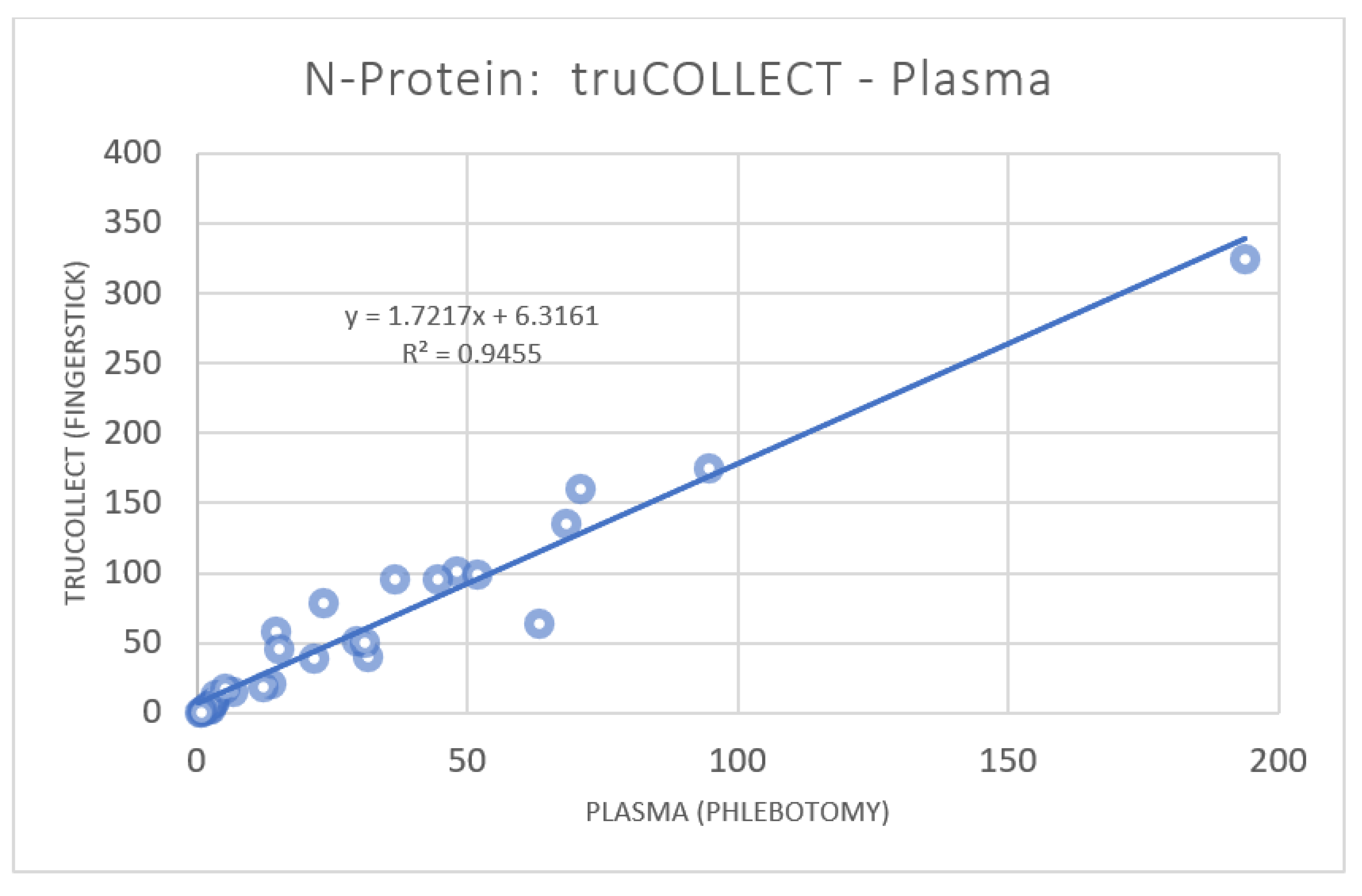
3.2. anti-SARS-CoV-2 Nucleocapsid Protein Titer Analysis
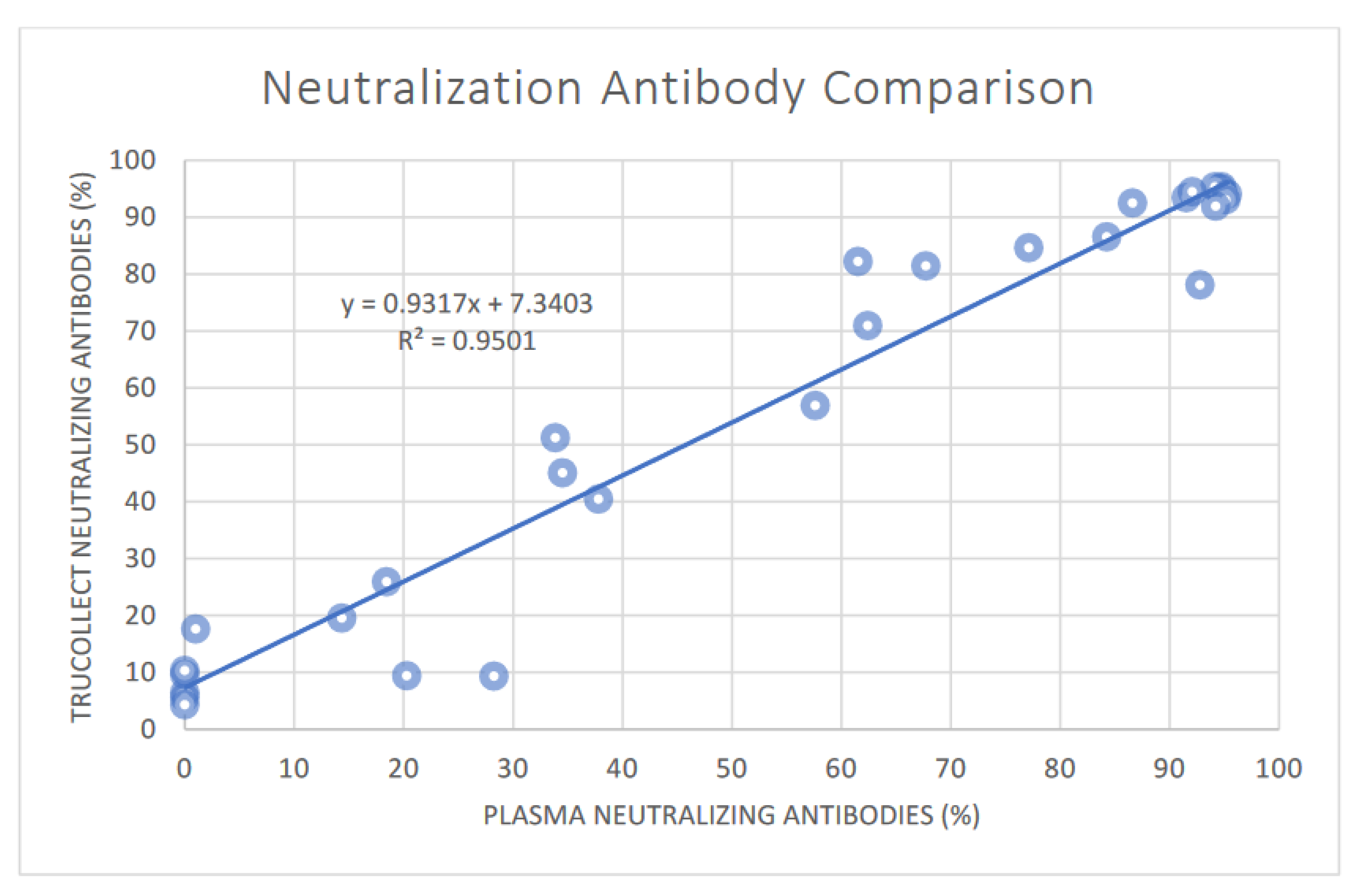
3.3. SARS-CoV-2 Neutralizing Antibody Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Symptoms of COVID-19; National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed on 26 September 2022).
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Long COVID or Post-COVID Conditions; National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html (accessed on 26 September 2022).
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic; World Health Organization: Geneva, Switzerland, 2022; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 26 September 2022).
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Stay Up to Date with Your COVID-19 Vaccines; National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/stay-up-to-date.html (accessed on 26 September 2022).
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Science Brief: SARS-CoV-2 Infection-induced and Vaccine-induced Immunity; National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases: Atlanta, GA, USA, 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/vaccine-induced-immunity.html (accessed on 26 September 2022).
- Haghi Ashtiani, M.T.; Sadeghi Rad, P.; Asnaashari, K.; Shahhosseini, A.; Berenji, F.; Mamishi, S. Role of serology tests in COVID-19 non-hospitalized patients: A cross-sectional study. PLoS ONE 2022, 17, e0266923. [Google Scholar] [CrossRef] [PubMed]
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. COVID-19 Serology Surveillance Strategy; National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases: Atlanta, GA, USA, 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/index.html (accessed on 26 September 2022).
- Wasniewski, M.; Laurentie, M.; Rizzo, F.; Servat, A.; Aubert, M.; Cliquet, F. Proficiency test for rabies serology: A design complying with international standards for a reliable assessment of participating laboratories. PLOS Negl. Trop. Dis. 2019, 13, e0007824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, M.H.; Yeo, Y.H.; So, S.; Gane, E.; Cheung, R.C.; Nguyen, M.H. Prevalence of Hepatitis B Vaccination Coverage and Serologic Evidence of Immunity Among US-Born Children and Adolescents From 1999 to 2016. JAMA Netw. Open 2020, 3, e2022388. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Sofer, D.; Bucris, E.D.; Mendelson, E. Surveillance and Diagnosis of West Nile Virus in the Face of Flavivirus Cross-Reactivity. Front. Microbiol. 2018, 9, 2421. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Murray, J.A.; Katzka, D.A. AGA Clinical Practice Update on Diagnosis and Monitoring of Celiac Disease-Changing Utility of Serology and Histologic Measures: Expert Review. Gastroenterology 2019, 156, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Nys, G.; Gallez, A.; Kok, M.; Cobraiville, G.; Servais, A.C.; Piel, G.; Pequeux, C.; Fillet, M. Whole blood microsampling for the quantitation of estetrol without derivatization by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2017, 140, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Pearce, N.; Vandenbroucke, J.P.; VanderWeele, T.J.; Greenland, S. Accurate Statistics on COVID-19 Are Essential for Policy Guidance and Decisions. Am. J. Public Health 2020, 110, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.U.W.; Prow, T.W. A review of microsampling techniques and their social impact. Biomed. Microdevices 2019, 21, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Zhang, D.; Pathakamuri, J.A.; Kuebler, D.; Yang, Y.; Loginova, Y.; Chu, E.; Madej, R.; Neves, J.V.; Singer, B.; et al. A new testing platform using fingerstick blood for quantitative antibody response evaluation after SARS-CoV-2 vaccination. Emerg. Microbes Infect. 2022, 11, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.M.; Flores, S.R.; De Jesús, V.R. Influence of Hematocrit and Total-Spot Volume on Performance Characteristics of Dried Blood Spots for Newborn Screening. Int. J. Neonatal Screen. 2015, 1, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Drucker, C.; O’Dwyer, B.; Hugo, P.; Fix, C.; Keane, C.; Rachmuth, G. Self-collection of Blood Specimens in Clinical Trials. Appl. Clin. Trials. 2021. Available online: https://www.appliedclinicaltrialsonline.com/view/self-collection-of-blood-specimens-in-clinical-trials (accessed on 26 September 2022).
- Brown, L.; Byrne, R.L.; Fraser, A.; Owen, S.I.; Cubas-Atienzar, A.I.; Williams, C.T.; Kay, G.A.; Cuevas, L.E.; Fitchett, J.; Fletcher, T.; et al. Self-sampling of capillary blood for SARS-CoV-2 serology. Sci. Rep. 2021, 11, 7754. [Google Scholar] [CrossRef] [PubMed]
- Matthews, M.M.; Kim, T.G.; Shibata, S.; Shibata, N.; Butcher, C.; Hyun, J.; Kim, K.Y.; Robb, T.; Jheng, S.S.; Narita, M.; et al. COVID-19 serological survey using micro blood sampling. Sci. Rep. 2021, 11, 9475. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.; Padmanabhan, N.; Romero, D.; Joe, J.; Gebremeskel, M.; Manjula, N.; Wohlstadter, N.; Wohlstadter, R.; Goodwin, P.; Quintero, L.; et al. Quantitative serology for SARS-CoV-2 using self-collected saliva and finger-stick blood. Sci. Rep. 2022, 12, 6560. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Miller, T.E.; Kirkpatrick, G.; Nixon, A.; Astudillo, M.G.; Yang, D.; Mahanta, L.M.; Murali, M.; Dighe, A.S.; Lennerz, J.; et al. Remote Fingerstick Blood Collection for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Antibody Testing. Arch. Pathol. Lab. Med. 2021, 145, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, C.; Croxatto, A.; Coste, A.T.; Pojer, F.; André, C.; Pellaton, C.; Farina, A.; Campos, J.; Hacker, D.; Lau, K.; et al. Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies. J. Virol. 2021, 95, e01828-20. [Google Scholar] [CrossRef]




| Demographics | S antigen | Percent Neutralization | N Antigen | Vaccination | COVID | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor ID | Age | Sex | truCOLLECT | Plasma | WB | truCOLLECT | Plasma | truCOLLECT | Plasma | State | Date | Date |
| 1 | 40–49 | F | 0.38 | 0.30 | 0.13 | 6.5 | 0.0 | 2.75 | 1.44 | UV | - | - |
| 2 | 20–29 | F | 0.48 | 0.36 | 0.14 | 5.6 | 0.0 | 2.03 | 1.40 | UV | - | - |
| 3 | 50–59 | F | 0.59 | 0.45 | 0.02 | 4.3 | 0.0 | 3.44 | 2.49 | UV | - | - |
| 13 | 20–29 | F | 0.70 | 0.53 | 0.20 | 17.7 | 1.0 | 15.93 | 6.69 | UV | - | - |
| 14 | 50–59 | F | 0.39 | 0.22 | 0.14 | 9.6 | 0.0 | 1.14 | 0.41 | UV | - | - |
| 15 | 20–29 | F | 0.73 | 0.55 | 0.18 | 10.4 | 0.0 | 7.76 | 3.16 | UV | - | - |
| 28 | 20–29 | F | 1.04 | 0.91 | 0.26 | 9.4 | 20.3 | 17.33 | 5.18 | UV | - | - |
| 30 | 30–39 | F | 0.39 | 0.23 | 0.11 | 26.0 | 18.5 | 1.94 | 1.02 | UV | - | - |
| 31 | 40–49 | M | 0.28 | 0.16 | 0.11 | 45.1 | 34.5 | 1.17 | 0.58 | UV | - | - |
| 4 | 30–39 | M | 13.77 | 17.60 | 6.24 | 94.6 | 94.6 | 325.33 | 193.70 | UV | - | Q2|2021 |
| 7 | 20–29 | M | 4.55 | 5.92 | 1.50 | 51.3 | 33.9 | 39.83 | 21.51 | UV | - | Q1|2021 |
| 12 | 20–29 | M | 4.69 | 7.87 | 1.91 | 84.6 | 77.1 | 175.10 | 94.42 | UV | - | Q4|2021 |
| 16 | 40–49 | F | 11.23 | 14.55 | 4.84 | 86.5 | 84.3 | 40.49 | 31.60 | UV | - | Q4|2021 |
| 19 | 40–49 | F | 12.52 | 10.97 | 3.47 | 92.5 | 86.6 | 58.80 | 14.42 | UV | - | Q1|2022 |
| 21 | 40–49 | M | 0.45 | 0.29 | 0.14 | 19.6 | 14.3 | 7.04 | 2.86 | UV | - | Q1|2022 |
| 22 | 20–29 | F | 4.88 | 6.09 | ND | 56.9 | 57.6 | 18.77 | 12.07 | UV | - | Q3|2021 |
| 24 | 20–29 | F | 5.67 | 5.82 | 1.55 | 71.0 | 62.4 | 50.43 | 30.95 | UV | - | Q1|2021 |
| 5 | 60–69 | F | 18.97 | 20.65 | 12.01 | 95.0 | 94.7 | 96.22 | 36.62 | B (M) | Q4|2021 | - |
| 9 | 20–29 | F | 5.76 | 9.65 | 2.34 | 81.5 | 67.7 | 7.96 | 3.07 | S (?) | Q2|2021 | - |
| 10 | 20–29 | F | 16.62 | 19.22 | 13.51 | 95.4 | 94.8 | 102.25 | 47.84 | S (?) | Q3|2021 | - |
| 17 | 50–59 | M | 20.54 | 20.75 | 16.62 | 95.3 | 94.2 | 96.02 | 44.52 | B (P) | Q4|2021 | - |
| 20 | 20–29 | F | 2.85 | 2.77 | ND | 40.5 | 37.8 | 2.65 | 2.15 | D (P) | Q2|2021 | - |
| 27 | 20–29 | F | 1.73 | 2.17 | 0.44 | 9.4 | 28.3 | 5.17 | 1.98 | S (P) | Q1|2021 | - |
| 6 | 20–29 | M | 10.96 | 12.43 | 3.61 | 93.4 | 91.5 | 20.73 | 13.71 | S (?) | Q1|2022 | Q3|2021 |
| 8 | 20–29 | F | 19.93 | 19.85 | 16.74 | 94.2 | 94.9 | 64.07 | 63.06 | D (M) | Q2|2021 | Q1|2022 |
| 11 | 60–69 | M | 7.92 | 12.12 | 3.06 | 82.2 | 61.5 | 99.31 | 51.63 | D (M) | Q2|2021 | Q1|2022 |
| 18 | 30–39 | M | 19.05 | 18.56 | 10.03 | 94.5 | 92.1 | 78.66 | 23.18 | S (?) | Q2|2021 | Q4|2021 |
| 23 | 20–29 | F | 8.95 | 10.73 | 3.49 | 94.1 | 95.3 | 52.15 | 29.39 | B (M) | Q4|2021 | Q1|2022 |
| 25 | 30–39 | M | 20.44 | 21.04 | ND | 93.0 | 95.2 | 135.36 | 68.22 | D (P) | Q1|2021 | Q1|2022 |
| 26 | 20–29 | M | 13.53 | 15.70 | ND | 78.1 | 92.8 | 12.78 | 3.37 | D (M) | Q3|2021 | Q3|2021 |
| 29 | 40–49 | M | 13.47 | 13.01 | ND | 91.9 | 94.2 | 161.17 | 70.93 | D (P) | Q2|2021 | Q4|2021 |
| 32 | 60–69 | F | ND | ND | ND | 6.4 | ND | 45.66 | 15.21 | D (P) | Q1|2021 | Q1|2022 |
| Average Neg | 0.55 | 0.41 | 0.14 | 5.77 | 2.46 | |||||||
| Stdev | 0.24 | 0.23 | 0.07 | 5.57 | 1.87 | |||||||
| Threshold Avg + 2 Stdev | 1.03 | 0.88 | 0.28 | 16.92 | 6.20 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCarthy, P.; Pathakamuri, J.A.; Kuebler, D.; Neves, J.; Krohn, M.; Rohall, M.; Archibeque, I.; Giese, H.; Werner, M.; Daviso, E.; et al. A Novel Dry-Stabilized Whole Blood Microsampling and Protein Extraction Method for Testing of SARS-CoV-2 Antibody Titers. Vaccines 2022, 10, 1760. https://doi.org/10.3390/vaccines10101760
McCarthy P, Pathakamuri JA, Kuebler D, Neves J, Krohn M, Rohall M, Archibeque I, Giese H, Werner M, Daviso E, et al. A Novel Dry-Stabilized Whole Blood Microsampling and Protein Extraction Method for Testing of SARS-CoV-2 Antibody Titers. Vaccines. 2022; 10(10):1760. https://doi.org/10.3390/vaccines10101760
Chicago/Turabian StyleMcCarthy, Patrick, Joseph A. Pathakamuri, Daniel Kuebler, Jocelyn Neves, Madison Krohn, Michael Rohall, Isaac Archibeque, Heidi Giese, Martina Werner, Eugenio Daviso, and et al. 2022. "A Novel Dry-Stabilized Whole Blood Microsampling and Protein Extraction Method for Testing of SARS-CoV-2 Antibody Titers" Vaccines 10, no. 10: 1760. https://doi.org/10.3390/vaccines10101760
APA StyleMcCarthy, P., Pathakamuri, J. A., Kuebler, D., Neves, J., Krohn, M., Rohall, M., Archibeque, I., Giese, H., Werner, M., Daviso, E., & Thomann, U. (2022). A Novel Dry-Stabilized Whole Blood Microsampling and Protein Extraction Method for Testing of SARS-CoV-2 Antibody Titers. Vaccines, 10(10), 1760. https://doi.org/10.3390/vaccines10101760






