Abstract
The maintenance of the mitochondrial function is essential in preventing and counteracting neurodegeneration. In particular, mitochondria of neuronal cells play a pivotal role in sustaining the high energetic metabolism of these cells and are especially prone to oxidative damage. Since overproduction of reactive oxygen species (ROS) is involved in the pathogenesis of neurodegeneration, dietary antioxidants have been suggested to counteract the detrimental effects of ROS and to preserve the mitochondrial function, thus slowing the progression and limiting the extent of neuronal cell loss in neurodegenerative disorders. In addition to their role in the redox-system homeostasis, mitochondria are unique organelles in that they contain their own genome (mtDNA), which acts at the interface between environmental exposures and the molecular triggers of neurodegeneration. Indeed, it has been demonstrated that mtDNA (including both genetics and, from recent evidence, epigenetics) might play relevant roles in modulating the risk for neurodegenerative disorders. This mini-review describes the link between the mitochondrial genome and cellular oxidative status, with a particular focus on neurodegeneration; moreover, it provides an overview on potential beneficial effects of antioxidants in preserving mitochondrial functions through the protection of mtDNA.
1. Introduction
Neurodegenerative diseases are characterized by the progressive degeneration of neurons and synapses of the brain cortex and some subcortical regions in the central nervous system. This process can be induced by various neurotoxic events, such as excessive inflammation, ROS production, and mitochondrial dysfunctions, among many others.
Indeed, due to their limited glycolytic capacity and extremely metabolically active nature, neurons are energetically demanding cells [1]. Since the energetic balance in nucleated eukaryotic cells strongly depends on mitochondria, neuronal health and survival require a delicate maintenance of the mitochondrial function. Mitochondria are cytoplasmic organelles that play a central role in energy generation by oxidative phosphorylation (OXPHOS) that occurs in the mitochondrial cristae (inner membrane’s convolutions) through five protein complexes (I–V). During OXPHOS, these complexes work along with electron carriers as an electron transport chain, also known as respiratory chain, receiving and donating electrons to the next complex until reaching complex V [2]. However, mitochondria do not only produce cellular adenosine triphosphate (ATP), but also regulate programmed and unprogrammed cell death, Ca2+ hemostasis and, especially, generate endogenous reactive oxygen species (ROS), whose overproduction may cause further damage to mitochondrial components and promote disease development. Coherently, a large body of evidence suggests that mitochondrial dysfunctions are prominent early features of neurons affected by neurodegenerative diseases [3,4].
Like for many progressive and multifactorial pathologies, environmental exposures strongly modulate the risk for these diseases’ onset, even in presence of a genetic predisposition. Since mitochondria are exquisitely sensitive to environmental threats, they act at the interface between inner and outer cellular environment. Hence, given the pivotal role of the mitochondrial function and ROS overproduction in the pathogenesis of neurodegeneration [5], antioxidants have been suggested to counteract the detrimental effects of ROS in neurodegenerative disorders [6,7]. In particular, mitochondria-targeted antioxidant therapies are suggested as promising strategies to promote mitochondrial function and neuronal health [8,9,10].
Remarkably, mitochondria are unique organelles since they contain their own genome (mitochondrial DNA, mtDNA), which directly regulates mitochondrial functions. Indeed, it has been highlighted that mtDNA genetics but also epigenetics might play relevant roles in modulating the risk for neurodegenerative disease, acting at the interface between environmental exposures and the molecular triggers of neurodegeneration. This review describes the link existing between the mitochondrial genome and cellular oxidative status, with a particular focus on neurodegeneration. Moreover, it discusses the potential beneficial effects of antioxidants in preserving the mitochondrial function through the protection of mtDNA genetics and epigenetics.
2. Mitochondrial DNA: From Genetics to Epigenetics
The human mtDNA is a double-stranded, circular molecule of 16,569 bp, where one strand is purine rich (i.e., heavy strand) and the complementary strand is rich in pyrimidines (i.e., light strand) [11]. Only about 3% of the mtDNA is noncoding (containing two replication origins, OH and OL, on the heavy and light strands, respectively), and the coding sequences can be transcribed on both DNA strands in a polycistronic manner into a large mitochondrial mRNA. The 37 genes contained in the mtDNA encode for 13 polypeptides (all subunits of enzyme complexes of the oxidative phosphorylation system), 22 transfer RNAs (tRNAs), and 2 ribosomal RNAs (rRNAs), required for the translation of each subunit within the mitochondrial compartment [12]. MtDNA regulates several, but not all, mitochondrial functions. Indeed, the mtDNA undergoes an intense crosstalk with the nuclear DNA (nDNA), which also regulates numerous mitochondrial tasks [13,14].
It is of common knowledge that mtDNA is maternally inherited. In fact, despite some evidence of a biparental inheritance of mtDNA having been proposed [15], this hypothesis lacks definite confirmation [16,17,18,19].
Notably, mitochondria are highly dynamic organelles: they undergo regular fusion, fission, and transport, the processes of which determines mitochondrial location, morphology, and number [20]. These processes are linked to the mitochondrial genome [21], which can be replicated and translated, and thus regulate the number and the functionality of mitochondria.
Typically, multiple copies of the mtDNA populate each individual mitochondrion and hundreds to thousands of mtDNA copies may be present in a single cell (i.e. polyploidy). Due to the presence of multiple copies of the mtDNA per cell, different alleles or mutations (either inherited or sporadically occurred) can be present in only a fraction of the total pool of mtDNA in a given cell or organism (i.e., heteroplasmy) [22]. Different levels of heteroplasmy may exist between tissues and cells of the same individual, resulting in complex phenotypic manifestations [23].
The mtDNA has a high mutation rate, and numerous mutations have been linked to a range of diseases [24,25]. In parallel, several mutations of nuclear genes essential for mtDNA maintenance have been identified as cause of mtDNA point mutations, multiple deletions, or mtDNA depletion [26].
In recent decades, it has been suggested that not only mtDNA genetics but also epigenetics can regulate mitochondrial functions. This regulation is called mitoepigenetics [27,28]. Since the mtDNA lacks histones (it is packaged by the mitochondrial transcription factor A, TFAM, into nucleoprotein complexes known as nucleoids), the focus has been mainly centered on DNA methylation. However, post-translational modification of the TFAM and non-coding RNAs can also contribute to regulation of gene expression and mtDNA replication [29]. The existence of an epigenetic regulation of mtDNA and its role have been heavily debated, particularly with regards to DNA methylation. In mtDNA, methyl groups donated from S-adenosyl-methionine (SAM) can be added to cytosines (5mC, 5-methyl cytosine) or adenine (6mA, 6-methyl adenine) by DNA methyltransferases enzymes (e.g., DNA methyl-transferase 1, DNA methyl-transferase 3a, and DNA methyl-transferase 3b) which have been identified also within mitochondria [30,31], suggesting that methylation occurs in mtDNA in CpGs but also in non-CpG contexts [32]. Also ten-eleven translocation proteins (TET1 and TET2) have been detected in the mitochondria [33], indicating that mtDNA demethylation can occur both via passive mechanisms or active oxidation-mediated demethylation [29,34,35]. An area of the mtDNA that has been frequently investigated for changes in methylation is the three-stranded displacement (D-loop) structure, that is located in the noncoding control region and regulates transcription and replication of mtDNA. The human mitochondrial D-loop region has 1122 base pairs, and its methylation may have a direct role in controlling mtDNA replication [36] and, possibly, transcription [37]. Indeed, since mitochondrial DNA polymerase γ requires an RNA primer, mtDNA replication is tightly linked to transcription [38]. Indeed, the D-loop locus implicated in the processing of the RNA primer for mitochondrial replication was found to be methylated in human and murine peripheral blood [33,39], suggesting a role for mtDNA methylation in the control of mtDNA replication [40]. Indeed, D-loop methylation alterations have been associated with numerous pathological conditions, ranging from cancer to CVD, neurodegeneration, and others [36,39,41,42,43,44].
An additional level of gene regulation might be provided by TFAM acetylation/phosphorylation, which modulates compaction of mtDNA into nucleoids and is thought to be crucial for regulating mtDNA segregation and expression. In particular, it has been shown that acetylation and phosphorylation of TFAM can fine-tune TFAM–DNA binding affinity to permit the discrete regulation of mtDNA dynamics. In particular, phosphorylation of TFMA has been demonstrated to regulate transcription by altering the ability of TFAM to locate promoter sites [45].
Furthermore, noncoding RNAs can also act at the mitochondrial level [29]. Indeed, mature micro-RNAs (miRNAs) are commonly present in the cytosol of cells, and it has been widely established that they are stable also in biological fluids (characteristic that makes them attractive biomarkers). Various studies have revealed that miRNAs can act also within the mitochondria (mito-miRNAs). In particular, mitomiRs are implicated in inflammaging [46], influencing the energetic, oxidative, and inflammatory status of senescent cells. These mito-miRNAs include miR-130a-3p [47], let-7b [48], miR-146a-5p [49], miR-181c-5p [50], miR-133a, and miR-1 [51]. Moreover, it has been demonstrated that they are differentially expressed in several pathologies [52], ranging from cardiovascular [53] to neurodegenerative diseases [54]. Hence, they are supposed to play a role in regulating mitochondrial gene expression and functions in both physiological and pathological conditions. However, despite changes of mito-miRNAs’ expression having been widely demonstrated, mechanistic details on their mode of action in the mitochondria are not clearly established. Most of the mitomiRs are encoded by the nuclear genome and translocate to the mitochondria [55], while some others can be encoded directly by the mitochondrial genome [56].
Furthermore, given the proximity of mtDNA to the sites of oxidative phosphorylation at the inner membrane of the mitochondria, it is not surprising that mtDNA is especially prone to oxidized DNA damage. As a result, oxidative damage and loss of mtDNA integrity are increasingly recognized to play a role in the development of many chronic age-related diseases [57]. Not only the protection from oxidative damage, but also the regulation of mtDNA copy number (although apparently in excess) has been revealed to be an important aspect of mitochondrial biogenesis, essential for normal cellular functions [58,59].
Therefore, the control of mtDNA replication, transcription and repair in the face of constant onslaught from endogenous and environmental agents has a central role in health [60,61]. Indeed, it has been demonstrated that environmental exposures, contributing to the onset of new mutations or oxidative damages or modulating epigenetic features of mtDNA, strongly impact mitochondrial health. In the light of this evidence, mtDNA depletion and malfunction have been implicated in cancer [62,63], metabolic and cardiovascular diseases [43,64,65,66,67], aging [68], and many human multifactorial disorders, including neurodegeneration [69,70].
3. Mitochondria in Neurodegeneration: The Interplay between Oxidative Stress and the Mitochondrial Genome
Neuron cells are particularly vulnerable to oxidative damage because of several reasons. Their membranes are rich in polyunsaturated fatty acids (PUFAs), that can be easily oxidized. They also contain abundant redox-active proteins with metals (e.g., iron, copper, etc.) as prosthetic groups, which can play a role in ROS metabolism [71,72]. The high oxygen consumption typical of these cells increases the risk of oxidative neuronal damage in particular at mitochondrial level, where oxygen is reduced to water during the oxidative phosphorylation. Indeed, at rest, about 1% of the oxygen consumed is partially reduced to superoxide anion in mitochondria of adult cells, and then reduced by the antioxidant system [73]. Hence, any deficiency in the antioxidant defense system that reduces ROS levels makes the brain more vulnerable to oxidative stress [5].
The oxidative stress resulting from both increased ROS production and/or the inadequacy of the antioxidant system’s response can lead to protein, lipid, and DNA oxidation. Interaction with oxygen atoms can induce oxidation of DNA bases, in particular guanine, which has the lowest reduction potential of the four DNA bases [74]. Guanine interacts with hydroxyl radicals and generates one of the major products of DNA oxidation, such as 8-oxo-2′-deoxyguanosine (8-oxo-dG), whose levels are increased in neurodegenerative disorders [75]. PUFAs located in the phospholipid bilayers are easily oxidized by ROS into fatty acid radicals. These unstable fatty acid radicals interact with oxygen, thus producing fatty acid peroxyl radicals, which interact with other PUFAs and trigger the production of other fatty acid radicals and lipid hydroperoxides. This repeated cycle of lipid peroxidation strongly affects structure and functions of biological membranes [76]. Moreover, it is well-known that ROS can also damage proteins by oxidation of amino acids, especially cysteine and methionine, thus triggering protein modifications—such as protein fragmentation by oxidation of the protein backbone, protein–protein cross-linking, amino acid side chain modification, and enzyme inactivation—resulting in activity loss [77].
Since the mitochondria are the center of metabolic pathways (i.e., citric acid cycle, fatty acids metabolism, urea cycle, etc.), increased mitochondrial ROS production can perturb the activity of enzymes involved in these paths, affecting both the ATP production and the metabolic responses. Given the previously explained effects of ROS on biological molecules, mitochondrial membranes are also susceptible to ROS oxidation, especially at the level of proteins and unsaturated fatty acids [78]. In particular, oxidation at the unsaturated acyl chains modifies the intermolecular interactions, changing the membrane fluidity and indirectly affecting protein functions. In addition, oxidation induces the formation of weak bonds between functional groups of proteins and other oxidized molecules, hence decreasing their dynamicity. Moreover, ROS control the expression of highly conserved heat-shock proteins, and the increased mitochondrial ROS production can perturb the folding of proteins synthesized by mitochondria [79]. Finally, oxidation of cardiolipin‘s acyl chains at the mitochondrial inner membrane can cause respiratory complex destabilization [79,80]. Cardiolipin can interact with the respiratory chain complexes and contribute to generate the electrochemical gradient. Additionally, since it can interact with death-inducing proteins, it is involved in the apoptotic process and oxidation of cardiolipin compromises apoptosis, promoting neuronal dysfunctions and the progression of neurodegeneration (e.g., Alzheimer’s and Parkinson’s disease) [80,81]. Coherently with this picture, plenty of studies highlight the involvement of mitochondrial dysfunctions in neurodegeneration [3,82,83,84], supporting the relevance of protecting systems, including antioxidant defenses, to maintain neuronal integrity and survival [85].
At the interface between oxidative stress and the mitochondrial function there is also the mtDNA. Indeed, it has been demonstrated that inhibition of mitochondrial dynamics affects the elimination of mtDNA damage and the transmission of mtDNA mutations [86,87]. DNA damage is a well-established trigger of apoptotic cell death in mitotic cells as well as in terminally differentiated cells such as neurons [88]. However, DNA repair enzymes can prevent the accumulation of mutation, and this has been well described in the nuclear DNA. Differently, mechanisms of damage and repair in the mitochondrial genome are poorly understood, despite the fact that mtDNA is subjected to higher levels of oxidative stress than is nuclear DNA [89,90,91]. This particular sensitivity to oxidative agents is likely due, as previously mentioned, to its proximity to the inner mitochondrial membrane, where oxidants are formed, but also to the lack of protective histones which lower the frequency of DNA strand breaks by scavenging the hydroxyl radicals [92,93]. Indeed, it has been proven that impairments in mitochondrial respiration and oxidative phosphorylation elicit an increase of mtDNA rearrangements and mutations [94], which has been linked to neurodegeneration [95,96,97]. On the other hand, it has been also demonstrated that mutations in the mtDNA exacerbate ROS production, contributing to disease onset and progression [98]. Moreover, alterations of the mitochondrial genome in neurodegeneration have been demonstrated also in terms of variation of copy number [99], that significantly varies in neurodegenerative diseases in respect to healthy controls, and has been proposed as an innovative biomarker [100,101,102,103]. This evidence supports the hypothesis that the mitochondrial genome significantly influences both pathological (e.g., neurodegeneration) and physiological processes (e.g., aging) [104].
Recently, in particular for neurodegenerative disorders, the new field of mitoepigenetics is grabbing a growing attention [6,40,42,105]. The role of epigenetic modifications in neurodegeneration has been widely discussed and recognized [106,107,108,109,110] both in animal models and humans [108,111,112,113,114], in particular in the case of early-life exposure to xenobiotics [110,115,116]. Nuclear epigenetics can drive the expression of genes involved in mitochondrial homeostasis, and, conversely, metabolic alterations occurring in mitochondria may affect the availability of substrates for epigenetic enzymes [117], thus leading to alterations of epigenetic signatures in the nuclear genome [13,117]. Currently, a large body of evidence supports the existence of a direct epigenetic regulation of mtDNA, though the mechanistic effect of epigenetic marks on gene expression in mitochondria is still debated [37,117,118,119]. Remarkably, impaired methylation levels of the D-loop region in animal models, postmortem brain regions, or circulating blood cells of patients with Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis, suggested that alterations of epigenetic regulations of mtDNA may mediate mitochondrial dysfunctions that are typical of neurodegenerative disorders [40]. Furthermore, the possibility that mtDNA methylation could act as a mediator between environmental exposure and the mitochondrial genome [120] might provide further insights on the complex environmentally linked etiology of neurodegenerative disorders. Finally, since epigenetic therapies have been proposed for treating neurodegenerative disorders [121,122], elucidating the effect of epigenetic regulations on the mitochondrial DNA might provide additional information in view of the future application of epigenetic therapies in clinical practice.
4. Antioxidants, Gene Expression, and Epigenetic Regulations: The Role of Nutriepigenomics in Neurodegenerative Diseases
Appropriate nutrition is an essential and modifiable factor that plays a key role in preventing and/or delaying the onset of neurodegeneration. Reducing risk factors linked to the lifestyle (i.e., smoking of cigarettes, limiting stress, avoiding toxins, promoting mental and physical training) but also preventing hypertension, type 2 diabetes, insulin resistance and obesity, increase of homocysteine, are important components of neurodegenerative disease prevention. For these reasons, the early protection from neurodegeneration starts with the choice of appropriate types and quality of food [110,123,124,125]. Indeed, positive clinical outcomes exerted by antioxidants supplementation. For example, vitamins C [126,127], E [128] and N-acetylcysteine [129] have been demonstrated active in oxidative-stress related pathologies [130,131]. A large body of evidence shows that many dietary antioxidants can help to delay their onset and progression [6,132,133,134], in particular when applied at the early-stages of the disease, and the application of dietary antioxidants for the prevention of neurodegeneration has also been proposed [71].
From the molecular point of view, numerous pathways are triggered by an appropriate nutrition, and a clear role of nutrigenomics in promoting neuronal health has been demonstrated [135]. Several vegetable foods rich in polyphenols (e.g., resveratrol [136], carotenoids [137], quercetin [138,139]) and other bioactive compounds (e.g., sulforaphane [140,141,142], melatonin [143], docosahexanoic acid [144], etc.), have been shown to be efficacious in preventing neurodegeneration by regulating gene expression and epigenetic regulation of anti-inflammatory and antioxidant pathways [110,135,145]. Studies on experimental models of neurodegeneration demonstrate that early treatment with antioxidants (i.e., coenzyme Q10, vitamin E, vitamin C) can prevent the progressive neuronal damage by reducing neuroinflammation, which is associated with increased production of proinflammatory cytokines (i.e., TNF-a, Rantes). These antioxidants counterbalance the progression of oxidative damage at protein, lipid and DNA level, as well as counteracting the decrease of glutathione (GSH) in the striatum [133,146]. These antioxidants also modulate the expression level of the pro-inflammatory nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB), as well as of the anti-inflammatory nuclear factor erythroid 2-related factor 2 (Nr-f2) [110]. In vitro studies on dopaminergic cell line show that GSH and tocotrienol can actively prevent oxidative neuronal damage [133,147]. Sulforaphane, contained in broccoli, modulates the antioxidant and anti-inflammatory responses by modulating the nuclear erythroid 2-related factor 2/ antioxidant response element (Nrf2/ARE) pathway, inhibiting NFkB and promoting cell detoxification by stimulation of phase II enzymes, that increase xenobiotics hydrophilicity and enhance their consequent excretion [142]. Besides, sulforaphane reduces the β-amyloid (Aβ) protein burden by upregulation of p75 neurotrophin receptor and histone deacetylases’ (HDAC1 and HDAC3) expression in an Alzheimer’s disease animal model [148]. Resveratrol is able to prevent microglia activation in Alzheimer’s disease due to its anti-inflammatory activity. It increases anti-inflammatory cytokine release and upregulates sirtuin 1 (SIRT1) expression which, due to its deacetylase activity, exerts a neuroprotective action [149,150]. In addition, resveratrol activates 5′-adenosine monophosphate-activated protein kinase (AMPK)-dependent signaling saving Aβ-mediated neurotoxicity in human neural stem cells [151]. Curcumin can cross the blood–brain barrier where it exerts a protective role against neurodegeneration in both Parkinson’s and Alzheimer’s diseases. In vitro and in vivo study show that curcumin has antioxidant and anti-inflammatory properties, together with the capacity to limit protein aggregation (i.e., a-synuclein, amyloid beta peptide, hyperphosphorylated tau) [152].
Moreover, an anti-inflammatory and antioxidant diet (especially during the early-life but also later) represents a central strategy for modulating brain plasticity and building an “epigenetic memory” that might promote neuronal resilience against stressors and prevent neurodegenerative phenomena [110,135]. Indeed, oxidative stress can also directly and indirectly influence the cellular epigenetic homeostasis [153,154]. Both DNA oxidation and TET-mediated hydroxymethylation affect the methylome [155]. ROS can directly convert 5mc to 5-hydroxymethylcytosine (5hmC) [156], that affects DNA methyl-transferase 1 (DNMT1) activity, thus leading to an improper methylation inheritance during mitosis and global hypomethylation [157]. Furthermore, they can modulate the methylome through the oxidization of guanosine to 8-oxo-20-deoxyguanosine (8-oxodG) thus inhibiting methylation of adjacent cytosine and further contributing to global hypomethylation of DNA [158,159]. At the same time, intracellular levels of metabolites such as acetyl-coenzyme A (acetyl-CoA), Fe, ketoglutarate, NAD+, and S-adenosylmethionine (SAM), which are essential for the epigenetic machinery (in particular for the activity of histone-modifying enzymes), depends on the global cellular metabolism and energy levels of the cell, thus they are strictly linked to the cellular oxidative status [160,161,162]. A tight interplay exists also between oxidative stress and miRNAs expression levels, since ROS affect miRNAs expression [163] and, conversely, miRNAs regulate the expression of numerous genes involved in an oxidative stress response [164,165]. Therefore, oxidative stress can globally influence the cell on multiple levels, including its epigenetic landscape [166], and can contribute to the pathogenesis of neurodegenerative disorders [111,167,168]. Given the crucial role of nutrition in the maintenance of the epigenetic regulation [169], and given the effects of dietary antioxidant in the modulation of the redox environment in the cell, an antioxidant and epigenetic diet contributes to preservation of cellular homeostasis [169] in both health and disease [170].
The existence of a link between nutrition and epigenetic reprogramming is well known [169]. Dietary intake of folate, vitamins B12, B2, B6, choline, betaine, and methionine impact the one carbon cycle and the cellular bioavailability of S-adenosylmethionine, which is the universal methyl-donor [169]. In addition, the mitochondrion, via its nucleo-cytoplasmic pools, is a major regulator of cellular levels of acetyl-CoA, the main acetyl donor [171]. Moreover, bioactive molecules contained in food [172], total caloric intake and specific dietary regimens [173] can also modulate the epigenome. Indeed, nutrition can affect the epigenome through at least two general mechanisms: either directly, by providing molecules which interact with the enzymes responsible for ‘writing’ or ‘erasing’ the epigenetic profiles, or indirectly through metabolic rewiring. Remarkably, both mechanisms converge on mitochondria, which produce a plethora of factors and substrates essential for epigenetic modifications and are at a crossroads of cellular energy metabolism [174].
Evidence about correlations between dietary antioxidants and epigenetic regulations in the nDNA and the associated molecular mechanisms has been recently and comprehensively reviewed by Beetch et al. [175]. It has been observed that compounds that decrease oxidative stress consequently impact DNA methylation landscapes. Despite most of evidence coming from in vitro or animal studies, vitamin C [176,177,178], A [179,180,181], and E [182], selenium [183], stilbenoids [184,185], epigallocatechin gallate [186], genistein [187], curcumin [188], and sulphoraphane [189,190] are all molecules that have been shown to mediate both epigenetic regulations and, as previously described, oxidative stress, demonstrating that these two pathways in the cell are strictly related [175]. Also curcumin has nutriepigenomics properties because it can inhibit DNA methyltransferases and regulate chromatin condensation by modulating histone acetyltransferases and histone deacetylases [191].
Summarizing, several dietary components and bioactive molecules can be used to contrast neurodegeneration due to their capacity to modulate oxidative stress, gene expression, and epigenetics regulations. Their employment has to be regular and in the first stages of neuronal damage.
5. Antioxidants and Mitochondria: From the General Function to the Mitochondrial Genomics
For a long time, a wide consensus has been given to the mitochondrial free radical theory of aging [192], which asserts that, by damaging mitochondria, external ROS can promote mtDNA damages and increase the endogenous production of free radicals, promoting mitochondrial dysfunctions. Thus, the usage of antioxidants and bioactive compounds that might act at the mitochondrial level has been proposed also in neurodegeneration to overcome excessive ROS production. For example, the manganese superoxide dismutase (MnSOD), encoded by genomic DNA, protects mitochondria from oxidative stress. This enzyme, located in the mitochondrial matrix, catalyzes the dismutation of superoxide anion, generated during the monoelectronic reduction of oxygen to water, into hydrogen peroxide. Clinical findings show that, in the early stage of neurodegeneration, a compensatory upregulation of MnSOD can be observed [193,194,195,196]. Oxidative stress upregulates MnSOD and its enzymatic activity can be modulated by several bioactive compounds (e.g., melatonin, dimethyl fumarate, etc.) and dietary antioxidants (e.g., tea catechin, curcumin, etc.). Melatonin promotes neuroprotection of Purkinje cells by MnSOD upregulation [197], as has been demonstrated for dimethyl fumarate in SH-SY5Y human neuroblastoma cell lines [198]. Longjing green tea catechin extract significantly upregulates MnSOD and increases its enzymatic activity in Drosophila melanogaster [199]. Curcumin can reduce mitochondrial superoxide anion accumulation and downregulate MnSOD in HT22 cells [200,201]. Pomegranate-derived polyphenols exert anti-inflammatory, antioxidant, and antimicrobial activity [202]. The pomegraniin A contained in the fruit extract reduces ROS by MnSOD activation. An increase of ROS production stimulates mitochondrial deacetylase sirtuin 3 (SIRT3) leading to MnSOD activation [203,204]. Quercetin supplementation prevents the aluminum-induced neurodegeneration in animal model by increasing MnSOD activity and glutathione levels and reducing lipid and protein oxidation [205]. Summarizing, the upregulation of MnSOD in the mitochondria exerted by antioxidants can help to counterbalance the disequilibrium in the redox homeostasis, preventing the ROS-induced neuronal damage and the progression of the neuronal disease.
Many of the previously mentioned bioactive molecules, which have both antioxidant and nutrigenomic effects, act at the interface with the mitochondrion [174]. Dietary supplements that can help to maintain mitochondrial homeostasis include, among others, coenzyme Q10, vitamins C, E, K1 and B, L-carnitine, and other mitochondrion-targeted antioxidants such as N-acetylcysteine, sodium pyruvate, and lipoic acid [206]. Also, flavonoids such as ampelopsin and pinocembrin can inhibit mitochondrial dysfunction and neuronal death through the regulation of gene expression of Nrf2 pathway. Moreover, supplementation with resveratrol also increases mitochondrial mass. It mediates the SIRT1-dependent deacetylation of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), thus resulting in stimulation of the mitochondria biogenesis, which in turn co-activates the nuclear respiratory factors (NRF-1 and NRF-2), that also promote the mitochondrial biogenesis [207,208,209]. Despite a lack of consensus about which is the general mechanism of action that could explain these effects induced by the resveratrol on mitochondria, some hints suggest the involvement of mitochondrial complex III [209,210]. Similarly, a role for mitochondria on the anti-inflammatory effects of curcumin has been also hypothesized, since a homozygous deletion of the mitochondrial uncoupling protein 2 is able to reverse the beneficial effects of this molecule in mice [211]. In addition, genistein can modulate the enzymatic activity of components of the oxidative phosphorylation system [174]; isoflavones can stimulate mitochondrial biogenesis and improves mitochondrial function in diabetes, as well as in aging of rodents [212,213]; sulphoraphane can enhance ROS and mitochondrial membrane depolarization in human ovarian cancer cell lines [214]. Interestingly, caloric restriction has been shown to exert many healthy effects (including neurodegeneration prevention [215,216]), consistent with reduced ROS generation by mitochondria, and many antioxidant compounds can also mimic such effects. For examples, N-acetyl cysteine (NAC) provides thiol groups to glutathione and to mitochondrial respiratory chain proteins. Thus, it may counteract both ROS generation and effects [217,218]. Of note, it has been widely demonstrated that the beneficial effects of caloric restriction are also mediated by epigenetic mechanisms [219,220], that, as previously described, are tightly linked with the oxidative status of the cell.
However, it must be also considered that ROS are generated by numerous biochemical reactions in the cell and have not only detrimental but also physiological roles. The general antioxidants do not have a selective action, and these substances are often used at quite high concentrations in order to obtain a therapeutic effect. Thus, mitochondria-targeted antioxidants has been proposed as a strategy to boost mitochondrial biogenesis and organelle bioenergetics, limiting mitochondrial ROS production and oxidative damage, without suppressing ROS that are important for cell signaling [221,222,223]. Promising results has been obtained on mitochondrial biogenesis and functions with this kind of specific antioxidants, including improving insulin sensitivity, muscle contractile function, and sarcopenia [224]. In particular, one of the most promising mitochondria-targeted antioxidant is the mitoquinone, or Mito-Q. Mito-Q consists of a quinone moiety linked to a triphenylphosphonium (TPP) moiety by a 10-carbon alkyl chain [225]. It is several-hundred-fold more potent than untargeted antioxidants in blocking ROS and preventing mitochondrial oxidative damage [226]. It is orally active, can cross mammalian membranes and has been revealed particular effective against lipid peroxidation [227]. Remarkably, it displayed a neuroprotective effect that make it a potential candidate against neuroinflammatory diseases, such as neurodegenerative disorders [228]. Additionally, it has been demonstrated to be effective also in the treatment of peripheral neuropathy in animal models of diet-induced obesity, increasing the possibility of its application in clinical practice [229].
Furthermore, taking into account the physiological role of ROS production in the cell, it has been recently suggested that mtDNA mutations may be generated by replication errors rather than by accumulated oxidative damage, so that mitochondrial dysfunction due to mtDNA alteration may rather be the major driver of the aging process [98,230]. This theory overturns the free-radical theory of aging, suggesting that the increase in ROS might be a consequence rather than a cause of aging and that ROS are mediators of the stress response to age-dependent damage [230,231]. This vision strengthens the hypothesis of a pivotal role of mitochondrial genome in driving pathological and physiological processes and might be reinforced also by the new discoveries on mitochondrial epigenetics. The interaction between mtDNA and oxidative stress have been proved also by demonstrating that increased mitochondrial oxidative stress derived from MnSOD mutant mice (Sod2−/+) promoted mtDNA glycation [232] and a 30% increase in the 8-OH deoxyguanosine levels of mtDNA [233], suggesting that aberrant regulations of this enzyme can affect also the mitochondrial DNA. On the other hand, unexpected evidence has been collected about absence of correlation between MnSOD activity and mtDNA mutation rate, that did not vary significantly in loss of function mutants in respect to controls [234,235].
Several bioactive molecules can act through interaction with the mtDNA. For example, CoQ10 diet is able to preserve mtDNA content and Tfam/OXPHOS complex protein expression in a mouse model of glaucoma [236]. In a case report, CoQ10 deficiency has been associated with a mitochondrial DNA depletion syndrome in a human case [237]. Remarkably, the mitochondrially-targeted Mito-Q has been demonstrated to protect the intestinal barrier by ameliorating mitochondrial DNA damage via the Nrf2/ARE signaling pathway [238]. Resveratrol can restore the suppressed SIRT1 expression and mtDNA expression in a cellular model of ovarian hypoxia [239]. Flavanones (in particular, naringin) are able to modulate mtDNA copy number in human leukocytes and has been identified as the key factor behind the association between fruit consumption and mtDNA copy number measured in a population based study [240]. Genistein alleviates the mitochondria-targeted DNA damage induced by β-amyloid peptides 25–35 in C6 glioma cells [241]. In particular, levels of 8-OHdG and mtDNA deletion decreased after a pre-treatement with genistein. Curcumin is able to suppress gastric tumor cell growth via decreased mtDNA content and ROS-mediated DNA polymerase γ depletion, thus reducing mitochondrial oxygen consumption and aerobic glycolysis [242]. In a rat model of myocardial ischemia reperfusion, EGCG may mediated cardioprotective effects also by inhibiting the release of mtDNA from damaged mitochondria, which have been proven to be a potent pro-inflammatory mediator [243]. Furthermore, EGCG may exert anti-obesity properties also by improving thermogenesis and mitochondria biogenesis in the brown adipose tissue (BAT). Indeed, EGCG-fed mice displayed higher body temperature and mtDNA content in BAT [244]. Of note, not only dietary antioxidants but other nutrients [245] or dietary regimen [246,247], as well as alteration of body composition [65], has shown association with mtDNA copy number, corroborating the hypothesis that diet and health status are strictly linked with the mtDNA function. Conversely, it has been recently demonstrated that alteration of mitochondrial DNA content can modulate antioxidant enzyme expressions and, consequentially, oxidative stress in the cell [248].
Mitochondria are also important for epigenetic regulations since these are strictly linked to the bioenergetic system, which provides the interface between the environment and the epigenome by converting environmental calories into ATP, acetyl-CoA, SAM, and reduced nicotinamide adenine dinucleotide (NAD+). Consistently, the clinical phenotypes of bioenergetic diseases are quite similar to those observed in epigenetic diseases (e.g., Rett, Angelman, Fragile X Syndromes, etc.), and an increasing number of epigenetic diseases are being associated with mitochondrial dysfunction [249]. Thus, the study of this bioenergetic–mitochondria–epigenomic network has broad implications for identifying the etiology, pathophysiology, and treatment of a wide range of common diseases.
While the correlation between nutrition, oxidative stress, and the nuclear epigenome is well described, to our knowledge, there are no molecular data on the effects of general antioxidants or mitochondria-targeted antioxidants on mtDNA epigenetics as yet. However, a certain responsiveness of the mitochondrial epigenome to external stimuli, including but not limited to dietary elements, has been postulated [65,67,120]. Remarkably, Corsi and colleagues recently demonstrated that mtDNA methylation can change accordingly to the adherence to the Mediterranean diet [250], corroborating the hypothesis of a potential role of nutrition in the modulation of mtDNA methylation.
In conclusion, although potential applications of this research field are wide, it must be considered that the body of literature investigating molecular effects of bioactive compounds and dietary antioxidants on the mtDNA are still mainly based on in vitro or in vivo studies. Few data have been collected in population-based studies involving dietary supplementations. For this reason, randomized clinical trials or well-controlled observational studies that could confirm or rather reject the evidence from current basic research are necessary before considering applications in clinics. In particular, only a limited number of studies have investigated mitoepigenetic changes in humans or animal models of neurodegenerative disorders so far. Hence, despite encouraging preliminary data, further molecular and functional studies are required to elucidate the impact of mtDNA methylation on the mitochondrial function in neurodegenerative diseases.
Moreover, concerning the field of mitochondrial epigenetics, the existence of this additional level of regulation on mtDNA is still questioned by some authors [251,252]. There are also discrepancies about where methylation occurs in mtDNA [32,37] and what methods and technologies should be used to study this mark [35,253]. It is still to be clarified, above all, what are the mechanistic aspects through which alterations of the mtDNA epigenome can lead to mitochondrial dysfunctions. Indeed, the mechanism through which mtDNA methylation is regulated and regulates mtDNA replication and transcription is under investigation [32,37,119,253,254]. Since there are still many aspects to be clarified, mitochondrial epigenetics represents a growing but also discussed research area that needs further confirmation to be definitely established and translated into clinical practice.
Finally, mechanistic aspects of how dietary antioxidants and bioactive molecules interact with the mitochondrial DNA (directly or indirectly either) are outstanding points toward which future research should be directed. Additional data are necessary to put the basis for the identification of specific mitochondrially-targeted DNA that could preserve mtDNA genetics and epigenetics and could be applied in neurodegenerative disorders prevention or treatment (Figure 1).
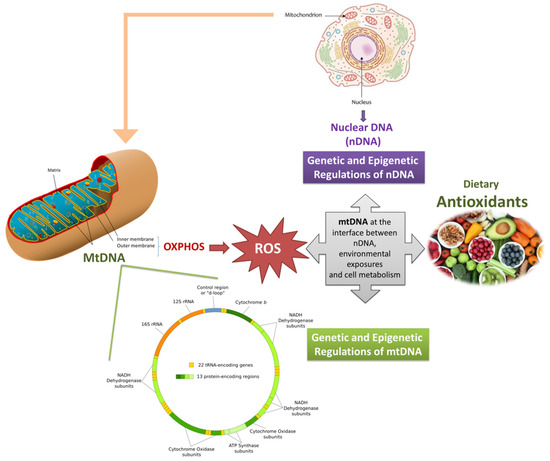
Figure 1.
Graphical representation of the interplay between mitochondrial genome (and epigenome), dietary antioxidants, and mitochondrial health. Mitochondria produces energy but also ROS, whose overproduction might induce mitochondrial dysfunctions, also through the alteration of the mitochondrial genome. Dietary antioxidants, can promote the mitochondrial function by reducing ROS production, modulating epigenetic pathways and, thus, restoring cellular homeostasis. Since mitochondrial dysfunctions (in particular those triggered by ROS) are typical of neurodegeneration, the role of dietary antioxidants in preventing these multifactorial diseases can be central, and it might pass through the protection of genetic and epigenetic features of the mitochondrial DNA.
6. Conclusions
Mitochondrial dynamics have been increasingly recognized as a common and early feature likely responsible for neuronal dysfunction in a wide range of neurodegenerative diseases. They strongly depend on ROS production, mtDNA genetics and epigenetics, which act at the interface with environmental exposures. Given the complex interactions between oxidative stress, epigenetics, and mitochondria, it can be postulated that both adequate nutrition [255,256,257] and physical activity [258,259,260] might positively impact the mitochondrial function, and that the effects might be mediated by molecular mechanisms that also include the intervention of epigenetic pathways [169,261].
Currently, therapeutic approaches for neurodegenerative diseases are limited and the prevention or alleviation of these multifactorial pathologies is a challenge for contemporary health services. Indeed, early detection and effective protection against neurodegeneration are key factors for a clinical success and healthy aging. Given the relevance of mitochondrial function on neurodegeneration, an epigenetic diet might have both preventive and therapeutic effects, acting at different levels, including the mitochondrial genome. Since the efficiency and number of mitochondria can be manipulated by pharmacological and dietary treatments, it has been suggested that mitochondria may be a potential target for preventive medicine by tailoring personalized dietary approaches and supplementations [174]. Moreover, the emerging usage of mtDNA methylation and copy number as biomarkers [64,65,262,263] emphasizes the relevance of studies on mtDNA genomics and epigenomics, and reinforces the need for further investigations of this aspect, particularly with a focus on neurodegenerative disorders. Further research aimed to test the effects of dietary antioxidants on the mitochondrial genome are warranted.
Author Contributions
Conceptualization, L.B.; Writing—original draft preparation, L.B. and R.G.; Writing—review and editing, L.B. and R.G.; Funding acquisition, R.G. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by FPA000033 to R.G.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kann, O.; Kovács, R. Mitochondria and neuronal activity. Am. J. Physiol. Cell Physiol. 2007, 292, C641–C657. [Google Scholar] [CrossRef] [PubMed]
- Burke, P.J. Mitochondria, Bioenergetics and Apoptosis in Cancer. Trends Cancer 2017, 3, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, L.; Liu, J.; Xie, F.; Su, B.; Wang, X. Abnormalities of Mitochondrial Dynamics in Neurodegenerative Diseases. Antioxidants 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, T.; Ziegler, A.C.; Dimitrion, P.; Zuo, L. Oxidative Stress in Neurodegenerative Diseases: From Molecular Mechanisms to Clinical Applications. Oxid. Med. Cell. Longev. 2017, 2017, 2525967. [Google Scholar] [CrossRef]
- Moosmann, B.; Behl, C. Antioxidants as treatment for neurodegenerative disorders. Expert Opin. Investig. Drugs 2002, 11, 1407–1435. [Google Scholar] [CrossRef]
- Albarracin, S.L.; Stab, B.; Casas, Z.; Sutachan, J.J.; Samudio, I.; Gonzalez, J.; Gonzalo, L.; Capani, F.; Morales, L.; Barreto, G.E. Effects of natural antioxidants in neurodegenerative disease. Nutr. Neurosci. 2012, 15, 1–9. [Google Scholar] [CrossRef]
- Dhanasekaran, A.; Kotamraju, S.; Kalivendi, S.V.; Matsunaga, T.; Shang, T.; Keszler, A.; Joseph, J.; Kalyanaraman, B. Supplementation of endothelial cells with mitochondria-targeted antioxidants inhibit peroxide-induced mitochondrial iron uptake, oxidative damage, and apoptosis. J. Biol. Chem. 2004, 279, 37575–37587. [Google Scholar] [CrossRef]
- Jauslin, M.L.; Meier, T.; Smith, R.A.J.; Murphy, M.P. Mitochondria-targeted antioxidants protect Friedreich Ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J. 2003, 17, 1972–1974. [Google Scholar] [CrossRef]
- Kelso, G.F.; Porteous, C.M.; Coulter, C.V.; Hughes, G.; Porteous, W.K.; Ledgerwood, E.C.; Smith, R.A.; Murphy, M.P. Selective targeting of a redox-active ubiquinone to mitochondria within cells: Antioxidant and antiapoptotic properties. J. Biol. Chem. 2001, 276, 4588–4596. [Google Scholar] [CrossRef]
- Asin-Cayuela, J.; Gustafsson, C.M. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007, 32, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.-W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Cagin, U.; Enriquez, J.A. The complex crosstalk between mitochondria and the nucleus: What goes in between? Int. J. Biochem. Cell Biol. 2015, 63, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Wiese, M.; Bannister, A.J. Two genomes, one cell: Mitochondrial-nuclear coordination via epigenetic pathways. Mol. Metab. 2020, 38, 100942. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental Inheritance of Mitochondrial DNA in Humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef] [PubMed]
- Lutz-Bonengel, S.; Parson, W. No further evidence for paternal leakage of mitochondrial DNA in humans yet. Proc. Natl. Acad. Sci. USA 2019, 116, 1821–1822. [Google Scholar] [CrossRef]
- Annis, S.; Fleischmann, Z.; Khrapko, M.; Franco, M.; Wasko, K.; Woods, D.; Kunz, W.S.; Ellis, P.; Khrapko, K. Quasi-Mendelian paternal inheritance of mitochondrial DNA: A notorious artifact, or anticipated behavior? Proc. Natl. Acad. Sci. USA 2019, 116, 14797–14798. [Google Scholar] [CrossRef]
- Vissing, J. Paternal comeback in mitochondrial DNA inheritance. Proc. Natl. Acad. Sci. USA 2019, 116, 1475–1476. [Google Scholar] [CrossRef]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Reply to Lutz-Bonengel et al.: Biparental mtDNA transmission is unlikely to be the result of nuclear mitochondrial DNA segments. Proc. Natl. Acad. Sci. USA 2019, 116, 1823–1824. [Google Scholar] [CrossRef]
- Ferree, A.; Shirihai, O. Mitochondrial dynamics: The intersection of form and function. Adv. Exp. Med. Biol. 2012, 748, 13–40. [Google Scholar] [CrossRef]
- Ishihara, T.; Ban-Ishihara, R.; Maeda, M.; Matsunaga, Y.; Ichimura, A.; Kyogoku, S.; Aoki, H.; Katada, S.; Nakada, K.; Nomura, M.; et al. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol. Cell. Biol. 2015, 35, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.F. Mitochondrial Genetics. Adv. Exp. Med. Biol. 2019, 1158, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Aryaman, J.; Johnston, I.G.; Jones, N.S. Mitochondrial Heterogeneity. Front. Genet. 2018, 9, 718. [Google Scholar] [CrossRef] [PubMed]
- Greaves, L.C.; Reeve, A.K.; Taylor, R.W.; Turnbull, D.M. Mitochondrial DNA and disease. J. Pathol. 2012, 226, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Greaves, L.C.; Turnbull, D.M. Mitochondrial DNA mutations and ageing. Biochim. Biophys. Acta 2009, 1790, 1015–1020. [Google Scholar] [CrossRef]
- Rusecka, J.; Kaliszewska, M.; Bartnik, E.; Tońska, K. Nuclear genes involved in mitochondrial diseases caused by instability of mitochondrial DNA. J. Appl. Genet. 2018, 59, 43–57. [Google Scholar] [CrossRef]
- Ghosh, S.; Singh, K.K.; Sengupta, S.; Scaria, V. Mitoepigenetics: The different shades of grey. Mitochondrion 2015, 25, 60–66. [Google Scholar] [CrossRef]
- Manev, H.; Dzitoyeva, S. Progress in mitochondrial epigenetics. Biomol. Concepts 2013, 4, 381–389. [Google Scholar] [CrossRef]
- Sharma, N.; Pasala, M.S.; Prakash, A. Mitochondrial DNA: Epigenetics and environment. Environ. Mol. Mutagen. 2019. [Google Scholar] [CrossRef]
- Shock, L.S.; Thakkar, P.V.; Peterson, E.J.; Moran, R.G.; Taylor, S.M. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 2011, 108, 3630–3635. [Google Scholar] [CrossRef]
- Wong, M.; Gertz, B.; Chestnut, B.A.; Martin, L.J. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front. Cell. Neurosci. 2013, 7, 279. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; Cuenin, C.; Chung, F.; Aguilera, J.R.R.; Fernandez-Jimenez, N.; Romero-Garmendia, I.; Bilbao, J.R.; Cahais, V.; Rothwell, J.; Herceg, Z. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 2019, 47, 10072–10085. [Google Scholar] [CrossRef] [PubMed]
- Bellizzi, D.; D’Aquila, P.; Scafone, T.; Giordano, M.; Riso, V.; Riccio, A.; Passarino, G. The Control Region of Mitochondrial DNA Shows an Unusual CpG and Non-CpG Methylation Pattern. DNA Res. 2013, 20, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Castegna, A.; Iacobazzi, V.; Infantino, V. The mitochondrial side of epigenetics. Physiol. Genomics 2015, 47, 299–307. [Google Scholar] [CrossRef]
- Liu, B.; Du, Q.; Chen, L.; Fu, G.; Li, S.; Fu, L.; Zhang, X.; Ma, C.; Bin, C. CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 2016, 6, 23421. [Google Scholar] [CrossRef]
- Gao, J.; Wen, S.; Zhou, H.; Feng, S. De-methylation of displacement loop of mitochondrial DNA is associated with increased mitochondrial copy number and nicotinamide adenine dinucleotide subunit 2 expression in colorectal cancer. Mol. Med. Rep. 2015, 12, 7033–7038. [Google Scholar] [CrossRef]
- Van der Wijst, M.G.P.; van Tilburg, A.Y.; Ruiters, M.H.J.; Rots, M.G. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 2017, 7, 177. [Google Scholar] [CrossRef]
- Kasiviswanathan, R.; Collins, T.R.L.; Copeland, W.C. The interface of transcription and DNA replication in the mitochondria. Biochim. Biophys. Acta 2012, 1819, 970–978. [Google Scholar] [CrossRef][Green Version]
- Ferreira, A.; Serafim, T.L.; Sardao, V.A.; Cunha-Oliveira, T. Role of mtDNA-related mitoepigenetic phenomena in cancer. Eur. J. Clin. Investig. 2015, 45 (Suppl. 1), 44–49. [Google Scholar] [CrossRef]
- Blanch, M.; Mosquera, J.L.; Ansoleaga, B.; Ferrer, I.; Barrachina, M. Altered Mitochondrial DNA Methylation Pattern in Alzheimer Disease-Related Pathology and in Parkinson Disease. Am. J. Pathol. 2016, 186, 385–397. [Google Scholar] [CrossRef]
- Mishra, M.; Kowluru, R.A. Epigenetic Modification of Mitochondrial DNA in the Development of Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5133–5142. [Google Scholar] [CrossRef] [PubMed]
- Stoccoro, A.; Siciliano, G.; Migliore, L.; Coppede, F. Decreased Methylation of the Mitochondrial D-Loop Region in Late-Onset Alzheimer’s Disease. J. Alzheimers Dis. 2017, 59, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.A.; Byun, H.-M. Platelet mitochondrial DNA methylation: A potential new marker of cardiovascular disease. Clin. Epigenet. 2015, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Stoccoro, A.; Mosca, L.; Carnicelli, V.; Cavallari, U.; Lunetta, C.; Marocchi, A.; Migliore, L.; Coppedè, F. Mitochondrial DNA copy number and D-loop region methylation in carriers of amyotrophic lateral sclerosis gene mutations. Epigenomics 2018, 10, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- King, G.A.; Hashemi Shabestari, M.; Taris, K.-K.H.; Pandey, A.K.; Venkatesh, S.; Thilagavathi, J.; Singh, K.; Krishna Koppisetti, R.; Temiakov, D.; Roos, W.H.; et al. Acetylation and phosphorylation of human TFAM regulate TFAM–DNA interactions via contrasting mechanisms. Nucleic Acids Res. 2018, 46, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, A.; Prattichizzo, F.; Micolucci, L.; Ceriello, A.; Procopio, A.D.; Rippo, M.R. Mitochondrial (Dys) Function in Inflammaging: Do MitomiRs Influence the Energetic, Oxidative, and Inflammatory Status of Senescent Cells? Mediat. Inflamm. 2017, 2017, 2309034. [Google Scholar] [CrossRef]
- Su, S.; Zhao, Q.; He, C.; Huang, D.; Liu, J.; Chen, F.; Chen, J.; Liao, J.-Y.; Cui, X.; Zeng, Y.; et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun. 2015, 6, 8523. [Google Scholar] [CrossRef]
- Teng, G.; Wang, W.; Dai, Y.; Wang, S.; Chu, Y.; Li, J. Let-7b Is Involved in the Inflammation and Immune Responses Associated with Helicobacter pylori Infection by Targeting Toll-Like Receptor 4. PLoS ONE 2013, 8, e56709. [Google Scholar] [CrossRef]
- Olivieri, F.; Lazzarini, R.; Babini, L.; Prattichizzo, F.; Rippo, M.R.; Tiano, L.; Di Nuzzo, S.; Graciotti, L.; Festa, R.; Brugè, F.; et al. Anti-inflammatory effect of ubiquinol-10 on young and senescent endothelial cells via miR-146a modulation. Free Radic. Biol. Med. 2013, 63, 410–420. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.-J.; Wu, X.-Y.; Hong, Z.; Wei, W.-S. MicroRNA-181c negatively regulates the inflammatory response in oxygen-glucose-deprived microglia by targeting Toll-like receptor 4. J. Neurochem. 2015, 132, 713–723. [Google Scholar] [CrossRef]
- Georgantas, R.W.; Streicher, K.; Greenberg, S.A.; Greenlees, L.M.; Zhu, W.; Brohawn, P.Z.; Higgs, B.W.; Czapiga, M.; Morehouse, C.A.; Amato, A.; et al. Inhibition of Myogenic MicroRNAs 1, 133, and 206 by Inflammatory Cytokines Links Inflammation and Muscle Degeneration in Adult Inflammatory Myopathies. Arthritis Rheumatol. 2014, 66, 1022–1033. [Google Scholar] [CrossRef]
- Duarte, F.V.; Palmeira, C.M.; Rolo, A.P. The Emerging Role of MitomiRs in the Pathophysiology of Human Disease. Adv. Exp. Med. Biol. 2015, 888, 123–154. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, H.; Das, S. Mitochondrial miRNA (MitomiR): A new player in cardiovascular health. Can. J. Physiol. Pharmacol. 2015, 93, 855–861. [Google Scholar] [CrossRef] [PubMed]
- John, A.; Kubosumi, A.; Reddy, P.H. Mitochondrial MicroRNAs in Aging and Neurodegenerative Diseases. Cells 2020, 9, 1345. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Rüberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chrétien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef]
- Ro, S.; Ma, H.-Y.; Park, C.; Ortogero, N.; Song, R.; Hennig, G.W.; Zheng, H.; Lin, Y.-M.; Moro, L.; Hsieh, J.-T.; et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013, 23, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sampath, H. Mitochondrial DNA Integrity: Role in Health and Disease. Cells 2019, 8, 100. [Google Scholar] [CrossRef]
- Clay Montier, L.L.; Deng, J.J.; Bai, Y. Number matters: Control of mammalian mitochondrial DNA copy number. J. Genet. Genomics 2009, 36, 125–131. [Google Scholar] [CrossRef]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef]
- Hahn, A.; Zuryn, S. The Cellular Mitochondrial Genome Landscape in Disease. Trends Cell Biol. 2019, 29, 227–240. [Google Scholar] [CrossRef]
- Copeland, W.C.; Longley, M.J. Mitochondrial genome maintenance in health and disease. DNA Repair 2014, 19, 190–198. [Google Scholar] [CrossRef]
- Hosgood, H.D., III; Liu, C.-S.; Rothman, N.; Weinstein, S.J.; Bonner, M.R.; Shen, M.; Lim, U.; Virtamo, J.; Cheng, W.; Albanes, D.; et al. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 2010, 31, 847–849. [Google Scholar] [CrossRef]
- Kirches, E. MtDNA As a Cancer Marker: A Finally Closed Chapter? Curr. Genomics 2017, 18, 255–267. [Google Scholar] [CrossRef][Green Version]
- Yue, P.; Jing, S.; Liu, L.; Ma, F.; Zhang, Y.; Wang, C.; Duan, H.; Zhou, K.; Hua, Y.; Wu, G.; et al. Association between mitochondrial DNA copy number and cardiovascular disease: Current evidence based on a systematic review and meta-analysis. PLoS ONE 2018, 13, e0206003. [Google Scholar] [CrossRef]
- Bordoni, L.; Smerilli, V.; Nasuti, C.; Gabbianelli, R. Mitochondrial DNA methylation and copy number predict body composition in a young female population. J. Transl. Med. 2019, 17, 399. [Google Scholar] [CrossRef]
- Umbria, M.; Ramos, A.; Aluja, M.P.; Santos, C. The role of control region mitochondrial DNA mutations in cardiovascular disease: Stroke and myocardial infarction. Sci. Rep. 2020, 10, 2766. [Google Scholar] [CrossRef]
- Bordoni, L.; Sawicka, A.K.; Szarmach, A.; Winklewski, P.J.; Olek, R.A.; Gabbianelli, R. A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women. Int. J. Mol. Sci. 2020, 21, 1047. [Google Scholar] [CrossRef]
- D’Aquila, P.; Bellizzi, D.; Passarino, G. Mitochondria in health, aging and diseases: The epigenetic perspective. Biogerontology 2015, 16, 569–585. [Google Scholar] [CrossRef]
- Antonyová, V.; Kejík, Z.; Brogyányi, T.; Kaplánek, R.; Pajková, M.; Talianová, V.; Hromádka, R.; Masařík, M.; Sýkora, D.; Mikšátková, L.; et al. Role of mtDNA disturbances in the pathogenesis of Alzheimer’s and Parkinson’s disease. DNA Repair 2020, 91, 102871. [Google Scholar] [CrossRef]
- Stanga, S.; Caretto, A.; Boido, M.; Vercelli, A. Mitochondrial Dysfunctions: A Red Thread across Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3719. [Google Scholar] [CrossRef]
- Kim, T.Y.; Leem, E.; Lee, J.M.; Kim, S.R. Control of Reactive Oxygen Species for the Prevention of Parkinson’s Disease: The Possible Application of Flavonoids. Antioxidants 2020, 9, 583. [Google Scholar] [CrossRef]
- Garza-Lombó, C.; Posadas, Y.; Quintanar, L.; Gonsebatt, M.E.; Franco, R. Neurotoxicity Linked to Dysfunctional Metal Ion Homeostasis and Xenobiotic Metal Exposure: Redox Signaling and Oxidative Stress. Antioxid. Redox Signal. 2018, 28, 1669–1703. [Google Scholar] [CrossRef]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Alam, Z.I.; Jenner, A.; Daniel, S.E.; Lees, A.J.; Cairns, N.; Marsden, C.D.; Jenner, P.; Halliwell, B. Oxidative DNA damage in the parkinsonian brain: An apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J. Neurochem. 1997, 69, 1196–1203. [Google Scholar] [CrossRef]
- Pamplona, R. Membrane phospholipids, lipoxidative damage and molecular integrity: A causal role in aging and longevity. Biochim. Biophys. Acta Bioenerg. 2008, 1777, 1249–1262. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Berlett, B.S. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab. Rev. 1998, 30, 225–243. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Hou, S.-C.; Way, T.-D.; Wong, C.-H.; Wang, I.-F. Heat-shock protein dysregulation is associated with functional and pathological TDP-43 aggregation. Nat. Commun. 2013, 4, 2757. [Google Scholar] [CrossRef]
- Ting, H.-C.; Chen, L.-T.; Chen, J.-Y.; Huang, Y.-L.; Xin, R.-C.; Chan, J.-F.; Hsu, Y.-H.H. Double bonds of unsaturated fatty acids differentially regulate mitochondrial cardiolipin remodeling. Lipids Health Dis. 2019, 18, 53. [Google Scholar] [CrossRef]
- Chen, W.-W.; Chao, Y.-J.; Chang, W.-H.; Chan, J.-F.; Hsu, Y.-H.H. Phosphatidylglycerol Incorporates into Cardiolipin to Improve Mitochondrial Activity and Inhibits Inflammation. Sci. Rep. 2018, 8, 4919. [Google Scholar] [CrossRef]
- Johri, A.; Beal, M.F. Mitochondrial Dysfunction in Neurodegenerative Diseases. J. Pharmacol. Exp. Ther. 2012, 342, 619–630. [Google Scholar] [CrossRef]
- Castelli, V.; Benedetti, E.; Antonosante, A.; Catanesi, M.; Pitari, G.; Ippoliti, R.; Cimini, A.; d’Angelo, M. Neuronal Cells Rearrangement During Aging and Neurodegenerative Disease: Metabolism, Oxidative Stress and Organelles Dynamic. Front. Mol. Neurosci. 2019, 12, 132. [Google Scholar] [CrossRef]
- Elfawy, H.A.; Das, B. Crosstalk between mitochondrial dysfunction, oxidative stress, and age related neurodegenerative disease: Etiologies and therapeutic strategies. Life Sci. 2019, 218, 165–184. [Google Scholar] [CrossRef]
- Cenini, G.; Lloret, A.; Cascella, R. Oxidative Stress in Neurodegenerative Diseases: From a Mitochondrial Point of View. Oxid. Med. Cell. Longev. 2019, 2019, 2105607. [Google Scholar] [CrossRef]
- Meyer, J.N.; Leuthner, T.C.; Luz, A.L. Mitochondrial fusion, fission, and mitochondrial toxicity. Toxicology 2017, 391, 42–53. [Google Scholar] [CrossRef]
- Mouli, P.K.; Twig, G.; Shirihai, O.S. Frequency and selectivity of mitochondrial fusion are key to its quality maintenance function. Biophys. J. 2009, 96, 3509–3518. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- De Souza-Pinto, N.C.; Bohr, V.A. The mitochondrial theory of aging: Involvement of mitochondrial DNA damage and repair. Int. Rev. Neurobiol. 2002, 53, 519–534. [Google Scholar] [CrossRef]
- Mandavilli, B.S.; Santos, J.H.; Van Houten, B. Mitochondrial DNA repair and aging. Mutat. Res. 2002, 509, 127–151. [Google Scholar] [CrossRef]
- Yang, J.-L.; Weissman, L.; Bohr, V.A.; Mattson, M.P. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair 2008, 7, 1110–1120. [Google Scholar] [CrossRef]
- Ljungman, M.; Hanawalt, P.C. Efficient protection against oxidative DNA damage in chromatin. Mol. Carcinog. 1992, 5, 264–269. [Google Scholar] [CrossRef]
- Enright, H.U.; Miller, W.J.; Hebbel, R.P. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 1992, 20, 3341–3346. [Google Scholar] [CrossRef][Green Version]
- Wei, Y.-H.; Lee, H.-C. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp. Biol. Med. 2002, 227, 671–682. [Google Scholar] [CrossRef]
- Nissanka, N.; Moraes, C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018, 592, 728–742. [Google Scholar] [CrossRef]
- Santos, R.X.; Correia, S.C.; Zhu, X.; Smith, M.A.; Moreira, P.I.; Castellani, R.J.; Nunomura, A.; Perry, G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer’s disease. Antioxid. Redox Signal. 2013, 18, 2444–2457. [Google Scholar] [CrossRef]
- Keogh, M.J.; Chinnery, P.F. Mitochondrial DNA mutations in neurodegeneration. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 1401–1411. [Google Scholar] [CrossRef]
- Hahn, A.; Zuryn, S. Mitochondrial Genome (mtDNA) Mutations that Generate Reactive Oxygen Species. Antioxidants 2019, 8, 392. [Google Scholar] [CrossRef]
- Soltys, D.T.; Pereira, C.P.M.; Rowies, F.T.; Farfel, J.M.; Grinberg, L.T.; Suemoto, C.K.; Leite, R.E.P.; Rodriguez, R.D.; Ericson, N.G.; Bielas, J.H.; et al. Lower mitochondrial DNA content but not increased mutagenesis associates with decreased base excision repair activity in brains of AD subjects. Neurobiol. Aging 2019, 73, 161–170. [Google Scholar] [CrossRef]
- Lowes, H.; Pyle, A.; Santibanez-Koref, M.; Hudson, G. Circulating cell-free mitochondrial DNA levels in Parkinson’s disease are influenced by treatment. Mol. Neurodegener. 2020, 15, 10. [Google Scholar] [CrossRef]
- Al-Kafaji, G.; Bakheit, H.F.; Alharbi, M.A.; Farahat, A.A.; Jailani, M.; Ebrahin, B.H.; Bakhiet, M. Mitochondrial DNA Copy Number in Peripheral Blood as a Potential Non-invasive Biomarker for Multiple Sclerosis. NeuroMol. Med. 2020, 22, 304–313. [Google Scholar] [CrossRef]
- Pyle, A.; Anugrha, H.; Kurzawa-Akanbi, M.; Yarnall, A.; Burn, D.; Hudson, G. Reduced mitochondrial DNA copy number is a biomarker of Parkinson’s disease. Neurobiol. Aging 2016, 38, 216.e7–216.e10. [Google Scholar] [CrossRef]
- Gambardella, S.; Limanaqi, F.; Ferese, R.; Biagioni, F.; Campopiano, R.; Centonze, D.; Fornai, F. Ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Front. Immunol. 2019, 10, 1064. [Google Scholar] [CrossRef]
- Pinto, M.; Moraes, C.T. Mitochondrial genome changes and neurodegenerative diseases. Biochim. Biophys. Acta 2014, 1842, 1198–1207. [Google Scholar] [CrossRef]
- Coppedè, F.; Stoccoro, A. Mitoepigenetics and Neurodegenerative Diseases. Front. Endocrinol. 2019, 10, 86. [Google Scholar] [CrossRef]
- Pavlou, M.A.S.; Outeiro, T.F. Epigenetics in Parkinson’s Disease. Adv. Exp. Med. Biol. 2017, 978, 363–390. [Google Scholar] [CrossRef]
- Sherwani, S.I.; Khan, H.A. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene 2015, 570, 17–24. [Google Scholar] [CrossRef]
- Miranda-Morales, E.; Meier, K.; Sandoval-Carrillo, A.; Salas-Pacheco, J.; Vázquez-Cárdenas, P.; Arias-Carrión, O. Implications of DNA Methylation in Parkinson’s Disease. Front. Mol. Neurosci. 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Lardenoije, R.; Iatrou, A.; Kenis, G.; Kompotis, K.; Steinbusch, H.W.M.; Mastroeni, D.; Coleman, P.; Lemere, C.A.; Hof, P.R.; van den Hove, D.L.A.; et al. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015, 131, 21–64. [Google Scholar] [CrossRef]
- Gabbianelli, R.; Damiani, E. Epigenetics and neurodegeneration: Role of early-life nutrition. J. Nutr. Biochem. 2018, 57, 1–13. [Google Scholar] [CrossRef]
- Zawia, N.H.; Lahiri, D.K.; Cardozo-Pelaez, F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009, 46, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, D.K.; Maloney, B.; Bayon, B.L.; Chopra, N.; White, F.A.; Greig, N.H.; Nurnberger, J.I. Transgenerational latent early-life associated regulation unites environment and genetics across generations. Epigenomics 2016, 8, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, D.; Montani, M.; Bordoni, L.; Galeazzi, R.; Nasuti, C.; Correia, S.L.; Domingues, V.F.; Jayant, M.; Brahmachari, V.; Massaccesi, L.; et al. In vivo and in silico studies to identify mechanisms associated with Nurr1 modulation following early life exposure to permethrin in rats. Neuroscience 2017, 340, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Kim-Ha, J.; Kim, Y.-J. Age-related epigenetic regulation in the brain and its role in neuronal diseases. BMB Rep. 2016, 49, 671–680. [Google Scholar] [CrossRef]
- Bordoni, L.; Nasuti, C.; Di Stefano, A.; Marinelli, L.; Gabbianelli, R. Epigenetic Memory of Early-Life Parental Perturbation: Dopamine Decrease and DNA Methylation Changes in Offspring. Oxid. Med. Cell. Longev. 2019, 2019, 1472623. [Google Scholar] [CrossRef]
- Bordoni, L.; Nasuti, C.; Fedeli, D.; Galeazzi, R.; Laudadio, E.; Massaccesi, L.; López-Rodas, G.; Gabbianelli, R. Early impairment of epigenetic pattern in neurodegeneration: Additional mechanisms behind pyrethroid toxicity. Exp. Gerontol. 2019, 124, 110629. [Google Scholar] [CrossRef]
- Mohammed, S.A.; Ambrosini, S.; Lüscher, T.; Paneni, F.; Costantino, S. Epigenetic Control of Mitochondrial Function in the Vasculature. Front. Cardiovasc. Med. 2020, 7, 28. [Google Scholar] [CrossRef]
- Van der Wijst, M.G.P.; Rots, M.G. Mitochondrial epigenetics: An overlooked layer of regulation? Trends Genet. 2015, 31, 353–356. [Google Scholar] [CrossRef]
- Mposhi, A.; Van der Wijst, M.G.; Faber, K.N.; Rots, M.G. Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. 2017, 22, 1099–1113. [Google Scholar]
- Lambertini, L.; Byun, H.-M. Mitochondrial Epigenetics and Environmental Exposure. Curr. Environ. Health Rep. 2016, 3, 214–224. [Google Scholar] [CrossRef]
- Coppedè, F. The potential of epigenetic therapies in neurodegenerative diseases. Front. Genet. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Irwin, M.H.; Moos, W.H.; Faller, D.V.; Steliou, K.; Pinkert, C.A. Epigenetic Treatment of Neurodegenerative Disorders: Alzheimer and Parkinson Diseases. Drug Dev. Res. 2016, 77, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Winiarska-Mieczan, A.; Baranowska-Wójcik, E.; Kwiecień, M.; Grela, E.R.; Szwajgier, D.; Kwiatkowska, K.; Kiczorowska, B. The Role of Dietary Antioxidants in the Pathogenesis of Neurodegenerative Diseases and Their Impact on Cerebral Oxidoreductive Balance. Nutrients 2020, 12, 435. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, C.; Bates, T.E.; Butterfield, D.A.; Calafato, S.; Cornelius, C.; De Lorenzo, A.; Kostova, A.T.D.; Calabrese, V. Natural antioxidants in Alzheimer’s disease. Expert Opin. Investig. Drugs 2007, 16, 1921–1931. [Google Scholar] [CrossRef]
- Remely, M.; Stefanska, B.; Lovrecic, L.; Magnet, U.; Haslberger, A.G. Nutriepigenomics: The role of nutrition in epigenetic control of human diseases. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 328–333. [Google Scholar] [CrossRef]
- Sasazuki, S.; Hayashi, T.; Nakachi, K.; Sasaki, S.; Tsubono, Y.; Okubo, S.; Hayashi, M.; Tsugane, S. Protective effect of vitamin C on oxidative stress: A randomized controlled trial. Int. J. Vitam. Nutr. Res. 2008, 78, 121–128. [Google Scholar] [CrossRef]
- Righi, N.C.; Schuch, F.B.; De Nardi, A.T.; Pippi, C.M.; de Almeida, R.G.; Puntel, G.O.; da Silva, A.M.V.; Signori, L.U. Effects of vitamin C on oxidative stress, inflammation, muscle soreness, and strength following acute exercise: Meta-analyses of randomized clinical trials. Eur. J. Nutr. 2020. [Google Scholar] [CrossRef]
- Niki, E. Evidence for beneficial effects of vitamin E. Korean J. Intern. Med. 2015, 30, 571–579. [Google Scholar] [CrossRef]
- Faghfouri, A.H.; Zarezadeh, M.; Tavakoli-Rouzbehani, O.M.; Radkhah, N.; Faghfuri, E.; Kord-Varkaneh, H.; Tan, S.C.; Ostadrahimi, A. The effects of N-acetylcysteine on inflammatory and oxidative stress biomarkers: A systematic review and meta-analysis of controlled clinical trials. Eur. J. Pharmacol. 2020, 173368. [Google Scholar] [CrossRef]
- Shen, Q.; Hiebert, J.B.; Hartwell, J.; Thimmesch, A.R.; Pierce, J.D. Systematic Review of Traumatic Brain Injury and the Impact of Antioxidant Therapy on Clinical Outcomes. Worldviews Evid. Based Nurs. 2016, 13, 380–389. [Google Scholar] [CrossRef]
- McCarty, M.F.; Lerner, A. Nutraceuticals Targeting Generation and Oxidant Activity of Peroxynitrite May Aid Prevention and Control of Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 3624. [Google Scholar] [CrossRef] [PubMed]
- Pinnen, F.; Cacciatore, I.; Cornacchia, C.; Sozio, P.; Cerasa, L.S.; Iannitelli, A.; Nasuti, C.; Cantalamessa, F.; Sekar, D.; Gabbianelli, R.; et al. Codrugs linking L-dopa and sulfur-containing antioxidants: New pharmacological tools against Parkinson’s disease. J. Med. Chem. 2009, 52, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Falcioni, M.L.; Nasuti, C.; Bergamini, C.; Fato, R.; Lenaz, G.; Gabbianelli, R. The primary role of glutathione against nuclear DNA damage of striatum induced by permethrin in rats. Neuroscience 2010, 168, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, L. Positive effect of an electrolyzed reduced water on gut permeability, fecal microbiota and liver in an animal model of Parkinson’s disease. PLoS ONE 2019, 14, e0223238. [Google Scholar] [CrossRef] [PubMed]
- Damiani, E.; Gabbianelli, R. Nutrigenomics as a Strategy for Neuronal Health. Healthy Aging Longev. 2019, 167–187. [Google Scholar] [CrossRef]
- Arbo, B.D.; André-Miral, C.; Nasre-Nasser, R.G.; Schimith, L.E.; Santos, M.G.; Costa-Silva, D.; Muccillo-Baisch, A.L.; Hort, M.A. Resveratrol Derivatives as Potential Treatments for Alzheimer’s and Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 103. [Google Scholar] [CrossRef]
- Cho, K.S.; Shin, M.; Kim, S.; Lee, S.B. Recent Advances in Studies on the Therapeutic Potential of Dietary Carotenoids in Neurodegenerative Diseases. Oxid. Med. Cell. Longev. 2018, 2018, 4120458. [Google Scholar] [CrossRef]
- Elumalai, P.; Lakshmi, S. Role of Quercetin Benefits in Neurodegeneration. Adv. Neurobiol. 2016, 12, 229–245. [Google Scholar] [CrossRef]
- Amanzadeh, E.; Esmaeili, A.; Rahgozar, S.; Nourbakhshnia, M. Application of quercetin in neurological disorders: From nutrition to nanomedicine. Rev. Neurosci. 2019, 30, 555–572. [Google Scholar] [CrossRef]
- Tarozzi, A.; Angeloni, C.; Malaguti, M.; Morroni, F.; Hrelia, S.; Hrelia, P. Sulforaphane as a potential protective phytochemical against neurodegenerative diseases. Oxid. Med. Cell. Longev. 2013, 2013, 415078. [Google Scholar] [CrossRef]
- Klomparens, E.A.; Ding, Y. The neuroprotective mechanisms and effects of sulforaphane. Brain Circ. 2019, 5, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Santín-Márquez, R.; Alarcón-Aguilar, A.; López-Diazguerrero, N.E.; Chondrogianni, N.; Königsberg, M. Sulforaphane—Role in aging and neurodegeneration. GeroScience 2019, 41, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, D.P. Melatonin: Clinical Perspectives in Neurodegeneration. Front. Endocrinol. 2019, 10, 480. [Google Scholar] [CrossRef] [PubMed]
- Bazan, N.G.; Molina, M.F.; Gordon, W.C. Docosahexaenoic acid signalolipidomics in nutrition: Significance in aging, neuroinflammation, macular degeneration, Alzheimer’s, and other neurodegenerative diseases. Annu. Rev. Nutr. 2011, 31, 321–351. [Google Scholar] [CrossRef]
- Singh, S.S.; Rai, S.N.; Birla, H.; Zahra, W.; Rathore, A.S.; Singh, S.P. NF-κB-Mediated Neuroinflammation in Parkinson’s Disease and Potential Therapeutic Effect of Polyphenols. Neurotox. Res. 2020, 37, 491–507. [Google Scholar] [CrossRef]
- Bernardo-Colón, A.; Vest, V.; Clark, A.; Cooper, M.L.; Calkins, D.J.; Harrison, F.E.; Rex, T.S. Antioxidants prevent inflammation and preserve the optic projection and visual function in experimental neurotrauma. Cell Death Dis. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Bordoni, L.; Fedeli, D.; Nasuti, C.; Capitani, M.; Fiorini, D.; Gabbianelli, R. Permethrin pesticide induces NURR1 up-regulation in dopaminergic cell line: Is the pro-oxidant effect involved in toxicant-neuronal damage? Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 201, 51–57. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, R.; Zhan, Z.; Li, X.; Zhou, F.; Xing, A.; Jiang, C.; Chen, Y.; An, L. Beneficial Effects of Sulforaphane Treatment in Alzheimer’s Disease May Be Mediated through Reduced HDAC1/3 and Increased P75NTR Expression. Front. Aging Neurosci. 2017, 9, 121. [Google Scholar] [CrossRef]
- Gomes, B.A.Q.; Silva, J.P.B.; Romeiro, C.F.R.; Dos Santos, S.M.; Rodrigues, C.A.; Gonçalves, P.R.; Sakai, J.T.; Mendes, P.F.S.; Varela, E.L.P.; Monteiro, M.C. Neuroprotective Mechanisms of Resveratrol in Alzheimer’s Disease: Role of SIRT1. Oxid. Med. Cell. Longev. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Fernandes, G.F.S.; Silva, G.D.B.; Pavan, A.R.; Chiba, D.E.; Chin, C.M.; Dos Santos, J.L. Epigenetic Regulatory Mechanisms Induced by Resveratrol. Nutrients 2017, 9, 1201. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Nicol, C.J.; Cheng, Y.-C. Resveratrol activation of AMPK-dependent pathways is neuroprotective in human neural stem cells against amyloid-beta-induced inflammation and oxidative stress. Neurochem. Int. 2018, 115, 1–10. [Google Scholar] [CrossRef]
- Xu, X.-Y.; Meng, X.; Li, S.; Gan, R.-Y.; Li, Y.; Li, H.-B. Bioactivity, Health Benefits, and Related Molecular Mechanisms of Curcumin: Current Progress, Challenges, and Perspectives. Nutrients 2018, 10, 1553. [Google Scholar] [CrossRef]
- Cyr, A.R.; Domann, F.E. The redox basis of epigenetic modifications: From mechanisms to functional consequences. Antioxid. Redox Signal. 2011, 15, 551–589. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, S.; Fischle, W. Oxidative stress signaling to chromatin in health and disease. Epigenomics 2016, 8, 843–862. [Google Scholar] [CrossRef] [PubMed]
- Chia, N.; Wang, L.; Lu, X.; Senut, M.-C.; Brenner, C.A.; Ruden, D.M. Hypothesis: Environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 2011, 6, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Jeschke, J.; Defrance, M.; Bachman, M.; Creppe, C.; Calonne, E.; Bizet, M.; Deplus, R.; Marroquí, L.; Libin, M.; et al. Genome-wide hydroxymethylcytosine pattern changes in response to oxidative stress. Sci. Rep. 2015, 5, 12714. [Google Scholar] [CrossRef]
- Madugundu, G.S.; Cadet, J.; Wagner, J.R. Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 2014, 42, 7450–7460. [Google Scholar] [CrossRef]
- Weitzman, S.A.; Turk, P.W.; Milkowski, D.H.; Kozlowski, K. Free radical adducts induce alterations in DNA cytosine methylation. Proc. Natl. Acad. Sci. USA 1994, 91, 1261–1264. [Google Scholar] [CrossRef] [PubMed]
- Turk, P.W.; Laayoun, A.; Smith, S.S.; Weitzman, S.A. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis 1995, 16, 1253–1255. [Google Scholar] [CrossRef]
- Mosharov, E.; Cranford, M.R.; Banerjee, R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry 2000, 39, 13005–13011. [Google Scholar] [CrossRef]
- Simpson, N.E.; Tryndyak, V.P.; Pogribna, M.; Beland, F.A.; Pogribny, I.P. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics 2012, 7, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Engedal, N.; Žerovnik, E.; Rudov, A.; Galli, F.; Olivieri, F.; Procopio, A.D.; Rippo, M.R.; Monsurrò, V.; Betti, M.; Albertini, M.C. From Oxidative Stress Damage to Pathways, Networks, and Autophagy via MicroRNAs. Oxid. Med. Cell. Longev. 2018, 2018, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 55. [Google Scholar] [CrossRef]
- Carbonell, T.; Gomes, A. V MicroRNAs in the regulation of cellular redox status and its implications in myocardial ischemia-reperfusion injury. Redox Biol. 2020, 36, 101607. [Google Scholar] [CrossRef]
- Guillaumet-Adkins, A.; Yañez, Y.; Peris-Diaz, M.D.; Calabria, I.; Palanca-Ballester, C.; Sandoval, J. Epigenetics and Oxidative Stress in Aging. Oxid. Med. Cell. Longev. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Romá-Mateo, C.; Pérez-Machado, G.; Peiró-Chova, L.; Pallardó, F.V. Role of Glutathione in the Regulation of Epigenetic Mechanisms in Disease. Free Radic. Biol. Med. 2017. [Google Scholar] [CrossRef]
- Delgado-Morales, R.; Agis-Balboa, R.C.; Esteller, M.; Berdasco, M. Epigenetic mechanisms during ageing and neurogenesis as novel therapeutic avenues in human brain disorders. Clin. Epigenet. 2017, 9, 67. [Google Scholar] [CrossRef]
- Bordoni, L.; Gabbianelli, R. Primers on nutrigenetics and nutri(epi)genomics: Origins and development of precision nutrition. Biochimie 2019, 160, 156–171. [Google Scholar] [CrossRef]
- Remely, M.; Lovrecic, L.; de la Garza, A.L.; Migliore, L.; Peterlin, B.; Milagro, F.I.; Martinez, A.J.; Haslberger, A.G. Therapeutic perspectives of epigenetically active nutrients. Br. J. Pharmacol. 2015, 172, 2756–2768. [Google Scholar] [CrossRef]
- Mariño, G.; Pietrocola, F.; Eisenberg, T.; Kong, Y.; Malik, S.A.; Andryushkova, A.; Schroeder, S.; Pendl, T.; Harger, A.; Niso-Santano, M.; et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 2014, 53, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Carlos-Reyes, Á.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; la Vega, H.; de la Cruz, O.N.; López-Camarillo, C. Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front. Genet. 2019, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Gensous, N.; Franceschi, C.; Santoro, A.; Milazzo, M.; Garagnani, P.; Bacalini, M.G. The Impact of Caloric Restriction on the Epigenetic Signatures of Aging. Int. J. Mol. Sci. 2019, 20, 2022. [Google Scholar] [CrossRef] [PubMed]
- Taormina, G.; Russo, A.; Latteri, M.A.; Mirisola, M.G. Mitochondrion at the Crossroad Between Nutrients and Epigenome. Front. Endocrinol. 2019, 10, 673. [Google Scholar] [CrossRef] [PubMed]
- Beetch, M.; Harandi-Zadeh, S.; Shen, K.; Lubecka, K.; Kitts, D.D.; O’Hagan, H.M.; Stefanska, B. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Br. J. Pharmacol. 2020, 177, 1382–1408. [Google Scholar] [CrossRef]
- Peng, D.; Ge, G.; Gong, Y.; Zhan, Y.; He, S.; Guan, B.; Li, Y.; Xu, Z.; Hao, H.; He, Z.; et al. Vitamin C increases 5-hydroxymethylcytosine level and inhibits the growth of bladder cancer. Clin. Epigenet. 2018, 10, 94. [Google Scholar] [CrossRef]
- Gerecke, C.; Schumacher, F.; Edlich, A.; Wetzel, A.; Yealland, G.; Neubert, L.K.; Scholtka, B.; Homann, T.; Kleuser, B. Vitamin C promotes decitabine or azacytidine induced DNA hydroxymethylation and subsequent reactivation of the epigenetically silenced tumour suppressor CDKN1A in colon cancer cells. Oncotarget 2018, 9, 32822–32840. [Google Scholar] [CrossRef]
- Starczak, M.; Zarakowska, E.; Modrzejewska, M.; Dziaman, T.; Szpila, A.; Linowiecka, K.; Guz, J.; Szpotan, J.; Gawronski, M.; Labejszo, A.; et al. In vivo evidence of ascorbate involvement in the generation of epigenetic DNA modifications in leukocytes from patients with colorectal carcinoma, benign adenoma and inflammatory bowel disease. J. Transl. Med. 2018, 16, 204. [Google Scholar] [CrossRef]
- Sun, X.; Xiao, Y.; Zeng, Z.; Shi, Y.; Tang, B.; Long, H.; Kanekura, T.; Wang, J.; Wu, H.; Zhao, M.; et al. All-Trans Retinoic Acid Induces CD4+CD25+FOXP3+ Regulatory T Cells by Increasing FOXP3 Demethylation in Systemic Sclerosis CD4+ T Cells. J. Immunol. Res. 2018, 2018, 1–7. [Google Scholar] [CrossRef]
- Stefanska, B.; Rudnicka, K.; Bednarek, A.; Fabianowska-Majewska, K. Hypomethylation and induction of retinoic acid receptor beta 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur. J. Pharmacol. 2010, 638, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Stefanska, B.; Salamé, P.; Bednarek, A.; Fabianowska-Majewska, K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br. J. Nutr. 2012, 107, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Khor, T.O.; Shu, L.; Saw, C.L.-L.; Wu, T.-Y.; Suh, N.; Yang, C.S.; Kong, A.-N.T. A γ-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J. Nutr. 2012, 142, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Speckmann, B.; Schulz, S.; Hiller, F.; Hesse, D.; Schumacher, F.; Kleuser, B.; Geisel, J.; Obeid, R.; Grune, T.; Kipp, A.P. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J. Nutr. Biochem. 2017, 48, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Lubecka, K.; Kurzava, L.; Flower, K.; Buvala, H.; Zhang, H.; Teegarden, D.; Camarillo, I.; Suderman, M.; Kuang, S.; Andrisani, O.; et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic NOTCH signaling through epigenetic regulation of MAML2 transcriptional activity. Carcinogenesis 2016, 37, 656–668. [Google Scholar] [CrossRef]
- Medina-Aguilar, R.; Pérez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; García-Mena, J.; Cuevas, S.R.; Ruíz-García, E.; de la Vega, H.A.; Juárez, J.H.; Flores-Pérez, A.; et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef]
- Zhang, B.-K.; Lai, Y.-Q.; Niu, P.-P.; Zhao, M.; Jia, S.-J. Epigallocatechin-3-gallate inhibits homocysteine-induced apoptosis of endothelial cells by demethylation of the DDAH2 gene. Planta Med. 2013, 79, 1715–1719. [Google Scholar] [CrossRef]
- Liu, X.; Sun, C.; Liu, B.; Jin, X.; Li, P.; Zheng, X.; Zhao, T.; Li, F.; Li, Q. Genistein mediates the selective radiosensitizing effect in NSCLC A549 cells via inhibiting methylation of the keap1 gene promoter region. Oncotarget 2016, 7, 27267–27279. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Hu, Y.; Wang, J.; Yuan, C. Curcumin inhibits growth of human breast cancer cells through demethylation of DLC1 promoter. Mol. Cell. Biochem. 2017, 425, 47–58. [Google Scholar] [CrossRef]
- Khan, M.A.; Sundaram, M.K.; Hamza, A.; Quraishi, U.; Gunasekera, D.; Ramesh, L.; Goala, P.; Alami, U.A.; Ansari, M.Z.; Rizvi, T.A.; et al. Sulforaphane Reverses the Expression of Various Tumor Suppressor Genes by Targeting DNMT3B and HDAC1 in Human Cervical Cancer Cells. Evid. Based Complement. Altern. Med. 2015, 2015, 412149. [Google Scholar] [CrossRef]
- Lubecka, K.; Kaufman-Szymczyk, A.; Fabianowska-Majewska, K. Inhibition of breast cancer cell growth by the combination of clofarabine and sulforaphane involves epigenetically mediated CDKN2A upregulation. Nucleosides Nucleotides Nucleic Acids 2018, 37, 280–289. [Google Scholar] [CrossRef]
- Hassan, F.-U.; Rehman, M.S.-U.; Khan, M.S.; Ali, M.A.; Javed, A.; Nawaz, A.; Yang, C. Curcumin as an Alternative Epigenetic Modulator: Mechanism of Action and Potential Effects. Front. Genet. 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Harman, D. Aging: A Theory Based on Free Radical and Radiation Chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Melov, S. SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 2013, 62, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Ilic, T.V.; Jovanovic, M.; Jovicic, A.; Tomovic, M. Oxidative stress indicators are elevated in de novo Parkinson’s disease patients. Funct. Neurol. 1999, 14, 141–147. [Google Scholar] [PubMed]
- Kocatürk, P.A.; Akbostanci, M.C.; Tan, F.; Kavas, G.O. Superoxide dismutase activity and zinc and copper concentrations in Parkinson’s disease. Pathophysiology 2000, 7, 63–67. [Google Scholar] [CrossRef]
- Younes-Mhenni, S.; Frih-Ayed, M.; Kerkeni, A.; Bost, M.; Chazot, G. Peripheral blood markers of oxidative stress in Parkinson’s disease. Eur. Neurol. 2007, 58, 78–83. [Google Scholar] [CrossRef]
- Cho, J.H.; Tae, H.-J.; Kim, I.-S.; Song, M.; Kim, H.; Lee, T.-K.; Kim, Y.-M.; Ryoo, S.; Kim, D.W.; Lee, C.-H.; et al. Melatonin alleviates asphyxial cardiac arrest-induced cerebellar Purkinje cell death by attenuation of oxidative stress. Exp. Neurol. 2019, 320, 112983. [Google Scholar] [CrossRef]
- Campolo, M.; Casili, G.; Lanza, M.; Filippone, A.; Paterniti, I.; Cuzzocrea, S.; Esposito, E. Multiple mechanisms of dimethyl fumarate in amyloid β-induced neurotoxicity in human neuronal cells. J. Cell. Mol. Med. 2018, 22, 1081–1094. [Google Scholar] [CrossRef]
- Li, Y.M.; Chan, H.Y.E.; Huang, Y.; Chen, Z.Y. Green tea catechins upregulate superoxide dismutase and catalase in fruit flies. Mol. Nutr. Food Res. 2007, 51, 546–554. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Yang, L.; Yuan, J.; Jia, J.; Yang, S. SOD2 Mediates Curcumin-Induced Protection against Oxygen-Glucose Deprivation/Reoxygenation Injury in HT22 Cells. Evid.-Based Complement. Altern. Med. 2019, 2019, 2160642. [Google Scholar] [CrossRef]
- Du, S.; Zhang, Y.; Yang, J.; Liu, X.; Wang, Y.; Xu, B.; Jia, J. Curcumin Alleviates β Amyloid-Induced Neurotoxicity in HT22 Cells via Upregulating SOD2. J. Mol. Neurosci. 2019, 67, 540–549. [Google Scholar] [CrossRef]
- Kandylis, P.; Kokkinomagoulos, E. Food Applications and Potential Health Benefits of Pomegranate and its Derivatives. Foods 2020, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.P.; Aryal, P.; Robinson, S.; Rafiu, R.; Obrenovich, M.; Perry, G. Polyphenols in Alzheimer’s Disease and in the Gut-Brain Axis. Microorganisms 2020, 8, 199. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.; Lin, Y.; Lei, Q.; Guan, K.-L.; Zhao, S.; Xiong, Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011, 12, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.R.; Wani, W.Y.; Sunkaria, A.; Kandimalla, R.J.L.; Verma, D.; Cameotra, S.S.; Gill, K.D. Quercetin protects against chronic aluminum-induced oxidative stress and ensuing biochemical, cholinergic, and neurobehavioral impairments in rats. Neurotox. Res. 2013, 23, 336–357. [Google Scholar] [CrossRef] [PubMed]
- Valero, T. Mitochondrial biogenesis: Pharmacological approaches. Curr. Pharm. Des. 2014, 20, 5507–5509. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef]
- Gledhill, J.R.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA 2007, 104, 13632–13637. [Google Scholar] [CrossRef] [PubMed]
- Zini, R.; Morin, C.; Bertelli, A.; Bertelli, A.A.; Tillement, J.P. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp. Clin. Res. 1999, 25, 87–97. [Google Scholar] [PubMed]
- Park, S.-J.; Ahmad, F.; Philp, A.; Baar, K.; Williams, T.; Luo, H.; Ke, H.; Rehmann, H.; Taussig, R.; Brown, A.L.; et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012, 148, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Zhang, H.; Wang, P.; Zhao, Y.; Li, Q.; Wei, X.; Cui, Y.; Sun, J.; Shang, Q.; Liu, D.; et al. Dietary curcumin ameliorates aging-related cerebrovascular dysfunction through the AMPK/uncoupling protein 2 pathway. Cell. Physiol. Biochem. 2013, 32, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Rasbach, K.A.; Schnellmann, R.G. Isoflavones promote mitochondrial biogenesis. J. Pharmacol. Exp. Ther. 2008, 325, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Taub, P.R.; Ramirez-Sanchez, I.; Ciaraldi, T.P.; Perkins, G.; Murphy, A.N.; Naviaux, R.; Hogan, M.; Maisel, A.S.; Henry, R.R.; Ceballos, G.; et al. Alterations in skeletal muscle indicators of mitochondrial structure and biogenesis in patients with type 2 diabetes and heart failure: Effects of epicatechin rich cocoa. Clin. Transl. Sci. 2012, 5, 43–47. [Google Scholar] [CrossRef]
- Biswas, R.; Ahn, J.-C.; Kim, J.-S. Sulforaphene Synergistically Sensitizes Cisplatin via Enhanced Mitochondrial Dysfunction and PI3K/PTEN Modulation in Ovarian Cancer Cells. Anticancer Res. 2015, 35, 3901–3908. [Google Scholar] [PubMed]
- Duan, W.; Ross, C.A. Potential therapeutic targets for neurodegenerative diseases: Lessons learned from calorie restriction. Curr. Drug Targets 2010, 11, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Kahn, M.; Samiei, A.; Gao, J.; Ota, K.T.; Rei, D.; Tsai, L.-H. A dietary regimen of caloric restriction or pharmacological activation of SIRT1 to delay the onset of neurodegeneration. J. Neurosci. 2013, 33, 8951–8960. [Google Scholar] [CrossRef] [PubMed]
- Banaclocha, M.M. Therapeutic potential of N-acetylcysteine in age-related mitochondrial neurodegenerative diseases. Med. Hypotheses 2001, 56, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, M.; Ito, Y. N-acetylcysteine and neurodegenerative diseases: Basic and clinical pharmacology. Cerebellum 2007, 6, 308–314. [Google Scholar] [CrossRef]
- Ng, Y.-G.; Fann, D.W.-F.; Jo, D.-G.; Sobey, C.G.; Arumugam, T.V. Dietary Restriction and Epigenetics: Part I. Cond. Med. 2019, 2, 284–299. [Google Scholar]
- Ng, Y.-G.; Fann, D.W.-F.; Jo, D.-G.; Sobey, C.G.; Arumugam, T.V. Epigenetic Regulation by Dietary Restriction: Part II. Cond. Med. 2019, 2, 300–310. [Google Scholar]
- Sheu, S.-S.; Nauduri, D.; Anders, M.W. Targeting antioxidants to mitochondria: A new therapeutic direction. Biochim. Biophys. Acta Mol. Basis Dis. 2006, 1762, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Stelmashook, E.V.; Isaev, N.K.; Genrikhs, E.E.; Novikova, S.V. Mitochondria-Targeted Antioxidants as Potential Therapy for the Treatment of Traumatic Brain Injury. Antioxidants 2019, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.; Deus, C.M.; Borges, F.; Oliveira, P.J. Mitochondria: Targeting mitochondrial reactive oxygen species with mitochondriotropic polyphenolic-based antioxidants. Int. J. Biochem. Cell Biol. 2018, 97, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Broome, S.C.; Woodhead, J.S.T.; Merry, T.L. Mitochondria-Targeted Antioxidants and Skeletal Muscle Function. Antioxidants 2018, 7, 107. [Google Scholar] [CrossRef]
- Braakhuis, A.J.; Nagulan, R.; Somerville, V. The Effect of MitoQ on Aging-Related Biomarkers: A Systematic Review and Meta-Analysis. Oxid. Med. Cell. Longev. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Escribano-Lopez, I.; Diaz-Morales, N.; Rovira-Llopis, S.; de Marañon, A.M.; Orden, S.; Alvarez, A.; Bañuls, C.; Rocha, M.; Murphy, M.P.; Hernandez-Mijares, A.; et al. The mitochondria-targeted antioxidant MitoQ modulates oxidative stress, inflammation and leukocyte-endothelium interactions in leukocytes isolated from type 2 diabetic patients. Redox Biol. 2016, 10, 200–205. [Google Scholar] [CrossRef]
- Feniouk, B.A.; Skulachev, V.P. Cellular and Molecular Mechanisms of Action of Mitochondria-Targeted Antioxidants. Curr. Aging Sci. 2017, 10, 41–48. [Google Scholar] [CrossRef]
- Miquel, E.; Cassina, A.; Martínez-Palma, L.; Souza, J.M.; Bolatto, C.; Rodríguez-Bottero, S.; Logan, A.; Smith, R.A.J.; Murphy, M.P.; Barbeito, L.; et al. Neuroprotective effects of the mitochondria-targeted antioxidant MitoQ in a model of inherited amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2014, 70, 204–213. [Google Scholar] [CrossRef]
- Fink, B.; Coppey, L.; Davidson, E.; Shevalye, H.; Obrosov, A.; Chheda, P.R.; Kerns, R.; Sivitz, W.; Yorek, M. Effect of mitoquinone (Mito-Q) on neuropathic endpoints in an obese and type 2 diabetic rat model. Free Radic. Res. 2020, 54, 311–318. [Google Scholar] [CrossRef]
- Lagouge, M.; Larsson, N.-G. The role of mitochondrial DNA mutations and free radicals in disease and ageing. J. Intern. Med. 2013, 273, 529–543. [Google Scholar] [CrossRef]
- Hekimi, S.; Lapointe, J.; Wen, Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011, 21, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Breyer, V.; Weigel, I.; Huang, T.-T.; Pischetsrieder, M. Endogenous mitochondrial oxidative stress in MnSOD-deficient mouse embryonic fibroblasts promotes mitochondrial DNA glycation. Free Radic. Biol. Med. 2012, 52, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.D.; Van Remmen, H.; Conrad, C.C.; Huang, T.T.; Epstein, C.J.; Richardson, A. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J. Biol. Chem. 1998, 273, 28510–28515. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.R.; Brown, K.E.; McCormick, M.L.; Coleman, M.C.; Case, A.J.; Watts, G.S.; Futscher, B.W.; Spitz, D.R.; Domann, F.E. Maintenance of mitochondrial genomic integrity in the absence of manganese superoxide dismutase in mouse liver hepatocytes. Redox Biol. 2013, 1, 172–177. [Google Scholar] [CrossRef]
- Itsara, L.S.; Kennedy, S.R.; Fox, E.J.; Yu, S.; Hewitt, J.J.; Sanchez-Contreras, M.; Cardozo-Pelaez, F.; Pallanck, L.J. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014, 10, e1003974. [Google Scholar] [CrossRef]
- Sim, M.S.; Lee, D.; Kim, K.-Y.; Weinreb, R.; Ju, W.-K. Effect of coenzyme Q10 on oxidative stress and mitochondrial DNA alteration in a mouse model of glaucoma. Investig. Ophthalmol. Vis. Sci. 2013, 54, 424. [Google Scholar]
- Montero, R.; Sánchez-Alcázar, J.A.; Briones, P.; Navarro-Sastre, A.; Gallardo, E.; Bornstein, B.; Herrero-Martín, D.; Rivera, H.; Martin, M.A.; Marti, R.; et al. Coenzyme Q10 deficiency associated with a mitochondrial DNA depletion syndrome: A case report. Clin. Biochem. 2009, 42, 742–745. [Google Scholar] [CrossRef]
- Hu, Q.; Ren, J.; Li, G.; Wu, J.; Wu, X.; Wang, G.; Gu, G.; Ren, H.; Hong, Z.; Li, J. The mitochondrially targeted antioxidant MitoQ protects the intestinal barrier by ameliorating mitochondrial DNA damage via the Nrf2/ARE signaling pathway. Cell Death Dis. 2018, 9, 403. [Google Scholar] [CrossRef]
- Nishigaki, A.; Kido, T.; Kida, N.; Kakita-Kobayashi, M.; Tsubokura, H.; Hisamatsu, Y.; Okada, H. Resveratrol protects mitochondrial quantity by activating SIRT1/PGC-1α expression during ovarian hypoxia. Reprod. Med. Biol. 2020, 19, 189–197. [Google Scholar] [CrossRef]
- Wu, S.; Li, X.; Meng, S.; Fung, T.; Chan, A.T.; Liang, G.; Giovannucci, E.; De Vivo, I.; Lee, J.H.; Nan, H. Fruit and vegetable consumption, cigarette smoke, and leukocyte mitochondrial DNA copy number. Am. J. Clin. Nutr. 2019, 109, 424–432. [Google Scholar] [CrossRef]
- Ma, W.; Hou, C.; Zhou, X.; Yu, H.; Xi, Y.; Ding, J.; Zhao, X.; Xiao, R. Genistein alleviates the mitochondria-targeted DNA damage induced by β-amyloid peptides 25–35 in C6 glioma cells. Neurochem. Res. 2013, 38, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Du, Z.; Li, G.; Chen, M.; Chen, X.; Liang, G.; Chen, T. Curcumin suppresses gastric tumor cell growth via ROS-mediated DNA polymerase γ depletion disrupting cellular bioenergetics. J. Exp. Clin. Cancer Res. 2017, 36, 47. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.-Y.; Zhang, H.-W.; Gu, J.; Xu, F.; Liang, H.-M.; Fan, K.-J.; Shen, J.-Y.; Xiao, Z.-H.; Zhang, E.-Y.; Hu, J. Mitochondrial DNA-induced inflammatory damage contributes to myocardial ischemia reperfusion injury in rats: Cardioprotective role of epigallocatechin. Mol. Med. Rep. 2017, 16, 7569–7576. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Shin, Y.; Jung, S.; Kim, Y. Effects of epigallocatechin-3-gallate on thermogenesis and mitochondrial biogenesis in brown adipose tissues of diet-induced obese mice. Food Nutr. Res. 2017, 61, 1325307. [Google Scholar] [CrossRef]
- Cioffi, F.; Senese, R.; Lasala, P.; Ziello, A.; Mazzoli, A.; Crescenzo, R.; Liverini, G.; Lanni, A.; Goglia, F.; Iossa, S. Fructose-Rich Diet Affects Mitochondrial DNA Damage and Repair in Rats. Nutrients 2017, 9, 323. [Google Scholar] [CrossRef]
- Bai, Y.; Carrillo, J.A.; Li, Y.; He, Y.; Song, J. Diet induced the change of mtDNA copy number and metabolism in Angus cattle. J. Anim. Sci. Biotechnol. 2020, 11, 84. [Google Scholar] [CrossRef]
- Liao, K.; Yan, J.; Mai, K.; Ai, Q. Dietary lipid concentration affects liver mitochondrial DNA copy number, gene expression and DNA methylation in large yellow croaker (Larimichthys crocea). Comp. Biochem. Physiol. Part B. Biochem. Mol. Biol. 2016, 193, 25–32. [Google Scholar] [CrossRef]
- Min, K.-H.; Lee, W. Alteration of mitochondrial DNA content modulates antioxidant enzyme expressions and oxidative stress in myoblasts. Korean J. Physiol. Pharmacol. 2019, 23, 519–528. [Google Scholar] [CrossRef]
- Wallace, D.C.; Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 2010, 10, 12–31. [Google Scholar] [CrossRef]
- Corsi, S.; Iodice, S.; Shannon, O.; Siervo, M.; Mathers, J.; Bollati, V.; Byun, H.-M. Mitochondrial DNA methylation is associated with Mediterranean diet adherence in a population of older adults with overweight and obesity. In Proceedings of the 13th European Nutrition Conference on Malnutrition in an Obese World: European Perspectives, Dublin, Ireland, 15–18 October 2019; Volume 79. [Google Scholar] [CrossRef]
- Mechta, M.; Ingerslev, L.R.; Fabre, O.; Picard, M.; Barrès, R. Evidence Suggesting Absence of Mitochondrial DNA Methylation. Front. Genet. 2017, 8, 166. [Google Scholar] [CrossRef]
- Hong, E.E.; Okitsu, C.Y.; Smith, A.D.; Hsieh, C.-L. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol. Cell. Biol. 2013, 33, 2683–2690. [Google Scholar] [CrossRef] [PubMed]
- Owa, C.; Poulin, M.; Yan, L.; Shioda, T. Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: Sources and avoidance of false-positive detection. PLoS ONE 2018, 13, e0192722. [Google Scholar] [CrossRef] [PubMed]
- Maresca, A.; Del Dotto, V.; Capristo, M.; Scimonelli, E.; Tagliavini, F.; Morandi, L.; Tropeano, C.V.; Caporali, L.; Mohamed, S.; Roberti, M.; et al. DNMT1 mutations leading to neurodegeneration paradoxically reflect on mitochondrial metabolism. Hum. Mol. Genet. 2020, 29, 1864–1881. [Google Scholar] [CrossRef] [PubMed]
- Wesselink, E.; Koekkoek, W.A.C.; Grefte, S.; Witkamp, R.F.; van Zanten, A.R.H. Feeding mitochondria: Potential role of nutritional components to improve critical illness convalescence. Clin. Nutr. 2019, 38, 982–995. [Google Scholar] [CrossRef]
- McClave, S.A.; Wischmeyer, P.E.; Miller, K.R.; van Zanten, A.R.H. Mitochondrial Dysfunction in Critical Illness: Implications for Nutritional Therapy. Curr. Nutr. Rep. 2019, 8, 363–373. [Google Scholar] [CrossRef]
- Guo, Q.; Li, F.; Duan, Y.; Wen, C.; Wang, W.; Zhang, L.; Huang, R.; Yin, Y. Oxidative stress, nutritional antioxidants and beyond. Sci. China Life Sci. 2020, 63, 866–874. [Google Scholar] [CrossRef]
- Cobley, J.N.; Moult, P.R.; Burniston, J.G.; Morton, J.P.; Close, G.L. Exercise improves mitochondrial and redox-regulated stress responses in the elderly: Better late than never! Biogerontology 2015, 16, 249–264. [Google Scholar] [CrossRef]
- Joseph, A.-M.; Adhihetty, P.J.; Leeuwenburgh, C. Beneficial effects of exercise on age-related mitochondrial dysfunction and oxidative stress in skeletal muscle. J. Physiol. 2016, 594, 5105–5123. [Google Scholar] [CrossRef]
- Huertas, J.R.; Casuso, R.A.; Agustín, P.H.; Cogliati, S. Stay Fit, Stay Young: Mitochondria in Movement: The Role of Exercise in the New Mitochondrial Paradigm. Oxid. Med. Cell. Longev. 2019, 2019, 1–18. [Google Scholar] [CrossRef]
- Petracci, I.; Gabbianelli, R.; Bordoni, L. The Role of Nutri(epi)genomics in Achieving the Body’s Full Potential in Physical Activity. Antioxidants 2020, 9, 498. [Google Scholar] [CrossRef]
- Corsi, S.; Iodice, S.; Vigna, L.; Cayir, A.; Mathers, J.C.; Bollati, V.; Byun, H.-M. Platelet mitochondrial DNA methylation predicts future cardiovascular outcome in adults with overweight and obesity. Clin. Epigenet. 2020, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Iacobazzi, V.; Castegna, A.; Infantino, V.; Andria, G. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol. Genet. Metab. 2013, 110, 25–34. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).