Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions
Abstract
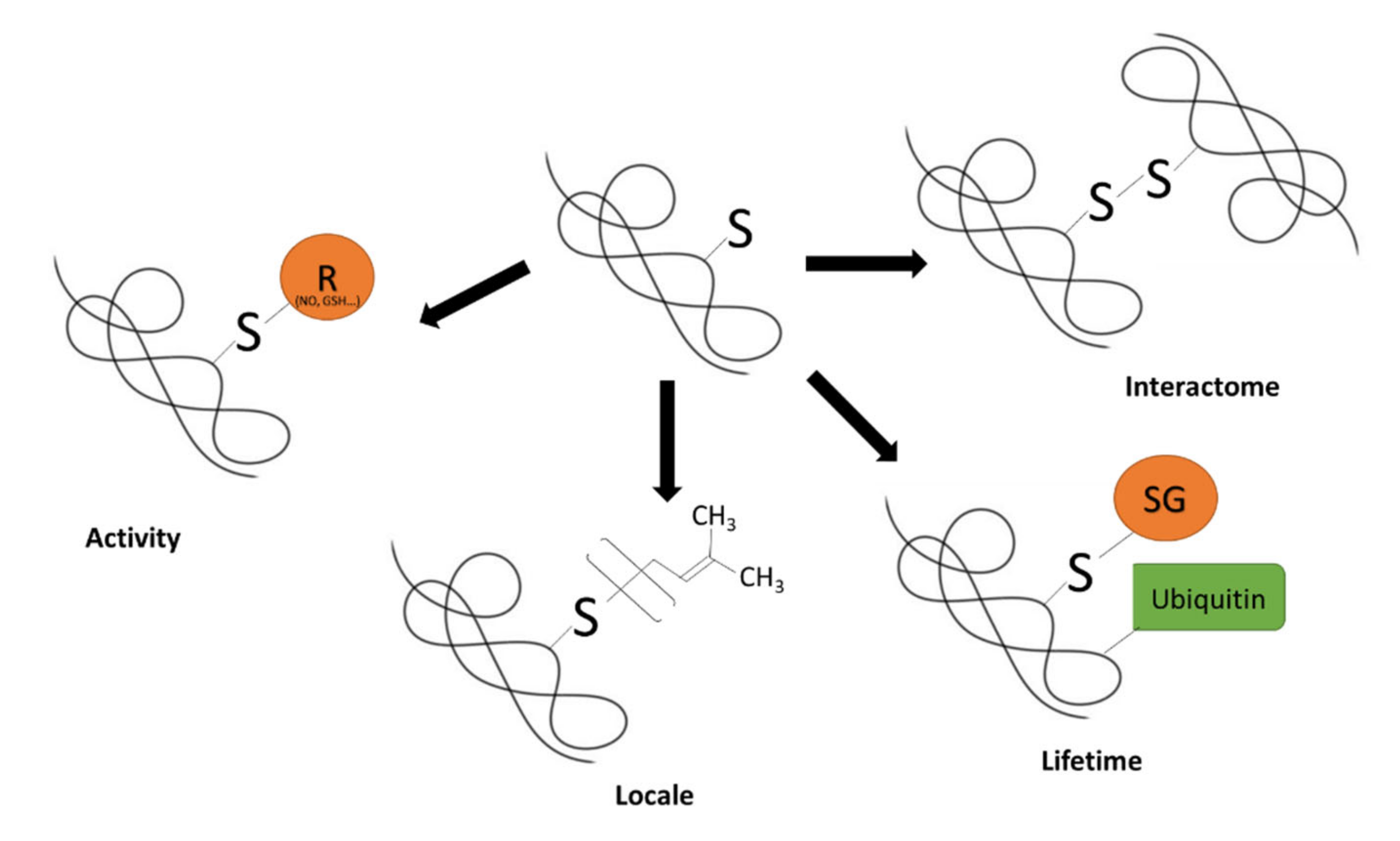
1. Protein Thiol Redox Biology: An Overview
| Modification | Example Reaction | Enzyme Regulation | Selected Examples |
|---|---|---|---|
| Sulfenic acid (SOH) | RS− + H2O2 → RSOH + H2O | Thioredoxin/PRDX. | EGF receptor SOH at Cys797 potentates tyrosine kinase activity [66]. Src Cys185 and 277 SOH enhances protein activity [67]. |
| Thiyl radical (RS•) | RS− + NO2• → RS• + NO2− | n/a | RS. play a role in the reversible oxidation of thiols to RSSG in NDUFV1 and NDUFS1 in complex I [68], which can inhibit enzyme activity [69,70,71,72,73]. A catalytic RS. enables ribonucleotide reductase to remove an oxygen atom from the 2-OH position in ribose to yield deoxyribose [74]. |
| Disulfide bonds (RSSR) | RSOH + RSH → RSSR + H2O | Thioredoxin isoforms. | Intermolecular RSSR moieties activate ATM dimers to initiate DNA double strand break repair [75]. RSSR exchange between PRDX2 and STAT3 represses STAT3 transcriptional activity [53]. RSSR of plasma membrane bound HPLAC1 enables plants to sense and respond to extracellular H2O2 [76]. |
| Glutathionylation (RSSG) | RS− + GSSG → RSSG + GS− | Glutaredoxin isoforms. | eNOS RSSG at multiple sites enhances uncoupling mediated superoxide production [77]. SMYD2 Cys13 RSSG dissociates SYMD2 from titin leading to sarcomere instability [78]. |
| S-nitrosation (RSNO) | RS• + NO• → RSNO | Protein mediated NO• transfer. | ND3 Cys39 RNSO holds complex I inactive to prevent oxidative damage in ischemia reperfusion injury [79,80,81]. LURE1 RSNO inhibits polyspermy in flowering plants [82]. |
2. The Case for Using Immunological Techniques to Assess Protein Thiol Redox State
3. Novel Immunological Techniques to Assess Protein Thiol Redox State
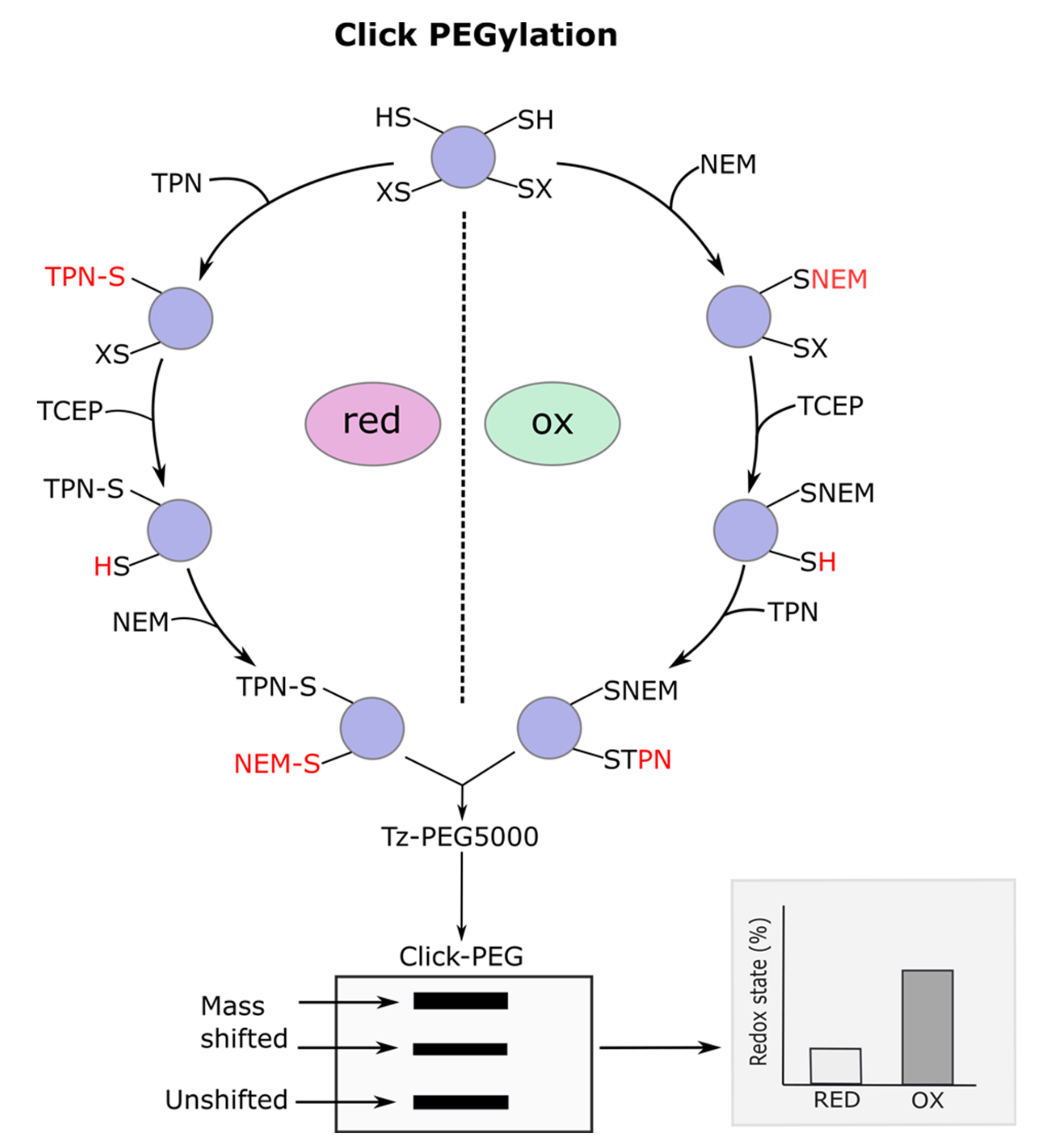
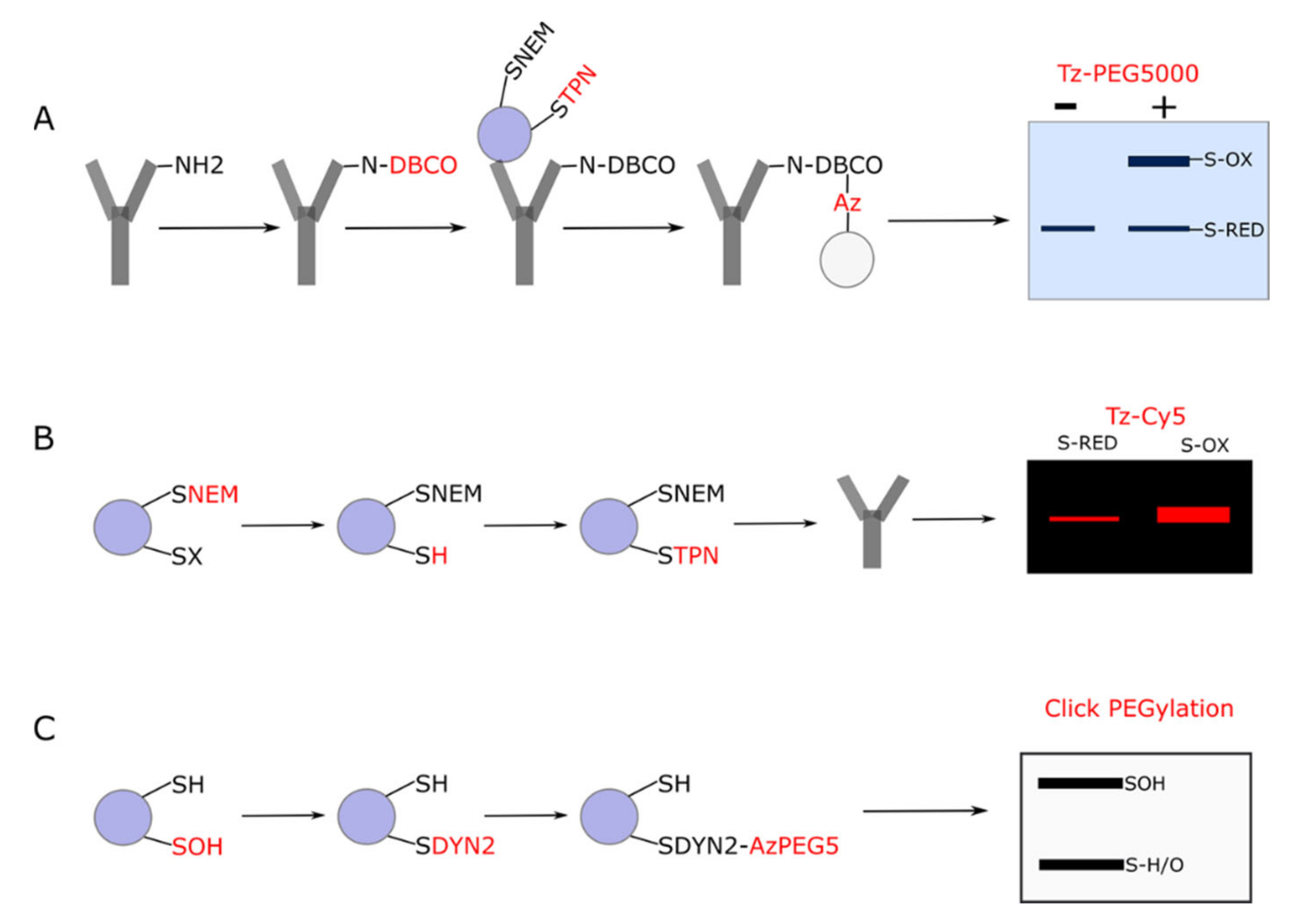
3.1. Click PEGylation
3.2. Underlying Principles
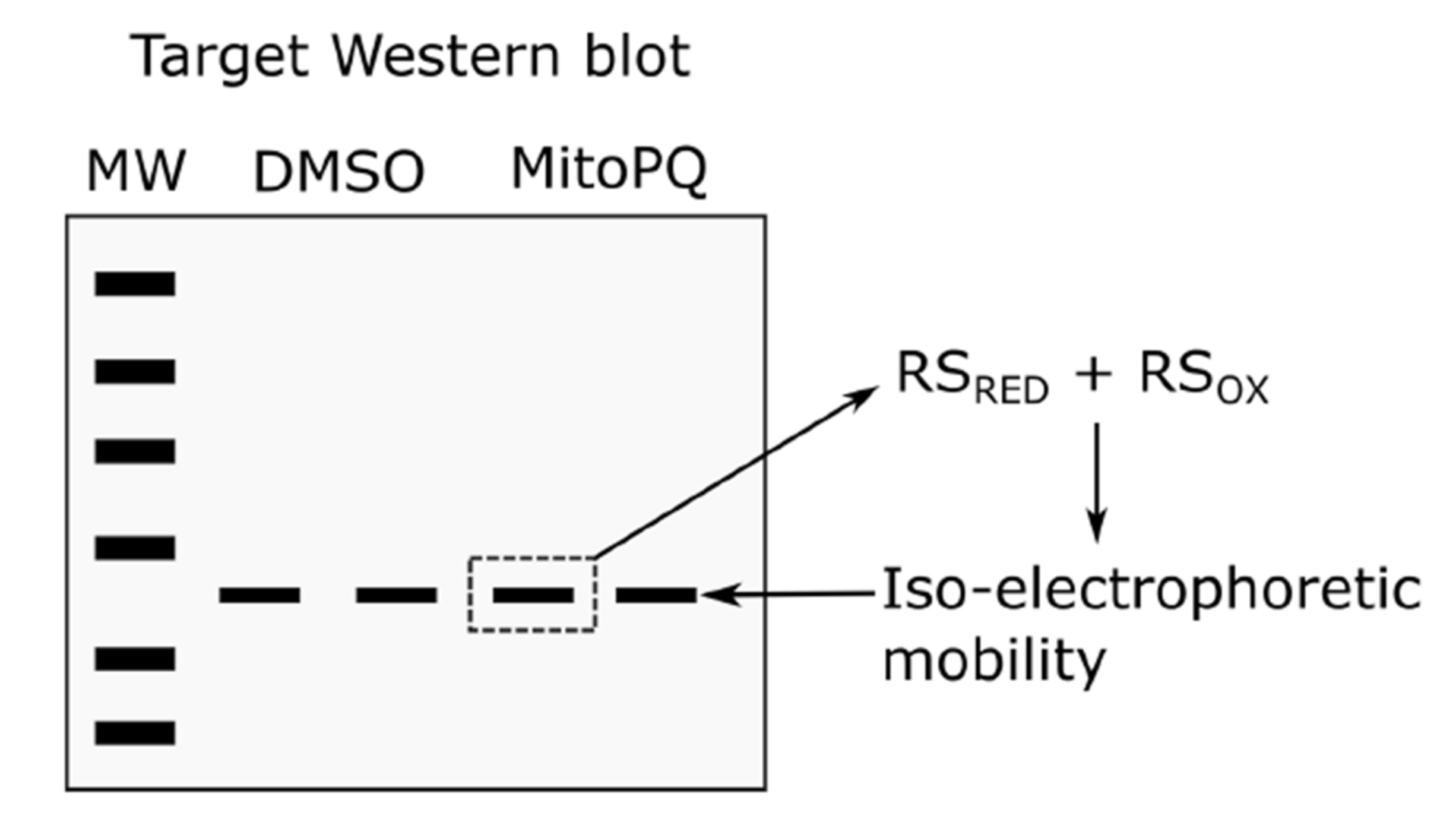
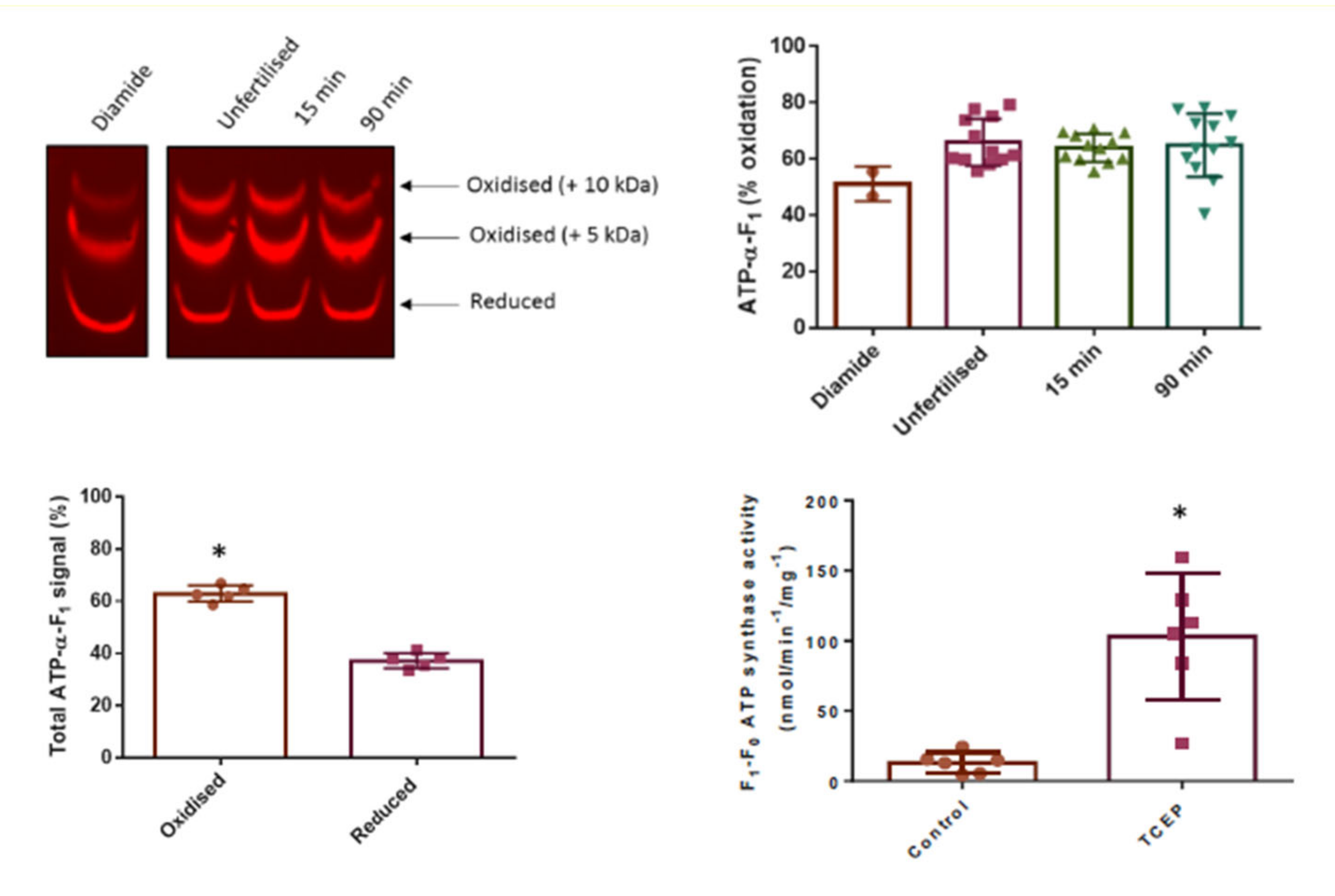
3.3. Click PEGylation Is A Useful Tool to Assess Protein Thiol Redox State: A Mitochondrial ATP Synthase Case Study
3.4. Challenges
3.5. Solutions
4. Opportunities: How to Use Click PEGylation to Advance Knowledge of Redox Biology
5. A Concluding Perspective and Recommendations
- Identify the number and solvent exposure (if structural models allow) of the target protein thiol, as well as, protein mass to rationally select the PEG size and gel percentage. Note some thiols can selectively become exposed to solvent (termed cryptic thiols).
- Select polyclonal antibodies if possible or monoclonal antibodies distal to the PEGylated thiols.
- Carefully prepare reaction buffers (e.g., pH in lysis buffer and SDS in Laemmli buffer) and consider using TCEP as a generic reductant to avoid sulfur autoxidation artefacts.
- Include an unreacted control with and without DTT in the Laemmli buffer to assess target recognition and RSSR mediated oligomeric mobility shifts, respectively.
- Consider direct reaction strategies to assess SOH occupancy.
- Judiciously appraise the merits of selective reduction strategies (they can affect other modification types).
- Perform reciprocal reduced and reversibly oxidized Click PEGylation analysis.
- If reciprocal reduced and reversibly oxidized Click PEGylation implies SO2/SO3 occupancy, then consider direct reaction strategies or mass shifting reduced and reversibly oxidized proteins together to confirm. Optionally, use a recombinant sulfiredoxin reduction system.
- Use complementary redox proteomics (e.g., MRM) and functional assays (e.g., enzyme assay) if possible.
- If Click PEGylation fails, consider antibody first workflows.
Author Contributions
Funding
Conflicts of Interest
References
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef]
- Murphy, M.P.; Holmgren, A.; Larsson, N.-G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.C.; Chang, C.J. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011, 7, 504–511. [Google Scholar] [CrossRef]
- Gutteridge, J.M.C.; Halliwell, B. Antioxidants: Molecules, medicines, and myths. Biochem. Biophys. Res. Commun. 2010, 393, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative Stress; Academic Press: London, UK, 1985. [Google Scholar]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Giorgio, M.; Trinei, M.; Migliaccio, E.; Pelicci, P. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007, 8, 722–728. [Google Scholar] [CrossRef]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Gardner, P.; Fridovich, I. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 1991, 266, 19328–19333. [Google Scholar]
- Gerschman, R.; Gilbert, D.L.; Nye, S.; Dwyer, P.; Fenn, W. Oxygen poisoning and x-irradiation: A mechanism in common. Science 1954, 119, 623–626. [Google Scholar] [CrossRef]
- Lane, N. Oxygen: The Molecule that Made the World; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Kettle, A. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 2013, 18, 642–660. [Google Scholar] [CrossRef] [PubMed]
- Babior, B.M.; Lambeth, J.D.; Nauseef, W. The neutrophil NADPH oxidase. Arch. Biochem. Biophys. 2002, 397, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Booth, D.M.; Enyedi, B.; Geiszt, M.; Varnai, P.; Hajnoczky, G. Redox Nanodomains Are Induced by and Control Calcium Signaling at the ER-Mitochondrial Interface. Mol. Cell 2016, 63, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Carroll, K.S. Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Long, M.J.C.; Poganik, J.R.; Aye, Y. Redox signaling by reactive electrophiles and oxidants. Chem. Rev. 2018, 118, 8798–8888. [Google Scholar] [CrossRef] [PubMed]
- Fukuto, J.M.; Carrington, S.J.; Tantillo, D.J.; Harrison, J.G.; Ignarro, L.J.; Freeman, B.A.; Chen, A.; Wink, D.A. Small molecule signaling agents: The integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 2012, 25, 769–793. [Google Scholar] [CrossRef]
- Li, Q.; Lancaster, J.R. Chemical foundations of hydrogen sulfide biology. Nitric Oxide Biol. Chem. 2013, 35, 21–34. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H. H 2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Cortese-Krott, M.M.; Koning, A.; Kuhnle, G.G.C.; Nagy, P.; Bianco, C.L.; Pasch, A.; Wink, D.A.; Fukuto, J.M.; Jackson, A.A.; Van Goor, H.; et al. The Reactive Species Interactome: Evolutionary Emergence, Biological Significance, and Opportunities for Redox Metabolomics and Personalized Medicine. Antioxidants Redox Signal. 2017, 27, 684–712. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. Mitochondrial Thiols in Antioxidant Protection and Redox Signaling: Distinct Roles for Glutathionylation and Other Thiol Modifications. Antioxid. Redox Signal. 2012, 16, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Collins, Y.; Chouchani, E.T.; James, A.M.; Menger, K.E.; Cocheme, H.M.; Murphy, M.P. Mitochondrial redox signalling at a glance. J. Cell Sci. 2012, 125, 1837. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Cobley, J.N.; Fiorello, M.-L.; Bailey, D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef]
- Ulrich, K.; Jakob, U. The role of thiols in antioxidant systems. Free Radic. Biol. Med. 2019, 140, 14–27. [Google Scholar] [CrossRef]
- Toledo, J.; Augusto, O. Connecting the Chemical and Biological Properties of Nitric Oxide. Chem. Res. Toxicol. 2012, 25, 975–989. [Google Scholar] [CrossRef]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin Evolution and the Regulation of Hydrogen Peroxide Signaling. Science (80) 2003, 300, 650–653. [Google Scholar] [CrossRef]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020. In press. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Pinto, S.; Getnet, D.; Nirujogi, R.; Manda, S.; Chaerkady, R.; Madugundu, A.; Kelkar, D.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.M.; Gladyshev, V.N. Cysteine Function Governs Its Conservation and Degeneration and Restricts Its Utilization on Protein Surfaces. J. Mol. Biol. 2010, 404, 902–916. [Google Scholar] [CrossRef] [PubMed]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.D.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Leichert, L.I.; Dick, T.P. Incidence and physiological relevance of protein thiol switches. Biol. Chem. 2015, 396, 389–399. [Google Scholar] [CrossRef]
- Miseta, A.; Csutora, P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef]
- Forman, H.J.; Augusto, O.; Brigelius-Flohe, R.; Dennery, P.A.; Kalyanaraman, B.; Ischiropoulos, H.; Mann, G.E.; Radi, R.; Roberts, L.J.; Vina, J.; et al. Even free radicals should follow some rules: A Guide to free radical research terminology and methodology. Free Radic. Biol. Med. 2015, 78, 233–235. [Google Scholar] [CrossRef]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The cysteine proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef]
- Requejo, R.; Hurd, T.R.; Costa, N.J.; Murphy, M.P. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 2010, 277, 1465–1480. [Google Scholar] [CrossRef]
- Forman, H.J.; Maiorino, M.; Ursini, F. Signaling functions of reactive oxygen species. Biochemistry 2010, 49, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Adimora, N.; Jones, D.P.; Kemp, M. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid. Redox Signal. 2010, 13, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Peralta, D.; Bronowska, A.K.; Morgan, B.; Dóka, É.; Van Laer, K.; Nagy, P.; Gräter, F.; Dick, T.P. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 2015, 11, 156–163. [Google Scholar] [CrossRef]
- Nagy, P.; Karton, A.; Betz, A.; Peskin, A.V.; Pace, P.; O’Reilly, R.J.; Hampton, M.B.; Radom, L.; Winterbourn, C.C. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: A kinetic and computational study. J. Biol. Chem. 2011, 286, 18048–18055. [Google Scholar] [CrossRef]
- Karplus, P.A. A primer on peroxiredoxin biochemistry. Free Radic. Biol. Med. 2015, 80, 183–190. [Google Scholar] [CrossRef]
- Zeida, A.; Trujillo, M.; Ferrer-Sueta, G.; Denicola, A.; Estrin, D.A.; Radi, R. Catalysis of Peroxide Reduction by Fast Reacting Protein Thiols. Chem. Rev. 2019, 119, 10829–10855. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohé, R.; Flohé, L. Basic Principles and Emerging Concepts in the Redox Control of Transcription Factors. Antioxid. Redox Signal. 2011, 15, 2335–2381. [Google Scholar] [CrossRef] [PubMed]
- Flohé, L.; Toppo, S.; Cozza, G.; Ursini, F. A comparison of thiol peroxidase mechanisms. Antioxid. Redox Signal. 2011, 15, 763–780. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.A.; Yim, S.H.; Shin, D.H.; Kang, D.; Yu, D.Y.; Rhee, S.G. Inactivation of Peroxiredoxin I by Phosphorylation Allows Localized H2O2 Accumulation for Cell Signaling. Cell 2010, 140, 517–528. [Google Scholar] [CrossRef]
- Sobotta, M.C.; Liou, W.; Stöcker, S.; Talwar, D.; Oehler, M.; Ruppert, T.; Scharf, A.N.D.; Dick, T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015, 11, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Stöcker, S.; Van Laer, K.; Mijuskovic, A.; Dick, T.P. The Conundrum of Hydrogen Peroxide Signaling and the Emerging Role of Peroxiredoxins as Redox Relay Hubs. Antioxid. Redox Signal. 2018, 28, 558–573. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Woo, H.A.; Kang, D. The Role of Peroxiredoxins in the Transduction. Antioxid. Redox Signal. 2018, 28, 537–557. [Google Scholar] [CrossRef] [PubMed]
- Travasso, R.D.M.; Sampaio dos Aidos, F.; Bayani, A.; Abranches, P.; Salvador, A. Localized Redox Relays as a Privileged Mode of Hydrogen Peroxide Signaling. Redox Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Are free radicals involved in thiol-based redox signaling? Free Radic. Biol. Med. 2015, 80, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 2013, 288, 26464–26472. [Google Scholar] [CrossRef]
- Hurd, T.R.; Costa, N.J.; Dahm, C.C.; Beer, S.M.; Brown, S.E.; Filipovska, A. Murphy Glutathionylation of Mitochondrial Proteins. Antioxid. Redox Signal. 2005, 7, 999–1010. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Superoxide as an intracellular radical sink. Free Radic. Biol. Med. 1993, 14, 85–90. [Google Scholar] [CrossRef]
- Reczek, C.R.; Chandel, N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33, 8–13. [Google Scholar] [CrossRef]
- Janssen-Heininger, Y.M.W.; Mossman, B.T.; Heintz, N.H.; Forman, H.J.; Kalyanaraman, B.; Finkel, T.; Stamler, J.S.; Rhee, S.G.; van der Vliet, A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 2008, 45, 1–17. [Google Scholar] [CrossRef]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Signal. 2015, 23, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Antunes, F.; Brito, P.M. Quantitative biology of hydrogen peroxide signaling. Redox Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- James, A.M.; Hoogewijs, K.; Logan, A.; Hall, A.R.; Ding, S.; Fearnley, I.M.; Murphy, M.P. Non-enzymatic N-acetylation of Lysine Residues by AcetylCoA Often Occurs via a Proximal S-acetylated Thiol Intermediate Sensitive to Glyoxalase II. Cell Rep. 2017, 18, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Truong, T.H.; Garcia, F.J.; Homann, A.; Gupta, V.; Leonard, S.E.; Carroll, K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2012, 8, 57–64. [Google Scholar] [CrossRef]
- Heppner, D.E.; Dustin, C.M.; Liao, C.; Hristova, M.; Veith, C.; Little, A.C.; Ahlers, B.A.; White, S.L.; Deng, B.; Lam, Y.W.; et al. Direct cysteine sulfenylation drives activation of the Src kinase. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Kang, P.T.; Zhang, L.; Chen, C.L.; Chen, J.; Green, K.B.; Chen, Y.R. Protein thiyl radical mediates S-glutathionylation of complex i. Free Radic. Biol. Med. 2012, 53, 962–973. [Google Scholar] [CrossRef]
- Taylor, E.R.; Hurrell, F.; Shannon, R.J.; Lin, T.K.; Hirst, J.; Murphy, M.P. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 2003, 278, 19603–19610. [Google Scholar] [CrossRef]
- Hurd, T.R.; Requejo, R.; Filipovska, A.; Brown, S.; Prime, T.A.; Robinson, A.J.; Fearnley, I.M.; Murphy, M.P. Complex I within oxidatively stressed bovine heart mitochondria is glutathionylated on Cys-531 and Cys-704 of the 75-kDa subunit: Potential role of Cys residues in decreasing oxidative damage. J. Biol. Chem. 2008, 283, 24801–24815. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Willmore, W.G. S-glutathionylation reactions in mitochondrial function and disease. Front. Cell Dev. Biol. 2014, 2, 1–17. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Treberg, J.R. Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 2016, 8, 110–118. [Google Scholar] [CrossRef]
- Mailloux, R.J. Protein S-glutathionylation reactions as a global inhibitor of cell metabolism for the desensitization of hydrogen peroxide signals. Redox Biol. 2020, 32. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology & Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Guo, Z.; Kozlov, S.; Lavin, M.F.; Person, M.D.; Paull, T.T. ATM activation by oxidative stress. Science (80) 2010, 330, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 2020, 578, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.A.; Wang, T.Y.; Varadharaj, S.; Reyes, L.A.; Hemann, C.; Talukder, M.A.H.; Chen, Y.R.; Druhan, L.J.; Zweier, J.L. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature 2010, 468, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Munkanatta Godage, D.N.P.; VanHecke, G.C.; Samarasinghe, K.T.G.; Feng, H.Z.; Hiske, M.; Holcomb, J.; Yang, Z.; Jin, J.P.; Chung, C.S.; Ahn, Y.H. SMYD2 glutathionylation contributes to degradation of sarcomeric proteins. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Methner, C.; Nadtochiy, S.M.; Logan, A.; Victoria, R.; Ding, S.; James, A.M.; Cochemé, H.M.; Reinhold, J.; Lilley, K.S.; et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013, 19, 753–759. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijević, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Emily, N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; James, A.M.; Work, L.M.; Saeb-parsy, K.; Frezza, C.; Krieg, T.; Murphy, M.P. A Unifying Mechanism for Mitochondrial Superoxide Production during Ischemia-Reperfusion Injury. Cell Metab. 2016, 23, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Liu, M.-C.J.; Kita, D.; Jordan, S.S.; Yeh, F.-L.J.; Yvon, R.; Carpenter, H.; Federico, A.N.; Garcia-Valencia, L.E.; Eyles, S.J.; et al. FERONIA controls pectin- and nitric oxide-mediated male–female interaction. Nature 2020, 579, 1–6. [Google Scholar] [CrossRef]
- Yang, J.; Carroll, K.S.; Liebler, D.C. The Expanding Landscape of the Thiol Redox Proteome. Mol. Cell. Proteomics 2016, 15, 1–11. [Google Scholar] [CrossRef]
- McDonagh, B. Detection of ROS induced proteomic signatures by mass spectrometry. Front. Physiol. 2017, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gould, N.S.; Evans, P.; Martínez-Acedo, P.; Marino, S.M.; Gladyshev, V.N.; Carroll, K.S.; Ischiropoulos, H. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem. Biol. 2015, 22, 965–975. [Google Scholar] [CrossRef]
- Leichert, L.I.; Gehrke, F.; Gudiseva, H.V.; Blackwell, T.; Ilbert, M.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Jakob, U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 8197–8202. [Google Scholar] [CrossRef]
- Go, Y.M.; Roede, J.R.; Walker, D.I.; Duong, D.M.; Seyfried, N.T.; Orr, M.; Liang, Y.; Pennell, K.D.; Jones, D.P. Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol. Cell. Proteomics 2013, 12, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ha, S.; Lee, H.; Lee, K. ROSics: Chemistry and proteomics of cysteine modifications in redox biology. Mass Spectrom. Rev. 2015, 34, 184–208. [Google Scholar] [CrossRef] [PubMed]
- Held, J.M. Redox Systems Biology: Harnessing the Sentinels of the Cysteine Redoxome. Antioxid. Redox Signal. 2020, 32, 659–676. [Google Scholar] [CrossRef]
- Cobley, J.N.; Sakellariou, G.K.; Husi, H.; McDonagh, B. Proteomic strategies to unravel age-related redox signalling defects in skeletal muscle. Free Radic. Biol. Med. 2019, 132, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging. Cell 2020, 180, 968–983. [Google Scholar] [CrossRef] [PubMed]
- Husi, H.; Ward, M.A.; Choudhary, J.S.; Blackstock, W.P.; Grant, S.G.N. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 2000, 3, 661–669. [Google Scholar] [CrossRef]
- Gracia, K.C.; Husi, H. Computational Approaches in Proteomics. In Computational Biology; Husi, H., Ed.; Codon Publications: Brisbane, Australia, 2019. [Google Scholar]
- Fernandes, M.; Sanches, B.; Husi, H. Cheminformatics and Computational Approaches in Metabolomics. In Computational Biology; Husi, H., Ed.; Codon Publications: Brisbane, Australia, 2019; pp. 143–159. [Google Scholar]
- Cervantes Gracia, K.; Llanas-Cornejo, D.; Husi, H. CVD and Oxidative Stress. J. Clin. Med. 2017, 6, 22. [Google Scholar] [CrossRef]
- Picotti, P.; Aebersold, R. Selected reaction monitoring–based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef]
- Held, J.M.; Danielson, S.R.; Behring, J.B.; Atsriku, C.; Britton, D.J.; Puckett, R.L.; Schilling, B.; Campisi, J.; Benz, C.C.; Gibson, B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteomics 2010, 9, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, L.A.G.; Hinchy, E.C.; Murphy, M.P.; Robb, E.L.; Cochemé, H.M. Click-PEGylation—A mobility shift approach to assess the redox state of cysteines in candidate proteins. Free Radic. Biol. Med. 2017, 108, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.R.; Zhu, J.; Hirst, J. Architecture of mammalian respiratory complex I. Nature 2014, 515, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Fandiño, A.S.; Rais, I.; Vollmer, M.; Elgass, H.; Schägger, H.; Karas, M. LC-nanospray-MS/MS analysis of hydrophobic proteins from membrane protein complexes isolated by blue-native electrophoresis. J. Mass Spectrom. 2005, 40, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.P.; Ganini, D. Immuno-spin trapping of macromolecules free radicals in vitro and in vivo – One stop shopping for free radical detection. Free Radic. Biol. Med. 2019, 131, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Low, F.M.; Hampton, M.B.; Peskin, A.V.; Winterbourn, C.C. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood 2007, 109, 2611–2617. [Google Scholar] [CrossRef]
- Woo, H.A.; Kang, S.W.; Kim, H.K.; Yang, K.S.; Chae, H.Z.; Rhee, S.G. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem. 2003, 278, 47361–47364. [Google Scholar] [CrossRef]
- Chae, H.Z.; Robison, K.; Poole, L.B.; Church, G.; Storz, G.; Rhee, S.G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: Alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 1994, 91, 7017–7021. [Google Scholar] [CrossRef]
- Robb, E.L.; Gawel, J.M.; Aksentijevic, D.; Cocheme, H.M.; Stewart, T.S.; Shchepinova, M.M.; Qiang, H.; Prime, T.A.; Bright, T.P.; James, A.M.; et al. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 2015, 89, 883–894. [Google Scholar] [CrossRef]
- Kussmaul, L.; Hirst, J. The mechanism of superoxide production by NADH: Ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 7607–7612. [Google Scholar] [CrossRef]
- Dröse, S.; Brandt, U.; Wittig, I. Mitochondrial respiratory chain complexes as sources and targets of thiol-based redox-regulation. Biochim. Biophys. Acta Proteins Proteomics 2014, 1844, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.G.; Winterbourn, C.C.; Hampton, M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2010, 425, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Brennan, J.P.; Miller, J.I.A.; Fuller, W.; Wait, R.; Begum, S.; Dunn, M.J.; Eaton, P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol. Cell. Proteomics 2006, 5, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Oviosu, O.; Eaton, P. The PEG-switch assay: A fast semi-quantitative method to determine protein reversible cysteine oxidation. J. Pharmacol. Toxicol. Methods 2013, 68, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Hurd, T.R.; Collins, Y.; Abakumova, I.; Chouchani, E.T.; Baranowski, B.; Fearnley, I.M.; Prime, T.A.; Murphy, M.P.; James, A.M. Inactivation of pyruvate dehydrogenase kinase 2 by mitochondrial reactive oxygen species. J. Biol. Chem. 2012, 287, 35153–35160. [Google Scholar] [CrossRef]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.T.; Dou, Y.; et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef]
- Rostovtsev, V.; Green, L.; Fokin, V.; Sharpless, K. A Stepwise Huisgen Cycloaddition Process Catalyzed by Copper (I): Regioselective Ligation of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Jewett, J.; Bertozzi, C. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010, 39, 1272–1279. [Google Scholar] [CrossRef]
- Cobley, J.N.; Noble, A.; Jimenez-Fernandez, E.; Valdivia Moya, M.T.; Guille, M.; Husi, H. Catalyst-free Click PEGylation reveals substantial mitochondrial ATP synthase sub-unit alpha oxidation before and after fertilisation. Redox Biol. 2019, 26, 101258. [Google Scholar] [CrossRef]
- Müller, T.; Winter, D. Systematic evaluation of protein reduction and alkylation reveals massive unspecific side effects by iodine-containing reagents. Mol. Cell. Proteomics 2017, 16, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Matsumoto, A.; Kim, S.-O.O.; Marshall, H.E.; Stamler, J.S. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.T.; Blackman, M.L.; Dmitrenko, O.; Fox, J.M. Design and synthesis of highly reactive dienophiles for the tetrazine-trans-cyclooctene ligation. J. Am. Chem. Soc. 2011, 133, 9646–9649. [Google Scholar] [CrossRef] [PubMed]
- Darko, A.; Wallace, S.; Dmitrenko, O.; Machovina, M.M.; Mehl, R.A.; Chin, J.W.; Fox, J.M. Conformationally strained trans-cyclooctene with improved stability and excellent reactivity in tetrazine ligation. Chem. Sci. 2014, 5, 3770–3776. [Google Scholar] [CrossRef] [PubMed]
- Ogilby, P.R. Singlet oxygen: There is indeed something new under the sun. Chem. Soc. Rev. 2010, 39, 3181–3209. [Google Scholar] [CrossRef]
- Diels, O.; Alder, K. Synthesen in der hydroaromatischen Reihe. Justus Liebigs Ann. Chem. 1928, 460, 98–122. [Google Scholar] [CrossRef]
- Blackman, M.L.; Royzen, M.; Fox, J.M. Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels—Alder Reactivity. J. Am. Chem. Soc. 2008, 130, 13518–13519. [Google Scholar] [CrossRef]
- Oliveira, B.L.; Guo, Z.; Bernardes, G.J.L. Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 2017, 46, 4895–4950. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef]
- Nicholls, D.G.; Ferguson, S.J. Bioenergetics 4, 4th ed.; Academic Press: London, UK, 2013. [Google Scholar]
- Walker, J.E. The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013, 41, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Hatefi, Y. Thiols in Oxidative Phosphorylation: Inhibition and Energy-Potentiated Uncoupling by Monothiol and Dithiol Modifiers. Biochemistry 1984, 23, 2449–2455. [Google Scholar] [CrossRef] [PubMed]
- Kaludercic, N.; Giorgio, V. The dual function of reactive oxygen/nitrogen species in bioenergetics and cell death: The role of ATP synthase. Oxid. Med. Cell. Longev. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.B.; Foster, D.B.; Rucker, J.; O’Rourke, B.; Kass, D.A.; Van Eyk, J.E. Redox regulation of mitochondrial ATP synthase: Implications for cardiac resynchronization therapy. Circ. Res. 2011, 109, 750–757. [Google Scholar] [CrossRef]
- Wang, S.B.; Murray, C.I.; Chung, H.S.; Van Eyk, J.E. Redox Regulation of Mitochondrial ATP Synthase. Trends Cardiovasc. Med. 2013, 23, 18. [Google Scholar] [CrossRef]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 1–15. [Google Scholar] [CrossRef]
- Harland, R.M.; Grainger, R.M. Xenopus research: Metamorphosed by genetics and genomics. Trends Genet. 2011, 27, 507–515. [Google Scholar] [CrossRef]
- Sidlauskaite, E.; Gibson, J.W.; Megson, I.L.; Whitfield, P.D.; Tovmasyan, A.; Batinic-Haberle, I.; Murphy, M.P.; Moult, P.R.; Cobley, J.N. Mitochondrial ROS cause motor deficits induced by synaptic inactivity: Implications for synapse pruning. Redox Biol. 2018, 16, 344–351. [Google Scholar] [CrossRef]
- Nalin, C.M.; McCarty, R.E. Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J. Biol. Chem. 1984, 259, 7275–7280. [Google Scholar]
- Garcia, J.; Han, D.; Sancheti, H.; Yap, L.P.; Kaplowitz, N.; Cadenas, E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J. Biol. Chem. 2010, 285, 39646–39654. [Google Scholar] [CrossRef]
- Cobley, J.; Noble, A.; Bessell, R.; Guille, M.; Husi, H. Reversible Thiol Oxidation Inhibits the Mitochondrial ATP Synthase in Xenopus laevis Oocytes. Antioxidants 2020, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.M.; Jones, D.P.; Harris, C. The Redox Theory of Development. Antioxid. Redox Signal. 2020, 32, 715–740. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.G.; Balin, A. Oxidative influence on development and differentiation: An overview of a free radical theory of development. Free Radic. Biol. Med. 1989, 6, 631–661. [Google Scholar] [CrossRef]
- Hitchler, M.J.; Domann, F.E. An epigenetic perspective on the free radical theory of development. Free Radic. Biol. Med. 2007, 43, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. Synapse pruning: Mitochondrial ROS with their hands on shears. BioEssays 2018, 40, e1800031. [Google Scholar] [CrossRef] [PubMed]
- Galkin, A.; Meyer, B.; Wittig, I.; Karas, M.; Schägger, H.; Vinogradov, A.; Brandt, U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 2008, 283, 20907–20913. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Chang, G.-D. Quantitative display of the redox status of proteins with maleimide-polyethylene glycol tagging. Electrophoresis 2019, 40, 491–498. [Google Scholar] [CrossRef]
- Zheng, C.Y.; Ma, G.; Su, Z. Native PAGE eliminates the problem of PEG-SDS interaction in SDS-PAGE and provides an alternative to HPLC in characterization of protein PEGylation. Electrophoresis 2007, 28, 2801–2807. [Google Scholar] [CrossRef] [PubMed]
- Wittig, I.; Braun, H.P.; Schägger, H. Blue native PAGE. Nat. Protoc. 2006, 1, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Fu, L.; Jung, Y.; Conte, M.L.; Lawson, J.R.; Lowther, W.T.; Sun, R.; Liu, K.; Yang, J.; Carroll, K.S. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003, 425, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Reisz, J.A.; Bechtold, E.; King, S.B.; Poole, L.B.; Furdui, C.M. Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J. 2013, 280, 6150–6161. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gupta, V.; Carroll, K.S.; Liebler, D.C. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 2014, 5, 4776. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Liu, K.; He, J.; Tian, C.; Yu, X.; Yang, J. Direct Proteomic Mapping of Cysteine Persulfidation. Antioxid. Redox Signal. 2019. [Google Scholar] [CrossRef]
- Heppner, D.E.; Hristova, M.; Ida, T.; Mijuskovic, A.; Dustin, C.M.; Bogdándi, V.; Fukuto, J.M.; Dick, T.P.; Nagy, P.; Li, J.; et al. Cysteine perthiosulfenic acid (Cys-SSOH): A novel intermediate in thiol-based redox signaling? Redox Biol. 2018, 14, 379–385. [Google Scholar] [CrossRef]
- Shi, Y.; Carroll, K.S. Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation. Acc. Chem. Res. 2020, 53, 20–31. [Google Scholar] [CrossRef]
- Burbulla, L.F.; Song, P.; Mazzulli, J.R.; Zampese, E.; Wong, Y.C.; Jeon, S.; Santos, D.P.; Blanz, J.; Obermaier, C.D.; Strojny, C.; et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 2017, 9080, 1–12. [Google Scholar] [CrossRef]
- Anderson, D.G.; Mariappan, S.V.S.; Buettner, G.R.; Doorn, J.A. Oxidation of 3,4-dihydroxyphenylacetaldehyde, a toxic dopaminergic metabolite, to a semiquinone radical and an ortho-quinone. J. Biol. Chem. 2011, 286, 26978–26986. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative stress and neurodegeneration: Where are we now? J. Neurochem. 2006, 97, 1634–1658. [Google Scholar] [CrossRef]
- Bersweiler, A.; D’Autréaux, B.; Mazon, H.; Kriznik, A.; Belli, G.; Delaunay-Moisan, A.; Toledano, M.B.; Rahuel-Clermont, S. A scaffold protein that chaperones a cysteine-sulfenic acid in H2O2 signaling. Nat. Chem. Biol. 2017, 13, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M. The Incomplete Glutathione Puzzle: Just Guessing at Numbers and Figures? Antioxidants Redox Signal. 2017, 27, 1130–1161. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.A.; Wang, B.; Miyagi, M.; Hess, D.T.; Stamler, J.S. Oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor/Ca2+ release channel (RyR1): Sites and nature of oxidative modification. J. Biol. Chem. 2013, 288, 22961–22971. [Google Scholar] [CrossRef] [PubMed]
- Place, N.; Ivarsson, N.; Venckunas, T.; Neyroud, D.; Brazaitis, M.; Cheng, A.J.; Ochala, J.; Kamandulis, S.; Girard, S.; Volungevičius, G.; et al. Ryanodine receptor fragmentation and sarcoplasmic reticulum Ca 2+ leak after one session of high-intensity interval exercise. Proc. Natl. Acad. Sci. USA 2015, 112, 201507176. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Yang, J.; Liebler, D.C.; Carroll, K.S. Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J. Am. Chem. Soc. 2017, 139, 5588–5595. [Google Scholar] [CrossRef] [PubMed]
- Scinto, S.L.; Ekanayake, O.; Seneviratne, U.; Pigga, J.E.; Boyd, S.J.; Taylor, M.T.; Liu, J.; Am Ende, C.W.; Rozovsky, S.; Fox, J.M. Dual-Reactivity trans-Cyclooctenol Probes for Sulfenylation in Live Cells Enable Temporal Control via Bioorthogonal Quenching. J. Am. Chem. Soc. 2019, 141, 10932–10937. [Google Scholar] [CrossRef]
- Samarasinghe, K.T.G.; Munkanatta Godage, D.N.P.; Vanhecke, G.C.; Ahn, Y.H. Metabolic synthesis of clickable glutathione for chemoselective detection of glutathionylation. J. Am. Chem. Soc. 2014, 136, 11566–11569. [Google Scholar] [CrossRef]
- Seneviratne, U.; Godoy, L.C.; Wishnok, J.S.; Wogan, G.N.; Tannenbaum, S.R. Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs. J. Am. Chem. Soc. 2013, 135, 7693–7704. [Google Scholar] [CrossRef]
- Shchepinova, M.M.; Cairns, A.G.; Prime, T.A.; Logan, A.; James, A.M.; Hall, A.R.; Vidoni, S.; Arndt, S.; Caldwell, S.T.; Prag, H.A.; et al. MitoNeoD: A Mitochondria-Targeted Superoxide Probe. Cell Chem. Biol. 2017, 3, 8–21. [Google Scholar] [CrossRef]
- Cochemé, H.M.; Quin, C.; McQuaker, S.J.; Cabreiro, F.; Logan, A.; Prime, T.A.; Abakumova, I.; Patel, J.V.; Fearnley, I.M.; James, A.M.; et al. Measurement of H2O2 within living drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011, 13, 340–350. [Google Scholar] [CrossRef]
- Logan, A.; Cochemé, H.M.; Li Pun, P.B.; Apostolova, N.; Smith, R.A.J.; Larsen, L.; Larsen, D.S.; James, A.M.; Fearnley, I.M.; Rogatti, S.; et al. Using exomarkers to assess mitochondrial reactive species in vivo. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, B.; Darley-Usmar, V.; Davies, K.J.A.; Dennery, P.A.; Forman, H.J.; Grisham, M.B.; Mann, G.E.; Moore, K.; Roberts, L.J.; Ischiropoulos, H. Measuring reactive oxygen and nitrogen species with fluorescent probes: Challenges and limitations. Free Radic. Biol. Med. 2012, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Orr, A.L.; Vargas, L.; Turk, C.N.; Baaten, J.E.; Matzen, J.T.; Dardov, V.J.; Attle, S.J.; Li, J.; Quackenbush, D.C.; Goncalves, R.L.S.; et al. Suppressors of superoxide production from mitochondrial complex III. Nat. Chem. Biol. 2015, 11, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Goncalves, R.L.S.; Orr, A.L.; Vargas, L.; Gerencser, A.A.; Borch Jensen, M.; Wang, Y.T.; Melov, S.; Turk, C.N.; Matzen, J.T.; et al. Suppressors of Superoxide-H2O2Production at Site IQof Mitochondrial Complex I Protect against Stem Cell Hyperplasia and Ischemia-Reperfusion Injury. Cell Metab. 2016, 24, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Bleier, L.; Wittig, I.; Heide, H.; Steger, M.; Brandt, U.; Drose, S. Generator-specific targets of mitochondrial reactive oxygen species. Free Radic. Biol. Med. 2015, 78, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–166. [Google Scholar] [CrossRef]
- Chandel, N.S.; Maltepe, E.; Goldwasser, E.; Mathieu, C.E.; Simon, M.C.; Schumacker, P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 11715–11720. [Google Scholar] [CrossRef]
- Guzy, R.D.; Hoyos, B.; Robin, E.; Chen, H.; Liu, L.; Mansfield, K.D.; Simon, M.C.; Hammerling, U.; Schumacker, P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005, 1, 401–408. [Google Scholar] [CrossRef]
- Schofield, C.J.; Ratcliffe, P.J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004, 5, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Sakellariou, G.K.; Owens, D.J.; Murray, S.; Waldron, S.; Gregson, W.; Fraser, W.D.; Burniston, J.G.; Iwanejko, L.A.; McArdle, A.; et al. Lifelong training preserves some redox-regulated adaptive responses after an acute exercise stimulus in aged human skeletal muscle. Free Radic. Biol. Med. 2014, 70, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; McHardy, H.; Morton, J.P.; Nikolaidis, M.G.; Close, G.L. Influence of vitamin C and vitamin E on redox signalling: Implications for exercise adaptations. Free Radic. Biol. Med. 2015, 84, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Moult, P.R.; Burniston, J.G.; Morton, J.P.; Close, G.L. Exercise improves mitochondrial and redox-regulated stress responses in the elderly: Better late than never! Biogerontology 2015, 16, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Margaritelis, N.V.; Morton, J.P.; Close, G.L.; Nikolaidis, M.G.; Malone, J.K. The basic chemistry of exercise-induced DNA oxidation: Oxidative damage, redox signaling, and their interplay. Front. Physiol. 2015, 6, 1–8. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Cobley, J.N.; Paschalis, V.; Veskoukis, A.S.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Principles for integrating reactive species into in vivo biological processes: Examples from exercise physiology. Cell. Signal. 2016, 28, 256–271. [Google Scholar] [CrossRef]
- Mason, S.A.; Trewin, A.J.; Parker, L.; Wadley, G.D. Antioxidant supplements and endurance exercise: Current evidence and mechanistic insights. Redox Biol. 2020, 101471. [Google Scholar] [CrossRef]
- Henríquez-Olguin, C.; Knudsen, J.R.; Raun, S.H.; Li, Z.; Dalbram, E.; Treebak, J.T.; Sylow, L.; Holmdahl, R.; Richter, E.A.; Jaimovich, E.; et al. Cytosolic ROS production by NADPH oxidase 2 regulates muscle glucose uptake during exercise. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Henriquez-Olguin, C.; Meneses-Valdes, R.; Jensen, T.E. Compartmentalized muscle redox signals controlling exercise metabolism—Current state, future challenges. Redox Biol. 2020, 101473. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Redox basis of exercise physiology. Redox Biol. 2020, 101499. [Google Scholar] [CrossRef]
- Cobley, J.N.; Bartlett, J.D.; Kayani, A.; Murray, S.W.; Louhelainen, J.; Donovan, T.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; et al. PGC-1?? transcriptional response and mitochondrial adaptation to acute exercise is maintained in skeletal muscle of sedentary elderly males. Biogerontology 2012, 13, 621–631. [Google Scholar] [CrossRef]
- Merry, T.L.; Ristow, M. Mitohormesis in exercise training. Free Radic. Biol. Med. 2016, 98, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. How exercise induces oxidative eustress. In Oxidative Stress; Sies, H., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 447–462. [Google Scholar]
- Wadley, A.J.; Aldred, S.; Coles, S.J. An unexplored role for Peroxiredoxin in exercise-induced redox signalling? Redox Biol. 2016, 8, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Malik, Z.A.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Burniston, J.G. Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle. Proteomes 2016, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.A.; Cobley, J.N.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Koch, L.G.; Britton, S.L.; Burniston, J.G. Label-Free LC-MS Profiling of Skeletal Muscle Reveals Heart-Type Fatty Acid Binding Protein as a Candidate Biomarker of Aerobic Capacity. Proteomes 2013, 1, 290–308. [Google Scholar] [CrossRef]
- Burniston, J.G.; Hoffman, E.P. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev. Proteomics 2011, 8, 361–377. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80) 2012, 337, 816–821. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J. A Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef]
- Van Montfort, R.; Congreve, M.; Tisi, D. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 2003, 423, 773–777. [Google Scholar] [CrossRef]
- Kramer, P.A.; Duan, J.; Gaffrey, M.J.; Shukla, A.K.; Wang, L.; Bammler, T.K.; Qian, W.J.; Marcinek, D.J. Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol. 2018, 17, 367–376. [Google Scholar] [CrossRef]
- Cobley, J.N.; Close, G.L.; Bailey, D.M.; Davison, G.W. Exercise redox biochemistry: Conceptual, methodological and technical recommendations. Redox Biol. 2017, 12, 540–548. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, S.; Sekhon, L.; Shah, R. Redox considerations in female reproductive function and assisted reproduction: From molecular mechanisms to health implications. Antioxidants Redox Signal. 2008, 10, 1375–1403. [Google Scholar] [CrossRef] [PubMed]
- Ruder, E.H.; Hartman, T.J.; Blumberg, J.; Goldman, M.B. Oxidative stress and antioxidants: Exposure and impact on female fertility. Hum. Reprod. Update 2008, 14, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Bontekoe, S.; Mantikou, E.; van Wely, M.; Seshadri, S.; Repping, S.; Mastenbroek, S. Low oxygen concentrations for embryo culture in assisted reproductive technologies. Cochrane Database Syst. Rev. 2012. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Oxidative stress in cell culture: An under-appreciated problem? FEBS Lett. 2003, 540, 3–6. [Google Scholar] [CrossRef]
- Halliwell, B. Cell culture, oxidative stress, and antioxidants: Avoiding pitfalls. Biomed. J. 2014. [Google Scholar] [CrossRef]
- Savitsky, P.A.; Finkel, T. Redox regulation of Cdc25C. J. Biol. Chem. 2002, 277, 20535–20540. [Google Scholar] [CrossRef]
- Han, Y.; Ishibashi, S.; Iglesias-Gonzalez, J.; Chen, Y.; Love, N.R.; Amaya, E. Ca2+-Induced Mitochondrial ROS Regulate the Early Embryonic Cell Cycle. Cell Rep. 2018, 22, 218–231. [Google Scholar] [CrossRef]
- Bell, K.F.S.; Al-Mubarak, B.; Martel, M.-A.; McKay, S.; Wheelan, N.; Hasel, P.; Márkus, N.M.; Baxter, P.; Deighton, R.F.; Serio, A.; et al. Neuronal development is promoted by weakened intrinsic antioxidant defences due to epigenetic repression of Nrf2. Nat. Commun. 2015, 6, 7066. [Google Scholar] [CrossRef]
- Dennery, P.A. Oxidative stress in development: Nature or nurture? Free Radic. Biol. Med. 2010, 49, 1147–1151. [Google Scholar] [CrossRef]
- Dick, T.P.; Ralser, M. Metabolic Remodeling in Times of Stress: Who Shoots Faster than His Shadow? Mol. Cell 2015, 59, 519–521. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [PubMed]
- Buettner, G.R. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anticancer. Agents Med. Chem. 2011, 11, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.; Zielonka, J.; Karoui, H.; Sikora, A.; Michalski, R.; Podsiadły, R.; Lopez, M.; Vasquez-Vivar, J.; Kalyanaraman, B.; Ouari, O. Detection and Characterization of Reactive Oxygen and Nitrogen Species in Biological Systems by Monitoring Species-Specific Products. Antioxid. Redox Signal. 2017, ars.2017.7398. [Google Scholar] [CrossRef]
- Zielonka, J.; Kalyanaraman, B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: Another inconvenient truth. Free Radic. Biol. Med. 2010, 48, 983–1001. [Google Scholar] [CrossRef]
- Miller, E.W.; Chang, C.J. Fluorescent probes for nitric oxide and hydrogen peroxide in cell signaling. Curr. Opin. Chem. Biol. 2007, 11, 620–625. [Google Scholar] [CrossRef]
- Belousov, V.; Fradkov, A.; Lukyanov, K.; Staroverov, D.; Shakhbazov, K.; Terskikh, A.; Lukyanov, S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. New tools for redox biology: From imaging to manipulation. Free Radic. Biol. Med. 2017, 109, 167–188. [Google Scholar] [CrossRef]
- Gutscher, M.; Sobotta, M.C.; Wabnitz, G.H.; Ballikaya, S.; Meyer, A.J.; Samstag, Y.; Dick, T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009, 284, 31532–31540. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Dick, T.P.; Meye, A.J.; Morgan, B. Dissecting Redox Biology using Fluorescent Protein Sensors. Antioxid. Redox Signal. 2016, 24, 680–712. [Google Scholar] [CrossRef] [PubMed]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Martínez Gache, S.A.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Whiteman, M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean? Br. J. Pharmacol. 2004, 142, 231–255. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P. Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008, 295, C849–C868. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Cobley, J.N.; Paschalis, V.; Veskoukis, A.S.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Going retro: Oxidative stress biomarkers in modern redox biology. Free Radic. Biol. Med. 2016, 98, 2–12. [Google Scholar] [CrossRef]
- Wardman, P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Radic. Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef]
- Bonini, M.G.; Rota, C.; Tomasi, A.; Mason, R.P. The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: A self-fulfilling prophesy? Free Radic. Biol. Med. 2006, 40, 968–975. [Google Scholar] [CrossRef] [PubMed]

| Advantage | Disadvantage |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cobley, J.N.; Husi, H. Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions. Antioxidants 2020, 9, 315. https://doi.org/10.3390/antiox9040315
Cobley JN, Husi H. Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions. Antioxidants. 2020; 9(4):315. https://doi.org/10.3390/antiox9040315
Chicago/Turabian StyleCobley, James Nathan, and Holger Husi. 2020. "Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions" Antioxidants 9, no. 4: 315. https://doi.org/10.3390/antiox9040315
APA StyleCobley, J. N., & Husi, H. (2020). Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions. Antioxidants, 9(4), 315. https://doi.org/10.3390/antiox9040315





