Visualization of the Redox Status of Cytosolic Glutathione Using the Organelle- and Cytoskeleton-Targeted Redox Sensors
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture and Transfection
2.3. Quantitative Analysis of Glutathione Oxidation
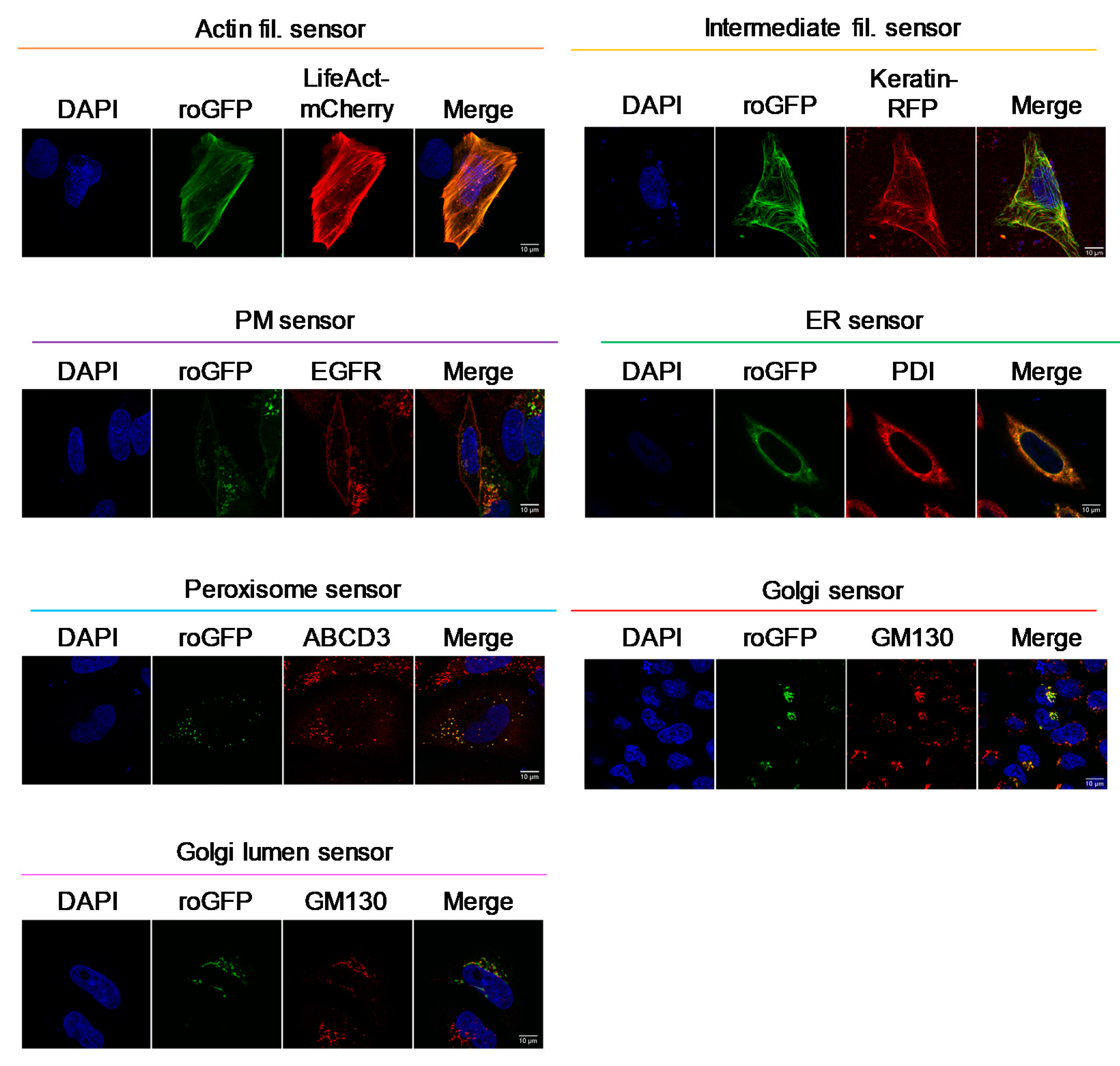
2.4. Immunostaining
3. Results
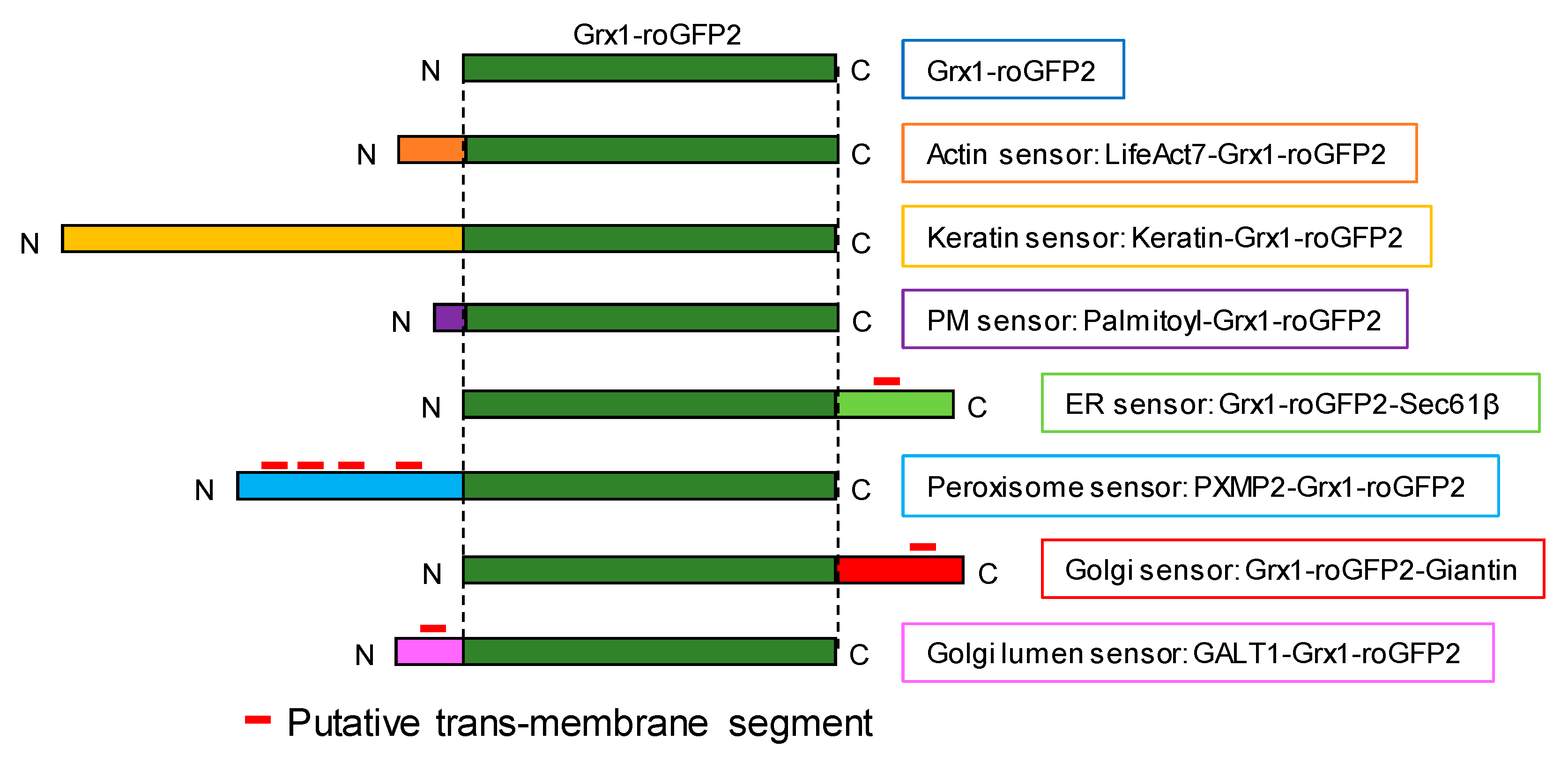
3.1. Targeting Redox Sensors to the Cytoplasmic Sides of Various Organelle Membranes
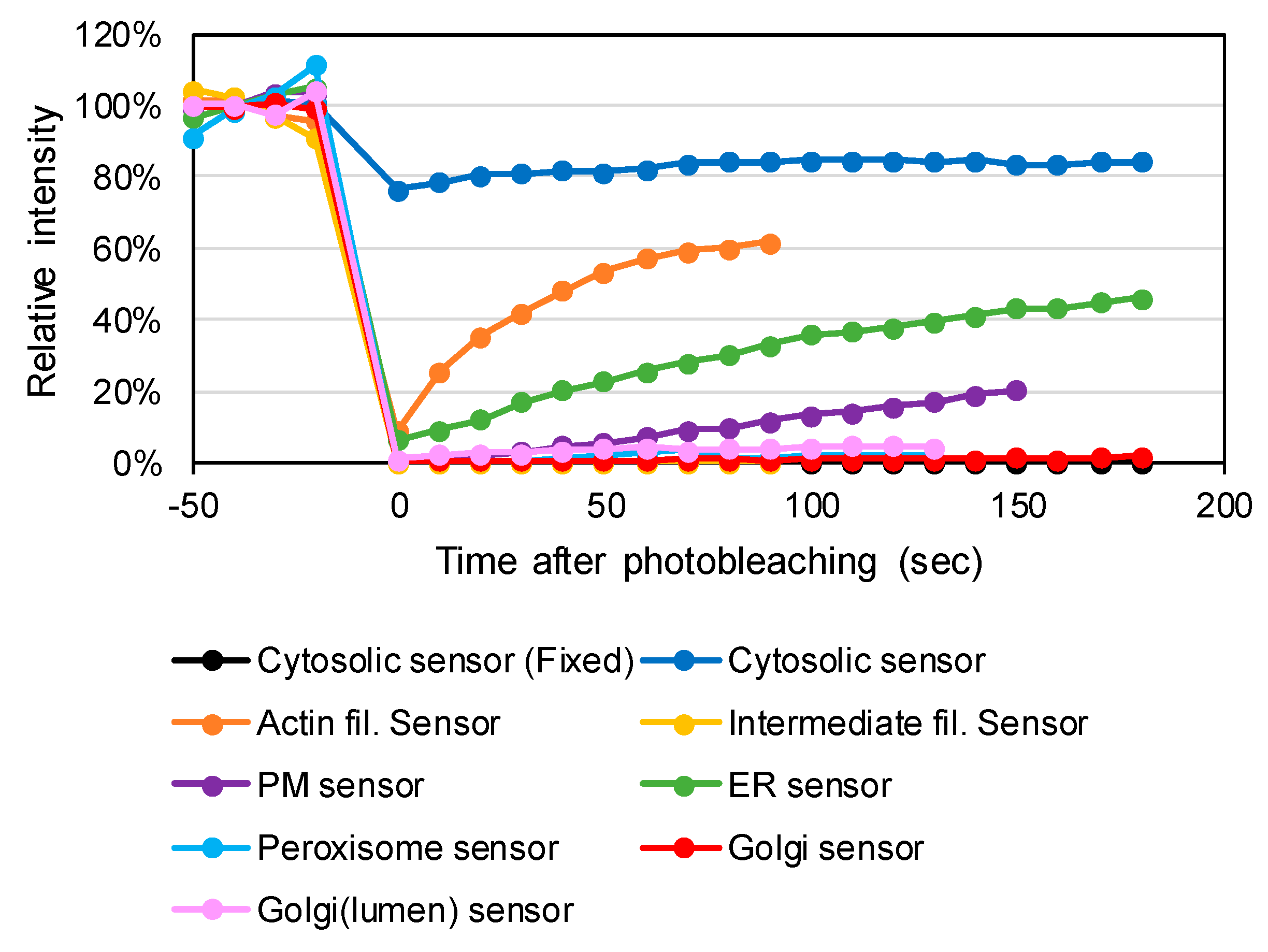
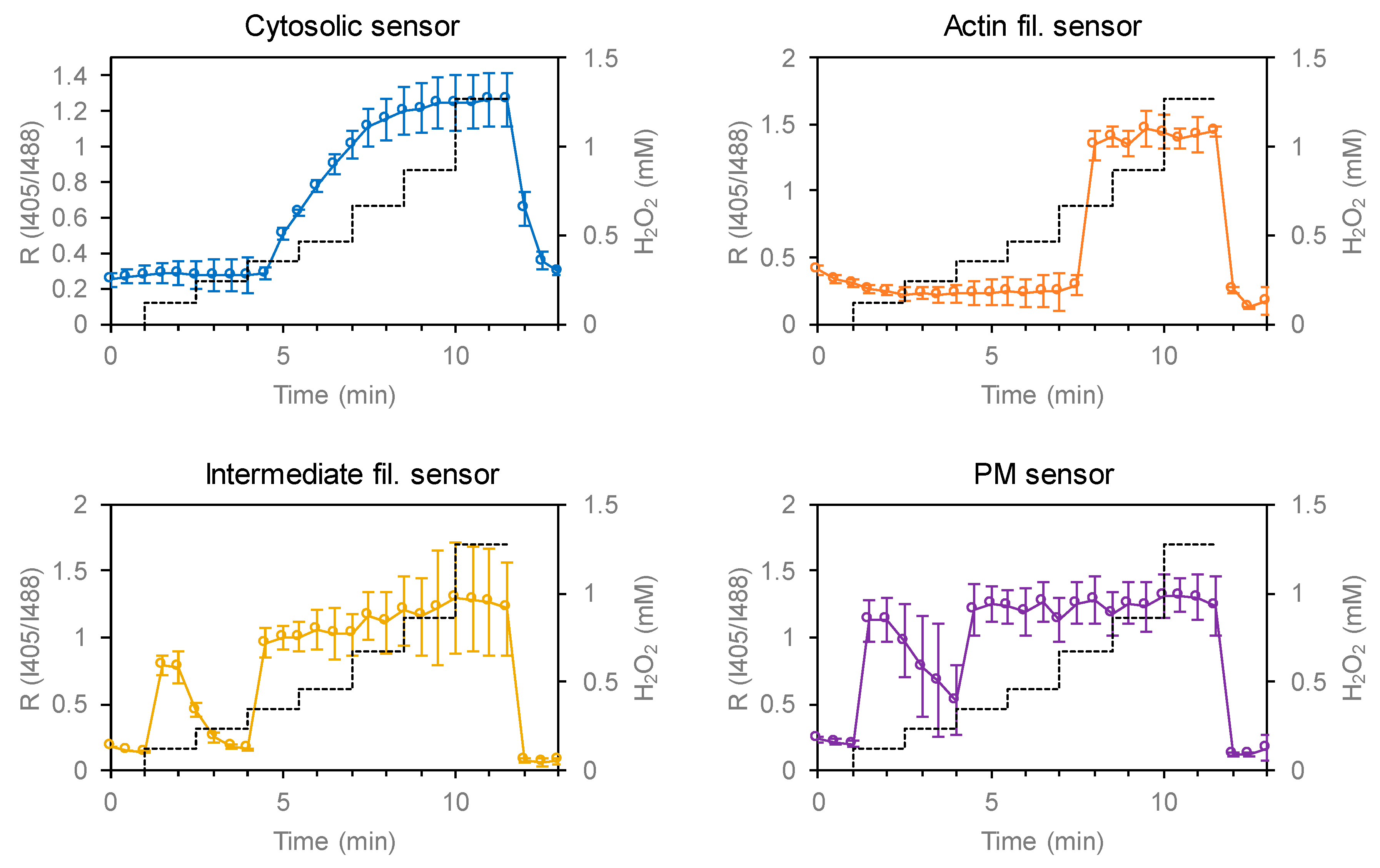
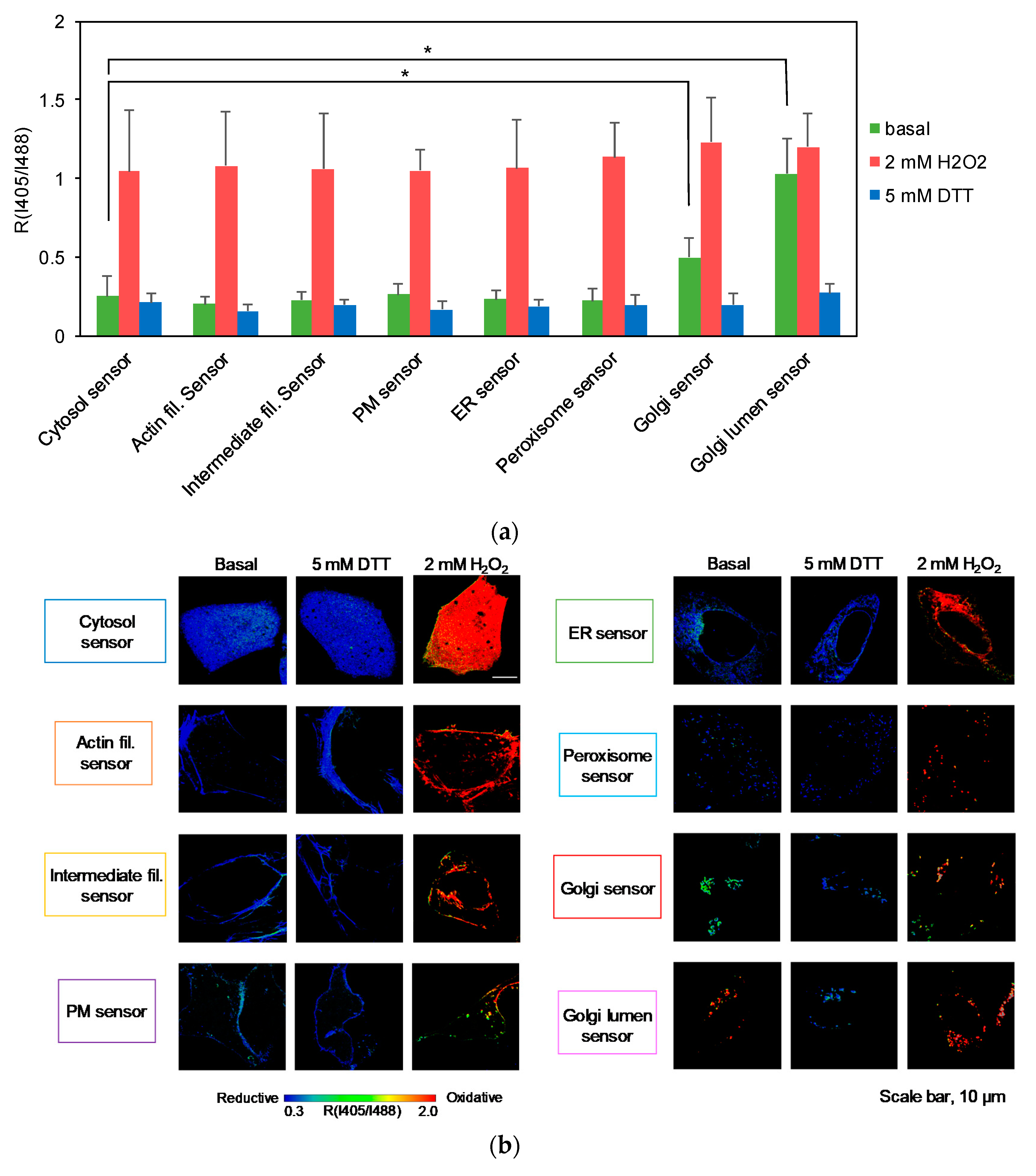
3.2. Testing the Redox Responses of the Developed Sensors
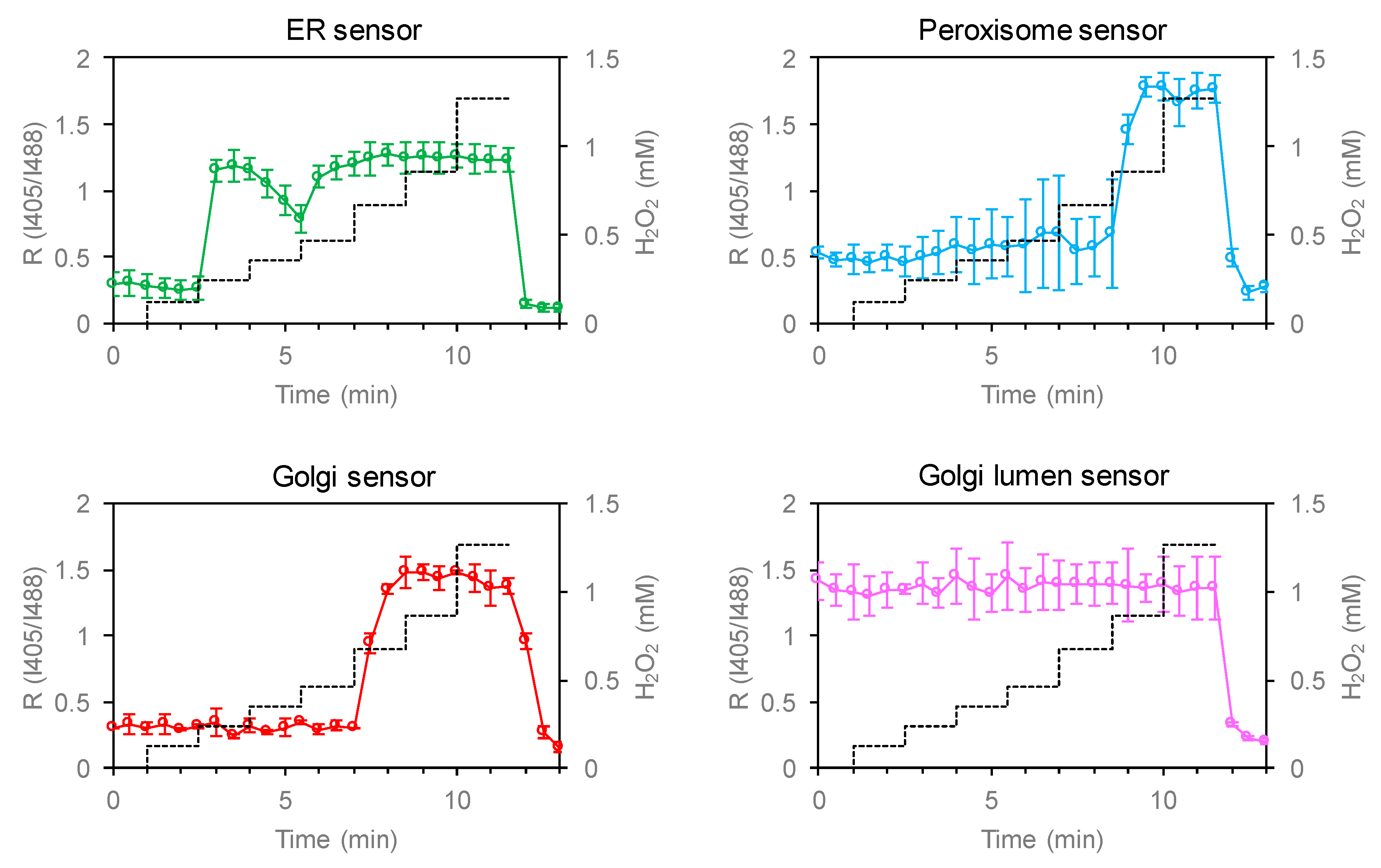
3.3. Unique Redox Environment near Organelle Membranes and Transmembrane Redox Gradient
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. J. Amino Acids 2012, 2012, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.R. The Glutathione Peroxidases. Cell. Mol. Life Sci. 2000, 57, 1825–1835. [Google Scholar] [CrossRef] [PubMed]
- Toppo, S.; Flohé, L.; Ursini, F.; Vanin, S.; Maiorino, M. Catalytic Mechanisms and Specificities of Glutathione Peroxidases: Variations of a Basic Scheme. Biochim. Biophys. Acta (BBA) Gen. Subj. 2009, 1790, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Gallogly, M.M.; Starke, D.W.; Mieyal, J.J. Mechanistic and Kinetic Details of Catalysis of Thiol-Disulfide Exchange by Glutaredoxins and Potential Mechanisms of Regulation. Antioxid. Redox Signal. 2009, 11, 1059–1081. [Google Scholar] [CrossRef]
- Hatori, Y.; Clasen, S.; Hasan, N.M.; Barry, A.N.; Lutsenko, S. Functional Partnership of the Copper Export Machinery and Glutathione Balance in Human Cells. J. Biol. Chem. 2012, 287, 26678–26687. [Google Scholar] [CrossRef]
- Hatori, Y.; Yan, Y.; Schmidt, K.; Furukawa, E.; Hasan, N.M.; Yang, N.; Liu, C.-N.; Sockanathan, S.; Lutsenko, S. Neuronal Differentiation is Associated with a Redox-Regulated Increase of Copper Flow to the Secretory Pathway. Nat. Commun. 2016, 7, 10640. [Google Scholar] [CrossRef]
- Singleton, W.C.J.; McInnes, K.T.; Cater, M.A.; Winnall, W.R.; McKirdy, R.; Yu, Y.; Taylor, P.E.; Ke, B.-X.; Richardson, D.R.; Mercer, J.F.B.; et al. Role of Glutaredoxin1 and Glutathione in Regulating the Activity of the Copper-transporting P-type ATPases, ATP7A and ATP7B. J. Biol. Chem. 2010, 285, 27111–27121. [Google Scholar] [CrossRef]
- Xiao, Z.; La Fontaine, S.; Bush, A.I.; Wedd, A.G. Molecular Mechanisms of Glutaredoxin Enzymes: Versatile Hubs for Thiol–Disulfide Exchange between Protein Thiols and Glutathione. J. Mol. Biol. 2019, 431, 158–177. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Yang, H.; Duffy, M.; Robinson, E.; Conrad-Antoville, A.; Lu, Y.-W.; Capps, T.; Braiterman, L.; Wolfgang, M.; Murphy, M.P.; et al. The Activity of Menkes Disease Protein ATP7A Is Essential for Redox Balance in Mitochondria. J. Biol. Chem. 2016, 291, 16644–16658. [Google Scholar] [CrossRef]
- Dardalhon, M.; Kumar, C.; Iraqui, I.; Vernis, L.; Kienda, G.; Banach-Latapy, A.; He, T.; Chanet, R.; Faye, G.; Outten, C.E.; et al. Redox-Sensitive YFP Sensors Monitor Dynamic Nuclear and Cytosolic Glutathione Redox Changes. Free Radic. Biol. Med. 2012, 52, 2254–2265. [Google Scholar] [CrossRef][Green Version]
- Gutscher, M.; Pauleau, A.-L.; Marty, L.; Brach, T.; Wabnitz, G.H.; Samstag, Y.; Meyer, A.J.; Dick, T.P. Real-Time Imaging of the Intracellular Glutathione Redox Potential. Nat. Methods 2008, 5, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.; Tachibana, C.; Rahr Winther, J.; Appenzeller-Herzog, C. Intracellular Glutathione Pools Are Heterogeneously Concentrated. Redox Biol. 2013, 1, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.; Sobotta, M.C.; Dick, T.P. Measuring EGSH and H2O2 with roGFP2-Based Redox Probes. Free. Radic. Biol. Med. 2011, 51, 1943–1951. [Google Scholar] [CrossRef] [PubMed]
- Giustarini, D.; Colombo, G.; Garavaglia, M.L.; Astori, E.; Portinaro, N.M.; Reggiani, F.; Badalamenti, S.; Aloisi, A.M.; Santucci, A.; Rossi, R.; et al. Assessment of Glutathione/Glutathione Disulphide Ratio and S -Glutathionylated Proteins in Human Blood, Solid Tissues, and Cultured Cells. Free. Radic. Biol. Med. 2017, 112, 360–375. [Google Scholar] [CrossRef]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.D.G.; Rossi, R. Detection of Glutathione in Whole Blood after Stabilization with N-ethylmaleimide. Anal. Biochem. 2011, 415, 81–83. [Google Scholar] [CrossRef]
- Moore, T.; Le, A.; Niemi, A.-K.; Kwan, T.; Cusmano-Ozog, K.; Enns, G.M.; Cowan, T.M. A new LC–MS/MS Method for the Clinical Determination of Reduced and Oxidized Glutathione from Whole Blood. J. Chromatogr. B 2013, 929, 51–55. [Google Scholar] [CrossRef]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.D.G.; Fanti, P.; Rossi, R. Analysis of GSH and GSSG after Derivatization with N-ethylmaleimide. Nat. Protoc. 2013, 8, 1660–1669. [Google Scholar] [CrossRef]
- Timur, S.; Odaci, D.; Dinçer, A.; Zihnioglu, F.; Telefoncu, A. Biosensing Approach for Glutathione Detection Using Glutathione Reductase and Sulfhydryl Oxidase Bienzymatic System. Talanta 2008, 74, 1492–1497. [Google Scholar] [CrossRef]
- Ezeriņa, D.; Morgan, B.; Dick, T.P. Imaging Dynamic Redox Processes with Genetically Encoded Probes. J. Mol. Cell. Cardiol. 2014, 73, 43–49. [Google Scholar] [CrossRef]
- Birk, J.; Meyer, M.; Aller, I.; Hansen, H.G.; Odermatt, A.; Dick, T.P.; Meyer, A.J.; Appenzeller-Herzog, C. Endoplasmic Reticulum: Reduced and Oxidized Glutathione Revisited. J. Cell Sci. 2013, 126, 1604–1617. [Google Scholar] [CrossRef]
- Schwarzländer, M.; Dick, T.P.; Meyer, A.J.; Morgan, B.; Meye, A.J. Dissecting Redox Biology Using Fluorescent Protein Sensors. Antioxid. Redox Signal. 2016, 24, 680–712. [Google Scholar] [CrossRef] [PubMed]
- Hatori, Y.; Inouye, S.; Akagi, R.; Seyama, T. Local Redox Environment Beneath Biological Membranes Probed by Palmitoylated-roGFP. Redox Biol. 2018, 14, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Bedard, K.; Krause, K.-H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Zurek, N.; Sparks, L.; Voeltz, G. Reticulon Short Hairpin Transmembrane Domains Are Used to Shape ER Tubules. Traffic (Cph. Den.) 2011, 12, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A Bright Monomeric Red Fluorescent Protein for Cellular Imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; von Stetten, D.; Noirclerc-Savoye, M.; Lelimousin, M.; Joosen, L.; Hink, M.A.; van Weeren, L.; Gadella, T.W., Jr.; Royant, A. Structure-Guided Evolution of Cyan Fluorescent Proteins towards a Quantum Yield of 93%. Nat. Commun. 2012, 3, 751. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright Monomeric Near-Infrared Fluorescent Proteins as Tags and Biosensors for Multiscale Imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef]
- Cline, D.J.; Redding, S.E.; Brohawn, S.G.; Psathas, J.N.; Schneider, J.P.; Thorpe, C. New Water-Soluble Phosphines as Reductants of Peptide and Protein Disulfide Bonds: Reactivity and Membrane Permeability†. Biochemistry 2004, 43, 15195–15203. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An Open Platform for Biomedical Image Analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Riedl, J.; Crevenna, A.H.; Kessenbrock, K.; Yu, J.H.; Neukirchen, D.; Bista, M.; Bradke, F.; Jenne, D.; Holak, T.A.; Werb, Z.; et al. Lifeact: A Versatile Marker to Visualize F-actin. Nat. Methods 2008, 5, 605–607. [Google Scholar] [CrossRef]
- Hirokawa, T.; Boon-Chieng, S.; Mitaku, S. SOSUI: Classification and Secondary Structure Prediction System for Membrane Proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef] [PubMed]
- Day, C.A.; Kraft, L.J.; Kang, M.; Kenworthy, A.K. Analysis of Protein and Lipid Dynamics Using Confocal Fluorescence Recovery after Photobleaching (FRAP). Curr. Protoc. Cytom. 2012, 62, 2–19. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, H.; Hailey, D.W.; Wunder, C.; Lippincott-Schwartz, J. The Fluorescence Protease Protection (FPP) Assay to Determine Protein Localization and Membrane Topology. Nat. Protoc. 2006, 1, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M. Glutathione Catalysis and the Reaction Mechanisms of Glutathione-Dependent Enzymes. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3217–3266. [Google Scholar] [CrossRef]
- Zou, Y.; Wang, A.; Shi, M.; Chen, X.; Liu, R.; Li, T.; Zhang, C.; Zhang, Z.; Zhu, L.; Ju, Z.; et al. Analysis of Redox Landscapes and Dynamics in Living Cells and in Vivo Using Genetically Encoded Fluorescent Sensors. Nat. Protoc. 2018, 13, 2362–2386. [Google Scholar] [CrossRef]
- Breckwoldt, M.; Pfister, F.M.J.; Bradley, P.M.; Marinković, P.; Williams, P.R.; Brill, M.S.; Plomer, B.; Schmalz, A.; Clair, D.K.S.; Naumann, R.; et al. Multiparametric Optical Analysis of Mitochondrial Redox Signals during Neuronal Physiology and Pathology in Vivo. Nat. Med. 2014, 20, 555–560. [Google Scholar] [CrossRef]
- Kiyohara, T.; Miyano, K.; Kamakura, S.; Hayase, J.; Chishiki, K.; Kohda, A.; Sumimoto, H. Differential Cell Surface Recruitment of the Superoxide-Producing NADPH Oxidases Nox1, Nox2 and Nox5: The Role of the Small GTPase Sar1. Genes Cells 2018, 23, 480–493. [Google Scholar] [CrossRef]
- Morgan, B.; Ezerina, D.; Amoako, T.N.; Riemer, J.; Seedorf, M.; Dick, T.P. Multiple Glutathione Disulfide Removal Pathways Mediate Cytosolic Redox Homeostasis. Nat. Chem. Biol. 2013, 9, 119–125. [Google Scholar] [CrossRef]
- Brose, J.; La Fontaine, S.; Wedd, A.G.; Xiao, Z. Redox Sulfur Chemistry of the Copper Chaperone Atox1 is Regulated by the Enzyme Glutaredoxin 1, the Reduction Potential of the Glutathione Couple GSSG/2GSH and the Availability of Cu(i). Metallomics 2014, 6, 793–808. [Google Scholar] [CrossRef]
- Lim, C.M.; Cater, M.A.; Mercer, J.F.; La Fontaine, S. Copper-Dependent Interaction of Glutaredoxin with the N Termini of the Copper-ATPases (ATP7A and ATP7B) Defective in Menkes and Wilson diseases. Biochem. Biophys. Res. Commun. 2006, 348, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Barrientos, G.C.; Cherednichenko, G.; Yang, T.; Padilla, I.T.; Truong, K.; Allen, P.D.; Lopez, J.R.; Pessah, I.N. Functional and Biochemical Properties of Ryanodine Receptor Type 1 Channels from Heterozygous R163C Malignant Hyperthermia-Susceptible Mice. Mol. Pharmacol. 2011, 79, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Ghazanfari, G.; Minaie, B.; Yasa, N.; Nakhai, L.A.; Mohammadirad, A.; Nikfar, S.; Dehghan, G.; Boushehri, V.S.; Jamshidi, H.; Khorasani, R.; et al. Biochemical and Histopathological Evidences for Beneficial Effects ofSatureja Khuzestanica JamzadEssential Oil on the Mouse Model of Inflammatory Bowel Diseases. Toxicol. Mech. Methods 2006, 16, 365–372. [Google Scholar] [CrossRef]
- Nieto, N.; Torres, M.; Fernández, M.; Girón, M.; Ríos, A.; Suárez, M.; Gil, A. Experimental Ulcerative Colitis Impairs Antioxidant Defense System in Rat Intestine. Dig. Dis. Sci. 2000, 45, 1820–1827. [Google Scholar] [CrossRef] [PubMed]
- Smeyne, M.; Smeyne, R.J. Glutathione Metabolism and Parkinson’s Disease. Free. Radic. Biol. Med. 2013, 62, 13–25. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatori, Y.; Kubo, T.; Sato, Y.; Inouye, S.; Akagi, R.; Seyama, T. Visualization of the Redox Status of Cytosolic Glutathione Using the Organelle- and Cytoskeleton-Targeted Redox Sensors. Antioxidants 2020, 9, 129. https://doi.org/10.3390/antiox9020129
Hatori Y, Kubo T, Sato Y, Inouye S, Akagi R, Seyama T. Visualization of the Redox Status of Cytosolic Glutathione Using the Organelle- and Cytoskeleton-Targeted Redox Sensors. Antioxidants. 2020; 9(2):129. https://doi.org/10.3390/antiox9020129
Chicago/Turabian StyleHatori, Yuta, Takanori Kubo, Yuichiro Sato, Sachiye Inouye, Reiko Akagi, and Toshio Seyama. 2020. "Visualization of the Redox Status of Cytosolic Glutathione Using the Organelle- and Cytoskeleton-Targeted Redox Sensors" Antioxidants 9, no. 2: 129. https://doi.org/10.3390/antiox9020129
APA StyleHatori, Y., Kubo, T., Sato, Y., Inouye, S., Akagi, R., & Seyama, T. (2020). Visualization of the Redox Status of Cytosolic Glutathione Using the Organelle- and Cytoskeleton-Targeted Redox Sensors. Antioxidants, 9(2), 129. https://doi.org/10.3390/antiox9020129




