Cell-Type Specific Analysis of Selenium-Related Genes in Brain
Abstract
1. Introduction
2. Materials and Methods
2.1. Allen Brain Atlas RNAseq Analysis
2.2. Antibodies and Mice
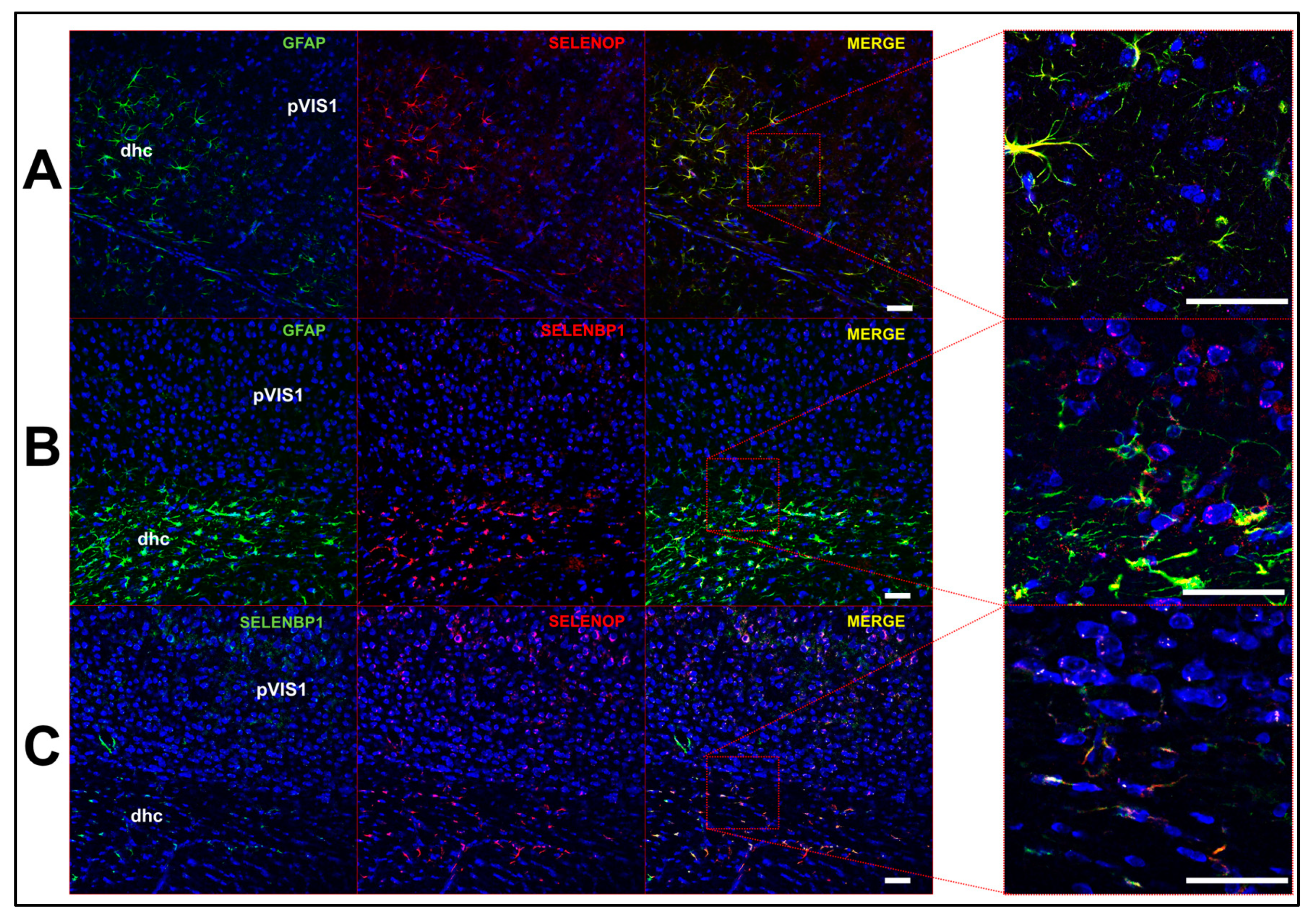
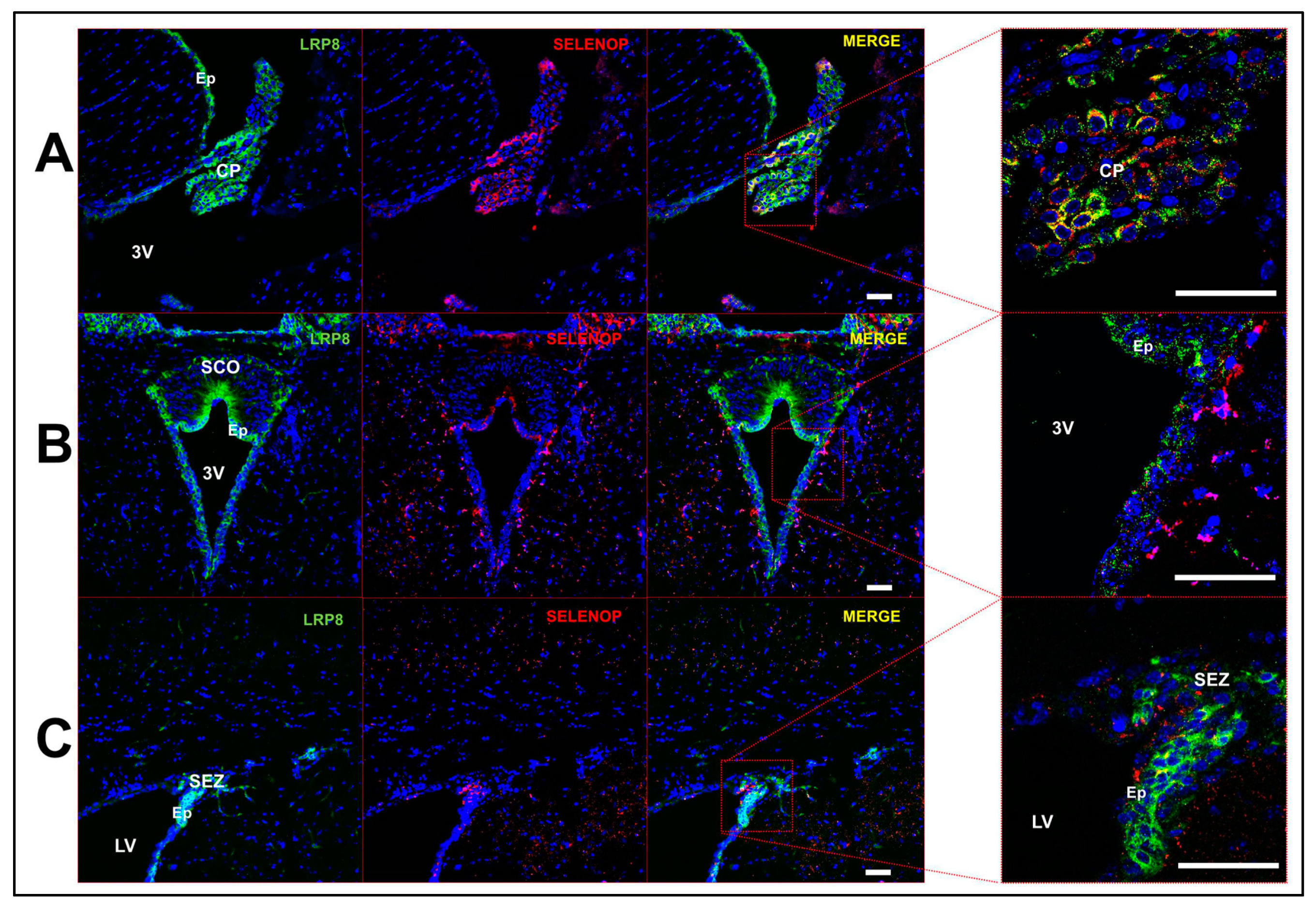
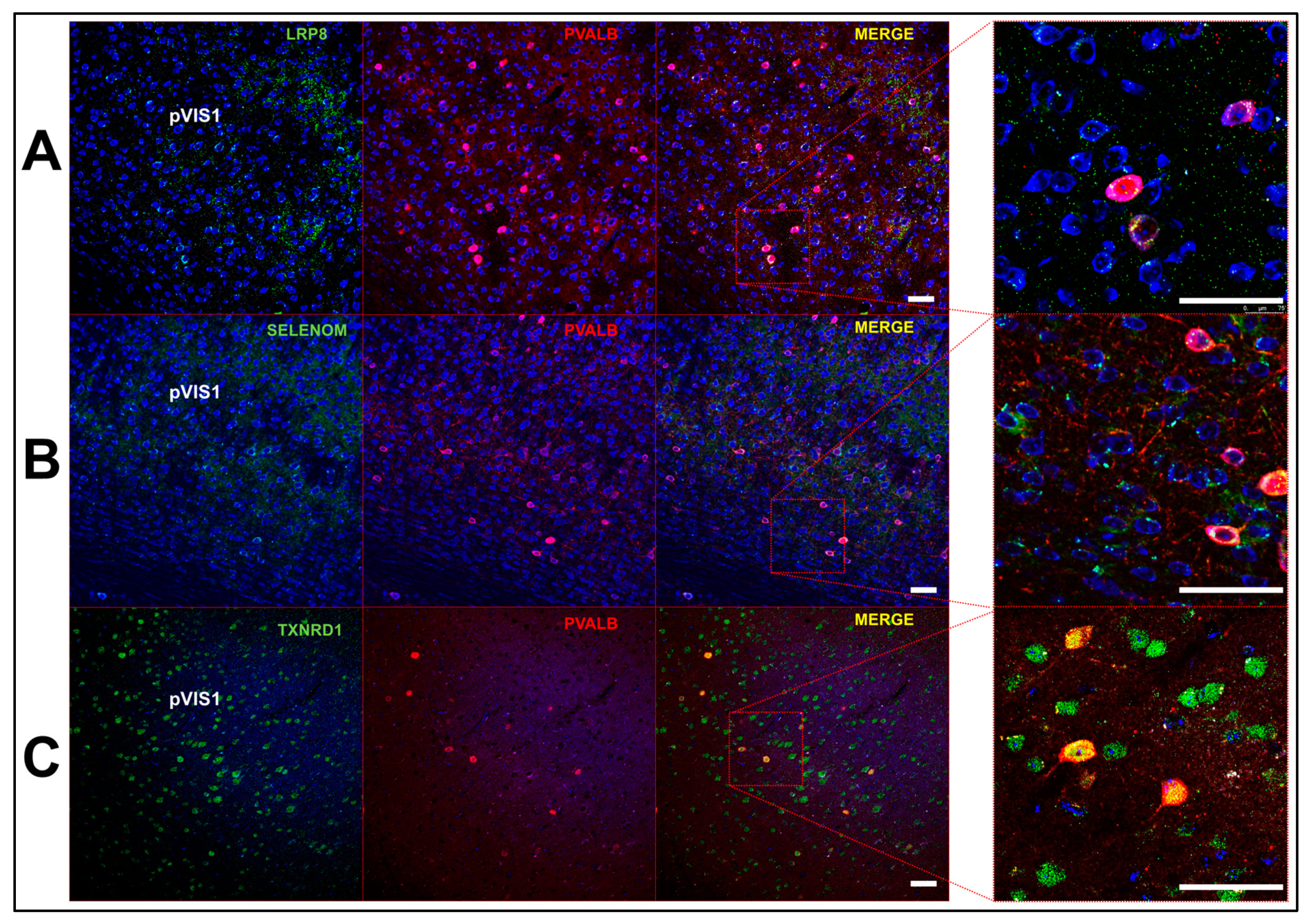
2.3. Multi-Label Immunofluorescent Microscopy
3. Results
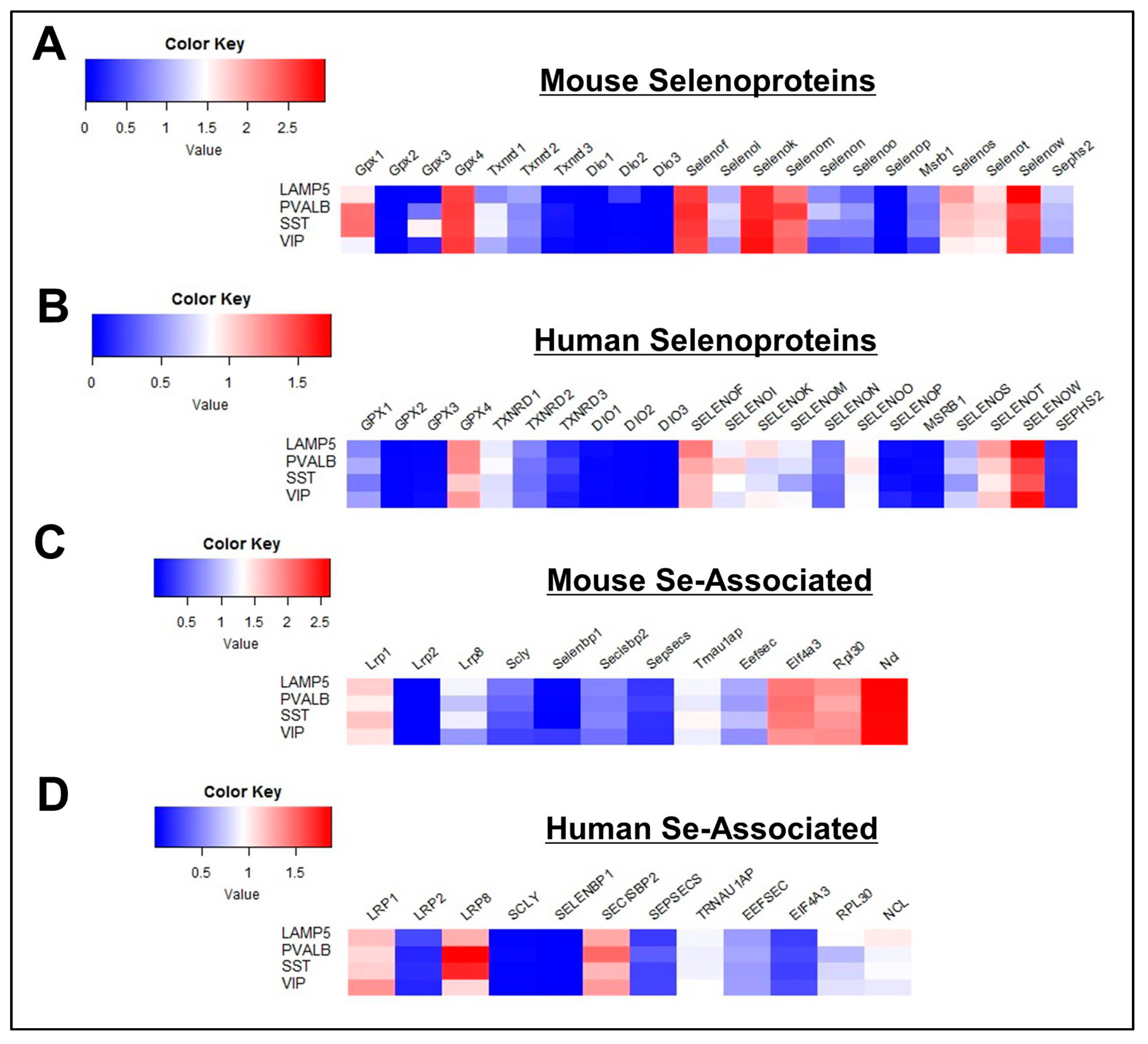
3.1. General Profile of Se-Related Transcriptome in Cortical Cells
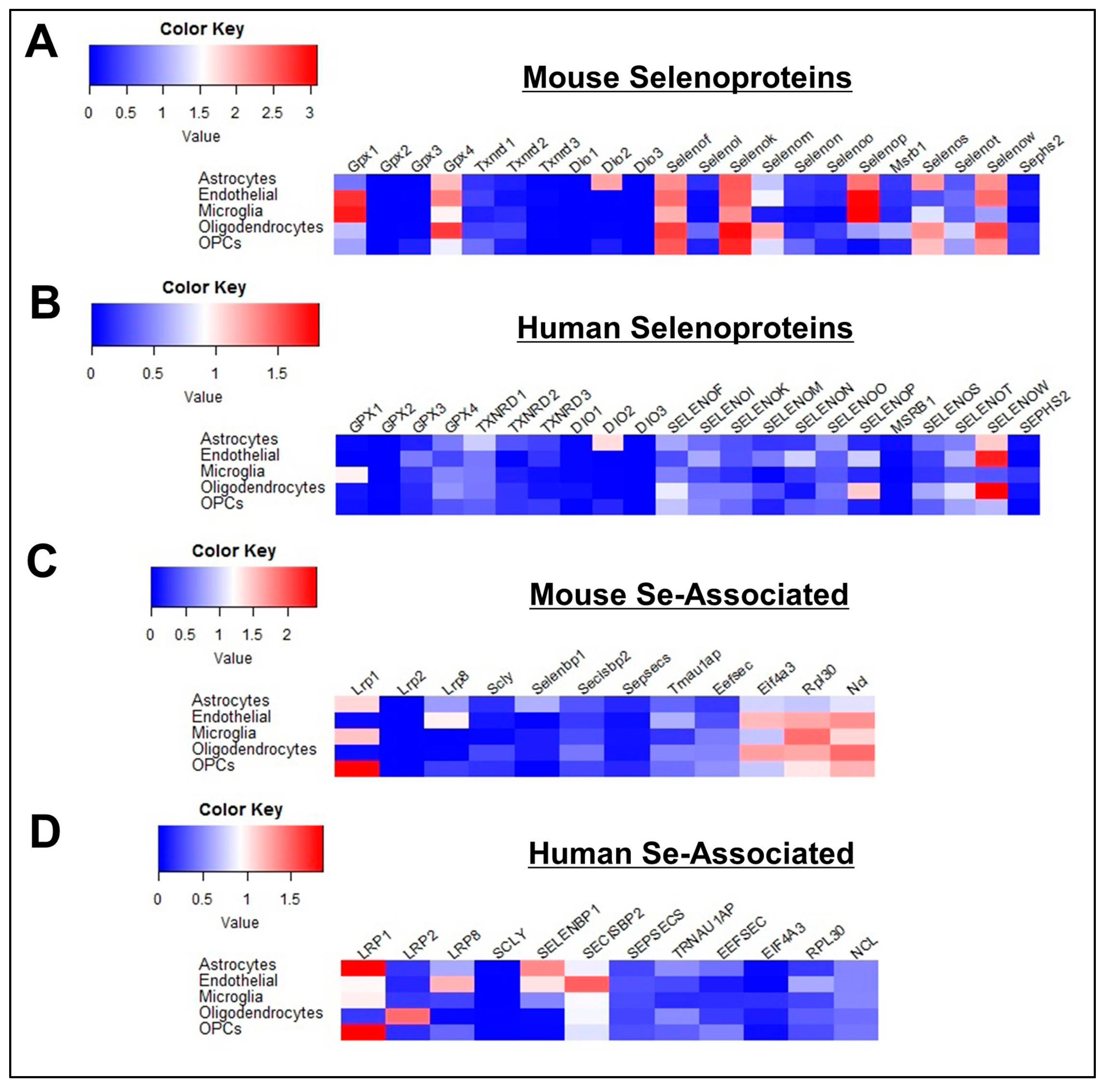
3.2. Profile of Se-Related Transcriptome in Non-Neuronal Cells
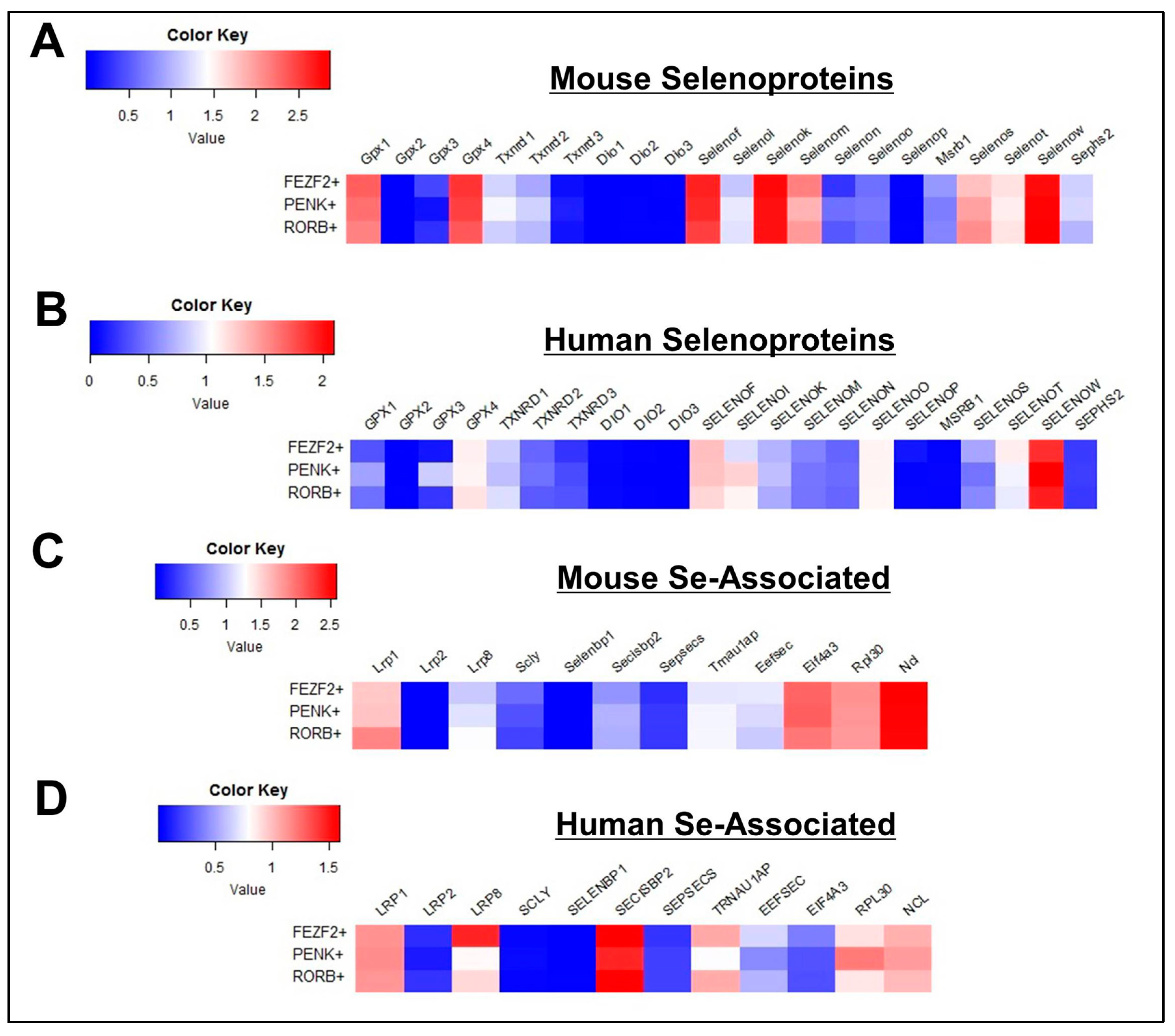
3.3. Profile of Se-Related Transcriptome in Glutamatergic Neurons
3.4. Profile of Se-Related Transcriptome in GABAergic Neurons
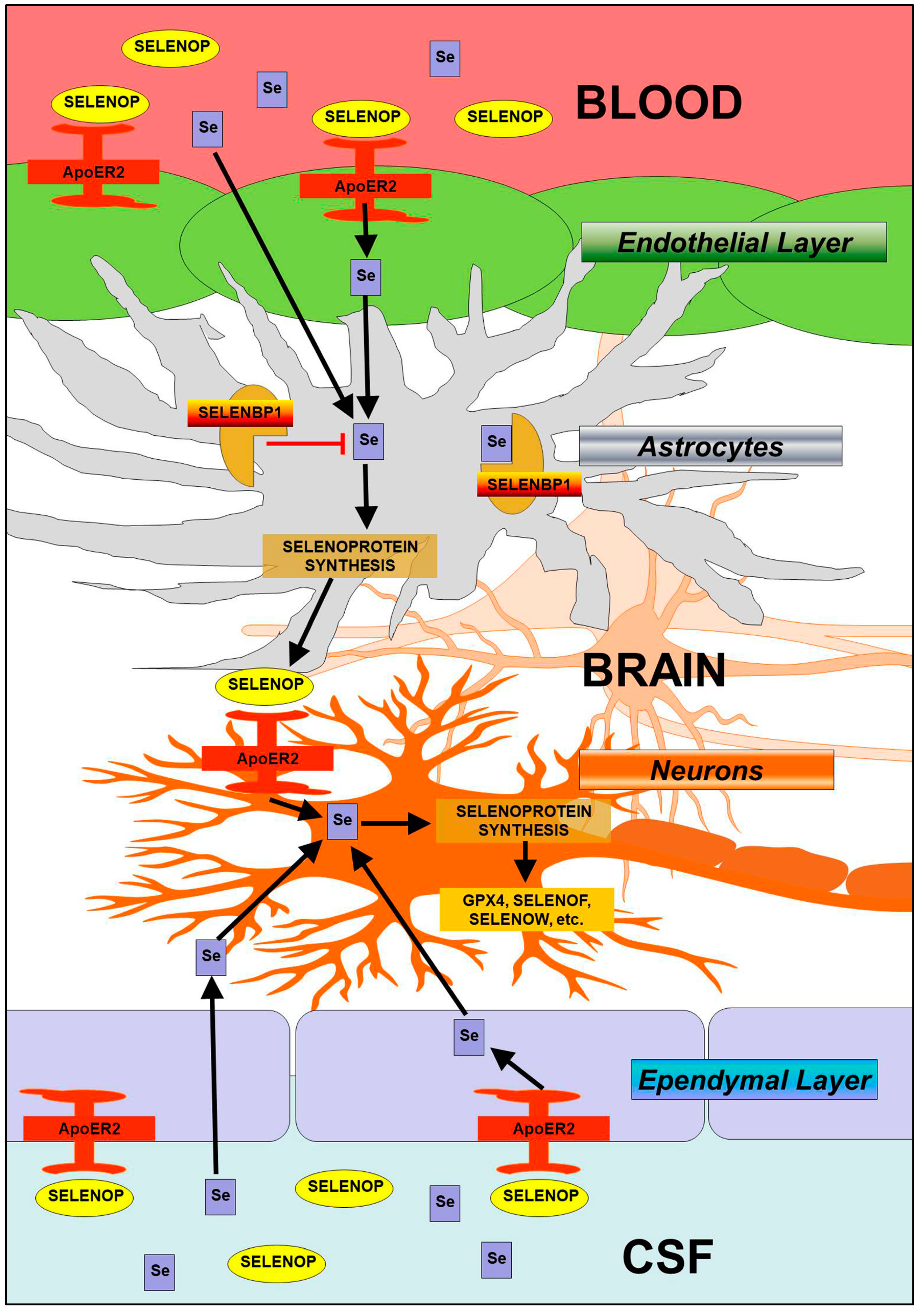
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allen Cell Types Database. Available online: http://celltypes.brain-map.org (accessed on 1 March 2019).
- Burk, R.F.; Hill, K.E. Regulation of Selenium Metabolism and Transport. Annu. Rev. Nutr. 2015, 35, 109–134. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Wirth, E.K.; Conrad, M.; Winterer, J.; Wozny, C.; Carlson, B.A.; Roth, S.; Schmitz, D.; Bornkamm, G.W.; Coppola, V.; Tessarollo, L.; et al. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010, 24, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Freitas, F.P.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Byrns, C.N.; Pitts, M.W.; Gilman, C.A.; Hashimoto, A.C.; Berry, M.J. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J. Biol. Chem. 2014, 289, 9662–9674. [Google Scholar] [CrossRef]
- Pitts, M.W.; Kremer, P.M.; Hashimoto, A.C.; Torres, D.J.; Byrns, C.N.; Williams, C.S.; Berry, M.J. Competition between the Brain and Testes under Selenium-Compromised Conditions: Insight into Sex Differences in Selenium Metabolism and Risk of Neurodevelopmental Disease. J. Neurosci. 2015, 35, 15326–15338. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E. Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta 2009, 1790, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E.; Olson, G.E.; Weeber, E.J.; Motley, A.K.; Winfrey, V.P.; Austin, L.M. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J. Neurosci. 2007, 27, 6207–6211. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.E.; Zhou, J.; McMahan, W.J.; Motley, A.K.; Atkins, J.F.; Gesteland, R.F.; Burk, R.F. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 2003, 278, 13640–13646. [Google Scholar] [CrossRef]
- Schomburg, L.; Schweizer, U.; Holtmann, B.; Flohé, L.; Sendtner, M.; Köhrle, J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003, 370, 397–402. [Google Scholar] [CrossRef]
- Valentine, W.M.; Abel, T.W.; Hill, K.E.; Austin, L.M.; Burk, R.F. Neurodegeneration in mice resulting from loss of functional selenoprotein P or its receptor apolipoprotein E receptor 2. J. Neuropathol. Exp. Neurol. 2008, 67, 68–77. [Google Scholar] [CrossRef]
- Hill, K.E.; Zhou, J.; McMahan, W.J.; Motley, A.K.; Burk, R.F. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J. Nutr. 2004, 134, 157–161. [Google Scholar] [CrossRef]
- Pitts, M.W.; Raman, A.V.; Hashimoto, A.C.; Todorovic, C.; Nichols, R.A.; Berry, M.J. Deletion of selenoprotein P results in impaired function of parvalbumin interneurons and alterations in fear learning and sensorimotor gating. Neurosci. 2012, 208, 58–68. [Google Scholar] [CrossRef]
- Donovan, J.; Copeland, P.R. Threading the needle: Getting selenocysteine into proteins. Antioxid. Redox Signal. 2010, 12, 881–892. [Google Scholar] [CrossRef]
- Squires, J.E.; Berry, M.J. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life 2008, 60, 232–235. [Google Scholar] [CrossRef]
- Hill, K.E.; Zhou, J.; Austin, L.M.; Motley, A.K.; Ham, A.J.L.; Olson, G.E.; Atkins, J.F.; Gesteland, R.F.; Burk, R.F. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J. Biol. Chem. 2007, 282, 10972–10980. [Google Scholar] [CrossRef]
- Olson, G.E.; Winfrey, V.P.; Hill, K.E.; Burk, R.F. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J. Biol. Chem. 2008, 283, 6854–6860. [Google Scholar] [CrossRef]
- Burk, R.F.; Olson, G.E.; Hill, K.E.; Winfrey, V.P.; Motley, A.K.; Kurokawa, S. Maternal-fetal transfer of selenium in the mouse. FASEB J. 2013, 27, 3249–3256. [Google Scholar] [CrossRef]
- Power, J.H.; Blumbergs, P.C. Cellular glutathione peroxidase in human brain: Cellular distribution, and its potential role in the degradation of Lewy bodies in Parkinson’s disease and dementia with Lewy bodies. Acta Neuropathol. 2009, 117, 63–73. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E.; Motley, A.K.; Winfrey, V.P.; Kurokawa, S.; Mitchell, S.L.; Zhang, W. Selenoprotein P and apolipoprotein E receptor-2 interact at the blood-brain barrier and also within the brain to maintain an essential selenium pool that protects against neurodegeneration. FASEB J. 2014, 28, 3579–3588. [Google Scholar] [CrossRef]
- Bansal, M.P.; Mukhopadhyay, T.; Scott, J.; Cook, R.G.; Mukhopadhyay, R.; Medina, D. DNA sequencing of a mouse liver protein that binds selenium: Implications for selenium’s mechanism of action in cancer prevention. Carcinogenesis 1990, 11, 2071–2073. [Google Scholar] [CrossRef]
- Bansal, M.P.; Oborn, C.J.; Danielson, K.G.; Medina, D. Evidence for two selenium-binding proteins distinct from glutathione peroxidase in mouse liver. Carcinogenesis 1989, 10, 541–546. [Google Scholar] [CrossRef]
- Elhodaky, M.; Diamond, A.M. Selenium-Binding Protein 1 in Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3437. [Google Scholar] [CrossRef]
- Kudin, A.P.; Baron, G.; Zsurka, G.; Hampel, K.G.; Elger, C.E.; Grote, A.; Weber, Y.; Lerche, H.; Thiele, H.; Nürnberg, P.; et al. Homozygous mutation in TXNRD1 is associated with genetic generalized epilepsy. Free Radic. Biol. Med. 2017, 106, 270–277. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Al-Khayat, A.; Al-Murshedi, F.; Al-Futaisi, A.; Chioza, B.A.; Pedro Fernandez-Murray, J.; Self, J.E.; Salter, C.G.; Harlalka, G.V.; Rawlins, L.E.; et al. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain 2017, 140, 547–554. [Google Scholar]
- Agamy, O.; Zeev, B.B.; Lev, D.; Marcus, B.; Fine, D.; Su, D.; Narkis, G.; Ofir, R.; Hoffmann, C.; Leshinsky-Silver, E.; et al. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am. J. Hum. Genet. 2010, 87, 538–544. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Schweizer, U.; Savaskan, N.E.; Hua, D.; Kipnis, J.; Hatfield, D.L.; Gladyshev, V.N. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J. Biol. Chem. 2008, 283, 2427–2438. [Google Scholar] [CrossRef]
- Yang, X.; Hill, K.E.; Maguire, M.J.; Burk, R.F. Synthesis and secretion of selenoprotein P by cultured rat astrocytes. Biochim. Biophys. Acta 2000, 1474, 390–396. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Alili, L.; Bilgic, E.; Sies, H.; Brenneisen, P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic. Biol. Med. 2006, 40, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Scharpf, M.; Schweizer, U.; Arzberger, T.; Roggendorf, W.; Schomburg, L.; Köhrle, J. Neuronal and ependymal expression of selenoprotein P in the human brain. J. Neural Transm. 2007, 114, 877–884. [Google Scholar] [CrossRef]
- Peters, M.M.; Hill, K.E.; Burk, R.F.; Weeber, E.J. Altered hippocampus synaptic function in selenoprotein P deficient mice. Mol. Neurodegener. 2006, 1, 12. [Google Scholar] [CrossRef]
- Chang, P.W.; Tsui, S.K.; Liew, C.C.; Lee, C.Y.; Waye, M.M.; Fung, K.P. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. J. Cell Biochem. 1997, 64, 217–224. [Google Scholar] [CrossRef]
- Fang, W.; Goldberg, M.L.; Pohl, N.M.; Bi, X.; Tong, C.; Xiong, B.; Koh, T.J.; Diamond, A.M.; Yang, W. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis 2010, 31, 1360–1366. [Google Scholar] [CrossRef]
- Pol, A.; Renkema, G.H.; Tangerman, A.; Winkel, E.G.; Engelke, U.F.; De Brouwer, A.P.; Lloyd, K.C.; Araiza, R.S.; Van Den Heuvel, L.; Omran, H.; et al. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018, 50, 120–129. [Google Scholar] [CrossRef]
- Glatt, S.J.; Everall, I.P.; Kremen, W.S.; Corbeil, J.; Šášik, R.; Khanlou, N.; Han, M.; Liew, C.C.; Tsuang, M.T. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 15533–15538. [Google Scholar] [CrossRef]
- Kanazawa, T.; Chana, G.; Glatt, S.J.; Mizuno, H.; Masliah, E.; Yoneda, H.; Tsuang, M.T.; Everall, I.P. The utility of SELENBP1 gene expression as a biomarker for major psychotic disorders: Replication in schizophrenia and extension to bipolar disorder with psychosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147B, 686–689. [Google Scholar] [CrossRef]
- Udawela, M.; Money, T.T.; Neo, J.; Seo, M.S.; Scarr, E.; Dean, B.; Everall, I.P. SELENBP1 expression in the prefrontal cortex of subjects with schizophrenia. Transl. Psychiatry 2015, 5, e615. [Google Scholar] [CrossRef]
- Eftekharpour, E.; Holmgren, A.; Juurlink, B.H. Thioredoxin reductase and glutathione synthesis is upregulated by t-butylhydroquinone in cortical astrocytes but not in cortical neurons. Glia 2000, 31, 241–248. [Google Scholar] [CrossRef]
- Savaskan, N.E.; Borchert, A.; Bräuer, A.U.; Kuhn, H. Role for glutathione peroxidase-4 in brain development and neuronal apoptosis: Specific induction of enzyme expression in reactive astrocytes following brain injury. Free Radic. Biol. Med. 2007, 43, 191–201. [Google Scholar] [CrossRef]
- Fradejas, N.; Pastor, M.D.; Mora-Lee, S.; Tranque, P.; Calvo, S. SEPS1 gene is activated during astrocyte ischemia and shows prominent antiapoptotic effects. J. Mol. Neurosci. 2008, 35, 259–265. [Google Scholar] [CrossRef]
- Fradejas, N.; Del Carmen Serrano-PÉREZ, M.; Tranque, P.; Calvo, S. Selenoprotein S expression in reactive astrocytes following brain injury. Glia 2011, 59, 959–972. [Google Scholar] [CrossRef]
- Gonzalez-Moreno, O.; Boque, N.; Redrado, M.; Milagro, F.; Campion, J.; Endermann, T.; Takahashi, K.; Saito, Y.; Catena, R.; Schomburg, L.; et al. Selenoprotein-P is down-regulated in prostate cancer, which results in lack of protection against oxidative damage. Prostate 2011, 71, 824–834. [Google Scholar] [CrossRef] [PubMed]

| Species | Cell Class | Number of Cells |
|---|---|---|
| Mouse | Non-Neuronal | 1383 |
| Mouse | Glutamatergic | 11,905 |
| Mouse | GABAergic | 10,534 |
| Human | Non-Neuronal | 914 |
| Human | Glutamatergic | 10,525 |
| Human | GABAergic | 4164 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasuclark, A.R.; Khadka, V.S.; Pitts, M.W. Cell-Type Specific Analysis of Selenium-Related Genes in Brain. Antioxidants 2019, 8, 120. https://doi.org/10.3390/antiox8050120
Sasuclark AR, Khadka VS, Pitts MW. Cell-Type Specific Analysis of Selenium-Related Genes in Brain. Antioxidants. 2019; 8(5):120. https://doi.org/10.3390/antiox8050120
Chicago/Turabian StyleSasuclark, Alexandru R., Vedbar S. Khadka, and Matthew W. Pitts. 2019. "Cell-Type Specific Analysis of Selenium-Related Genes in Brain" Antioxidants 8, no. 5: 120. https://doi.org/10.3390/antiox8050120
APA StyleSasuclark, A. R., Khadka, V. S., & Pitts, M. W. (2019). Cell-Type Specific Analysis of Selenium-Related Genes in Brain. Antioxidants, 8(5), 120. https://doi.org/10.3390/antiox8050120





