Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry
Abstract
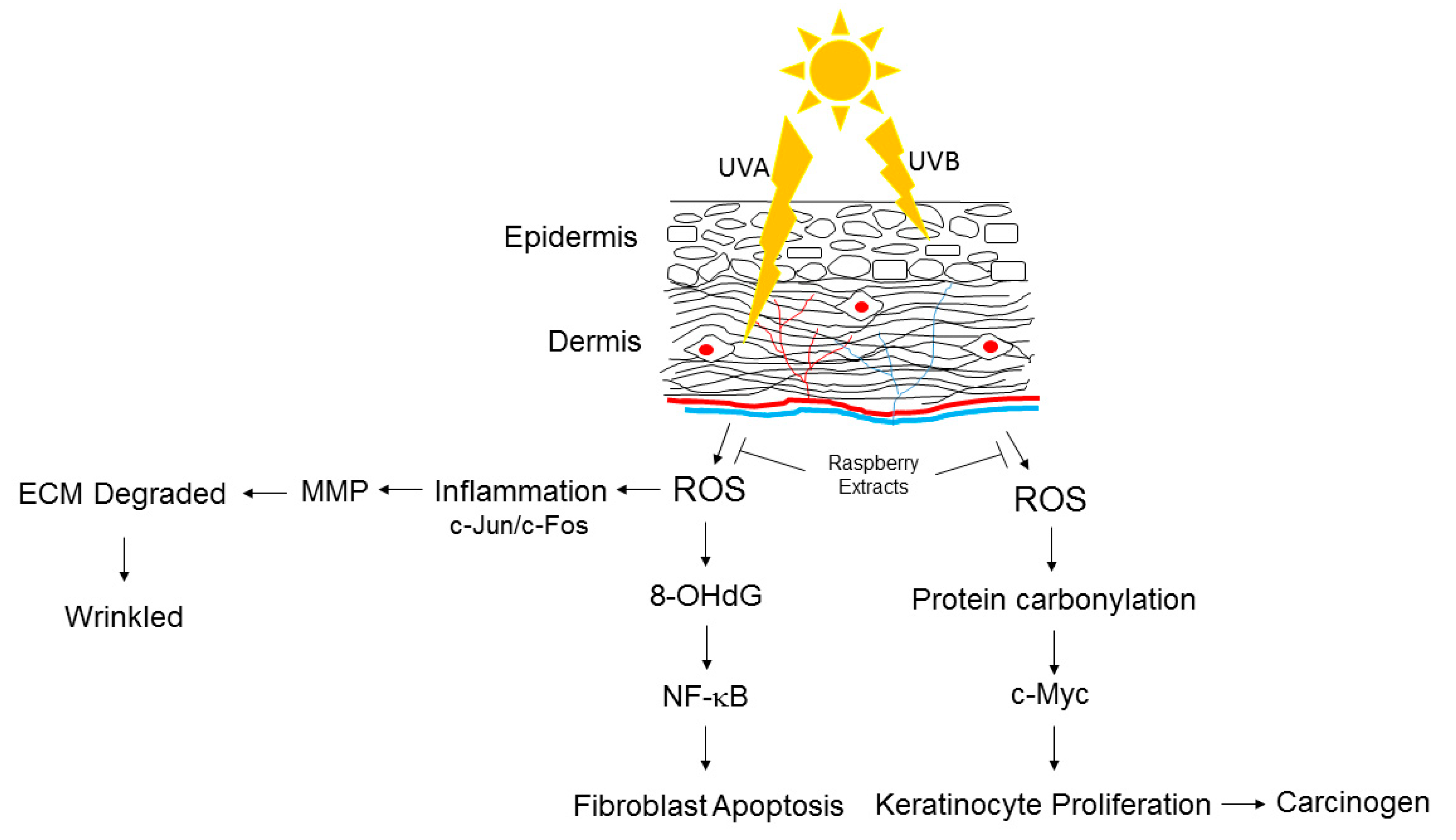
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Histological Study of the Mouse Skin
2.3. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assays
2.4. Gelatin Zymography
2.5. Two-Dimensional Electrophoresis (2-DE) Analysis
2.6. Determination of Protein Carbonyls with DNP Immunostaining
2.7. In-Gel Digestion and Mass Spectrometric Analysis
2.8. Network Analysis with MetaCore™
2.9. Western Blot Experiments
2.10. Evaluation of Cell Viability by MTT Analysis
2.11. Statistical Analysis
3. Results
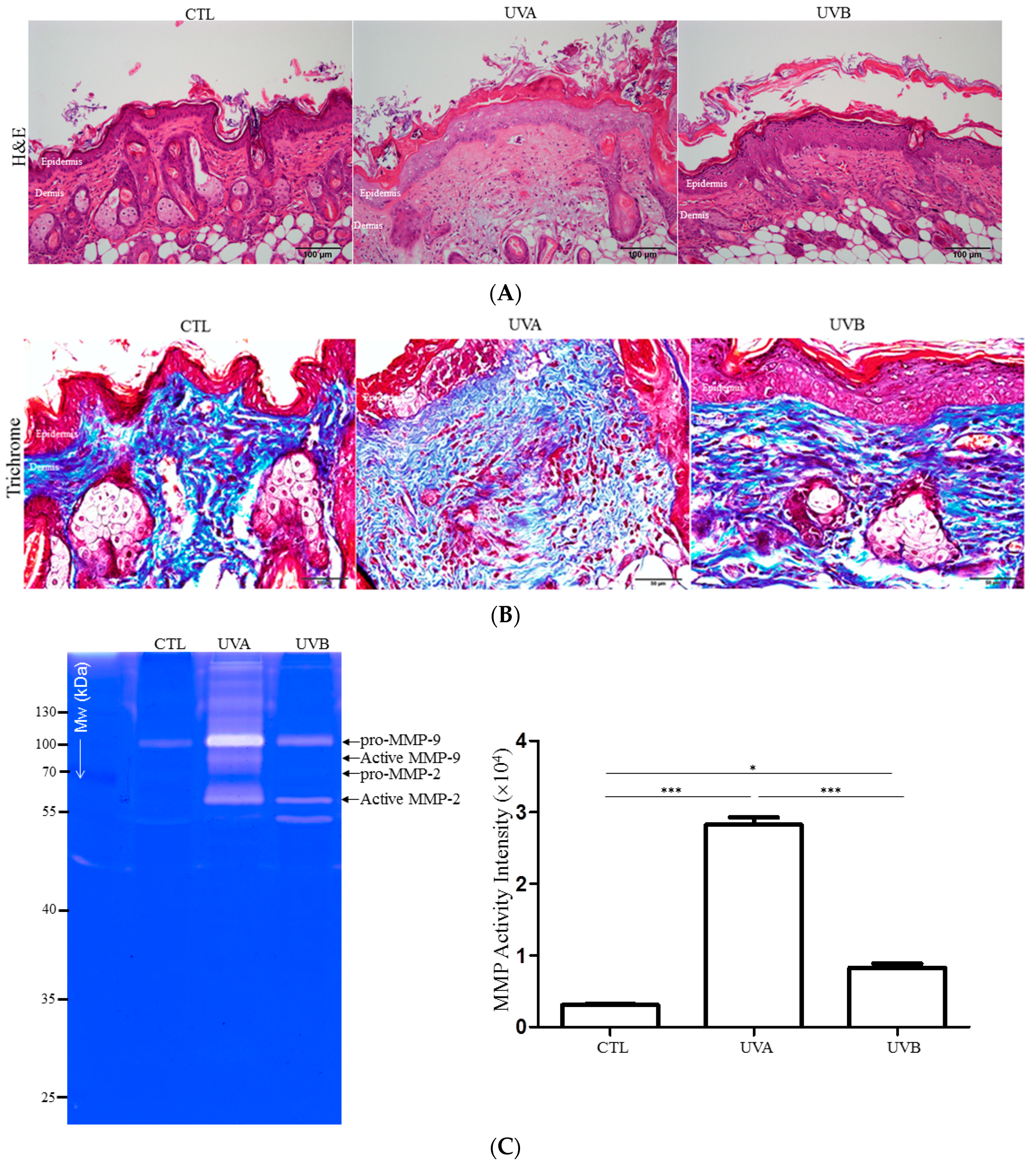
3.1. Histological Assessment
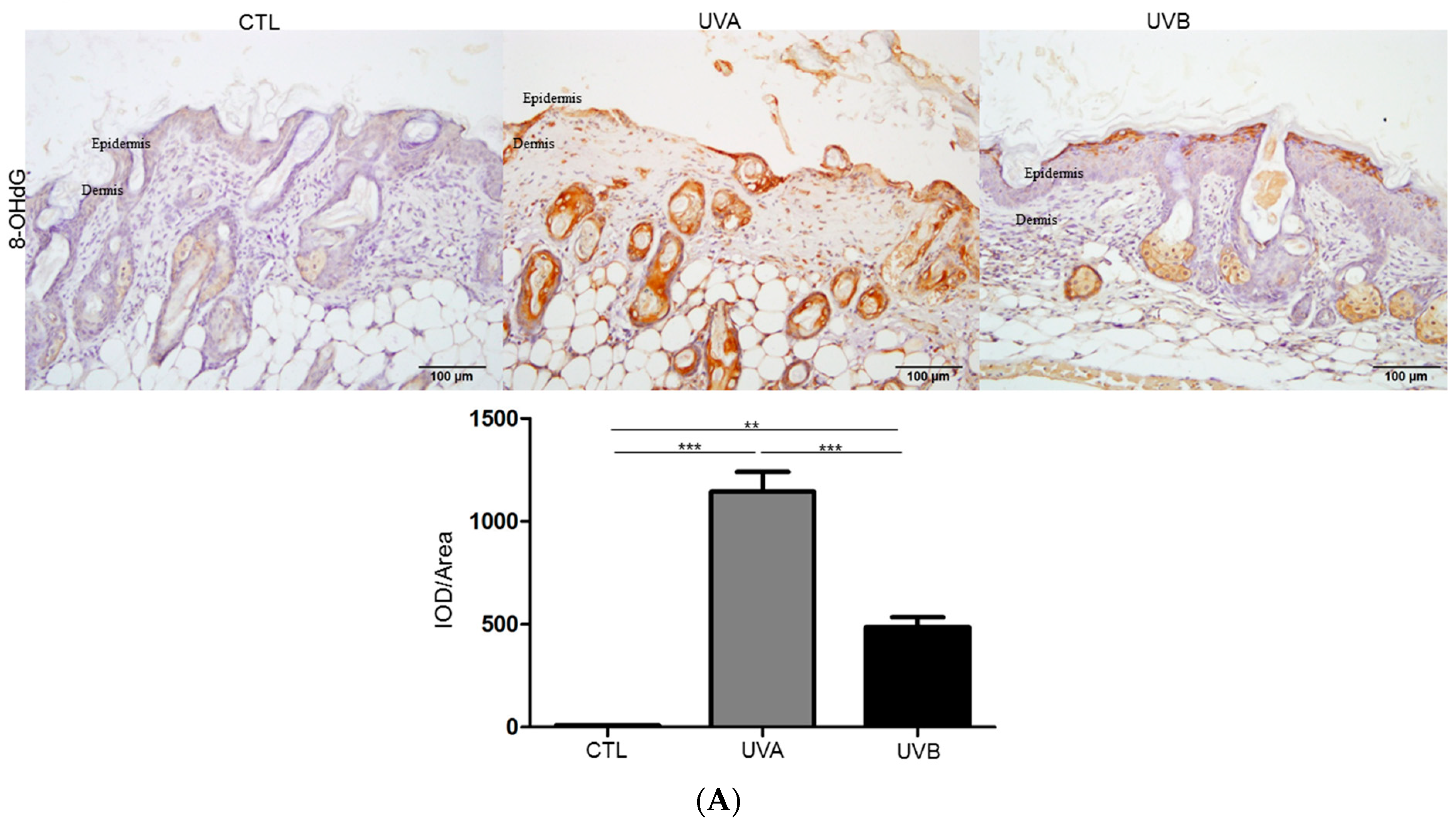
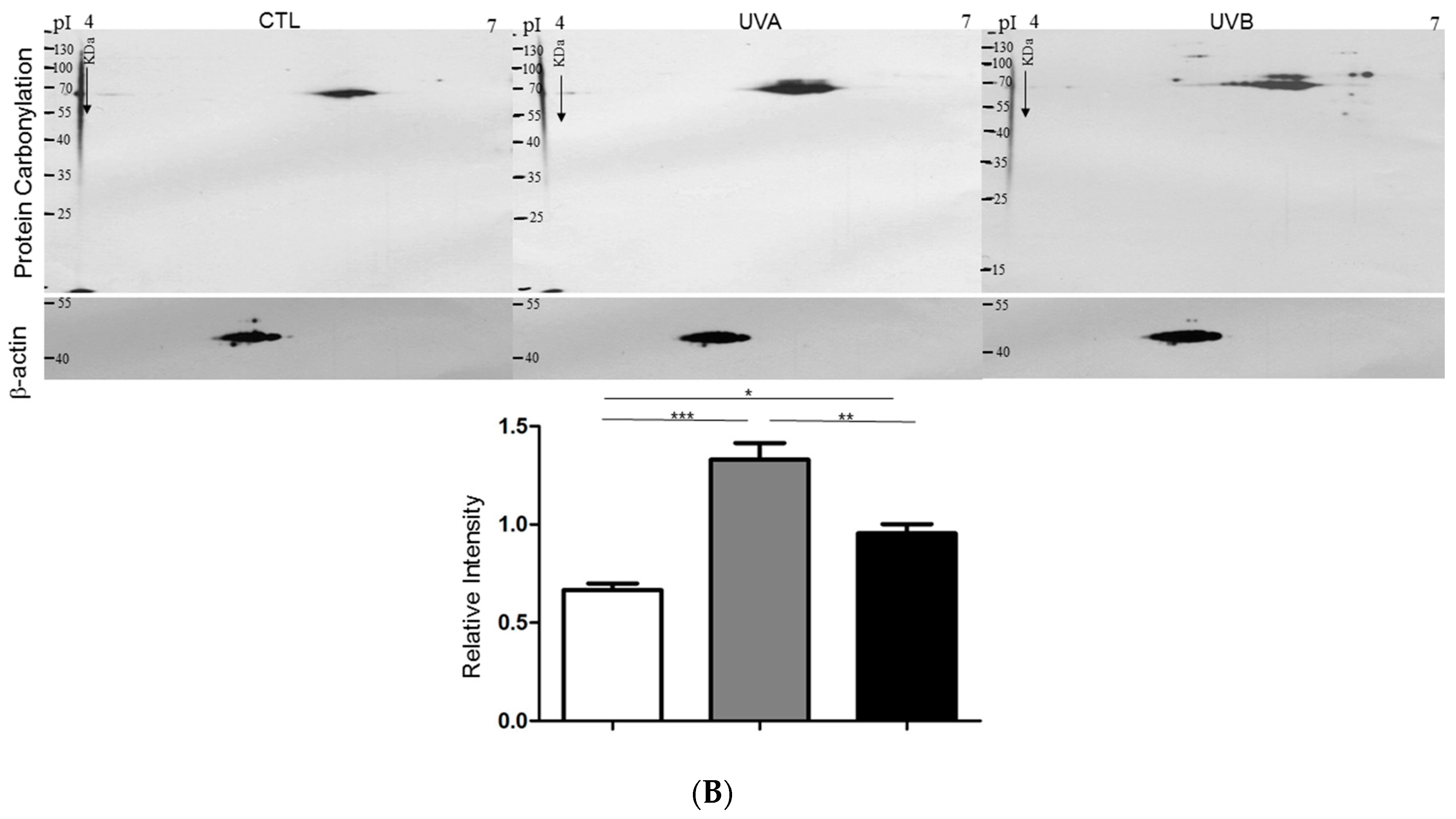
3.2. Effects of UVA and UVB Exposure in the Generation of Oxidative Stress
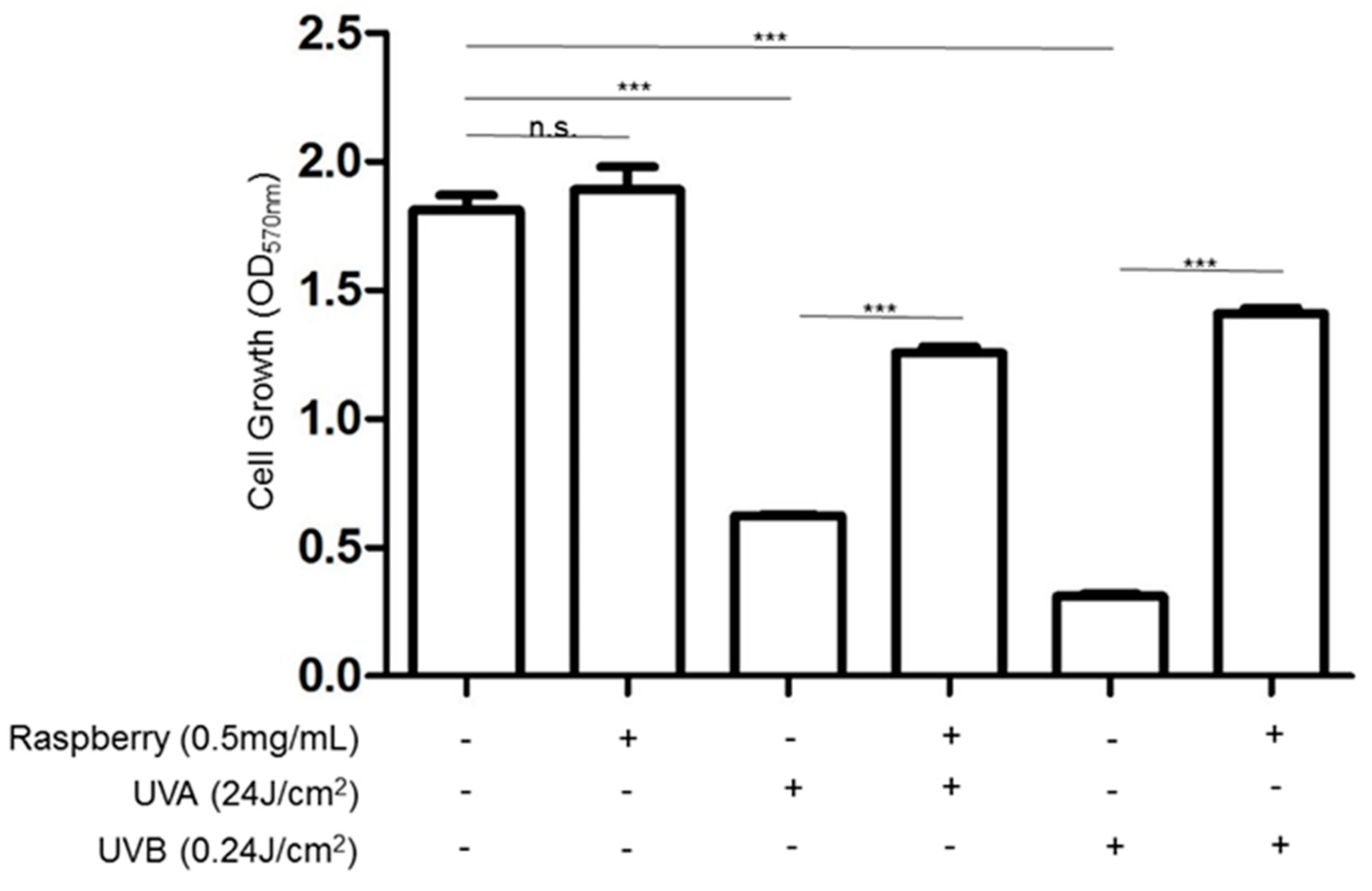
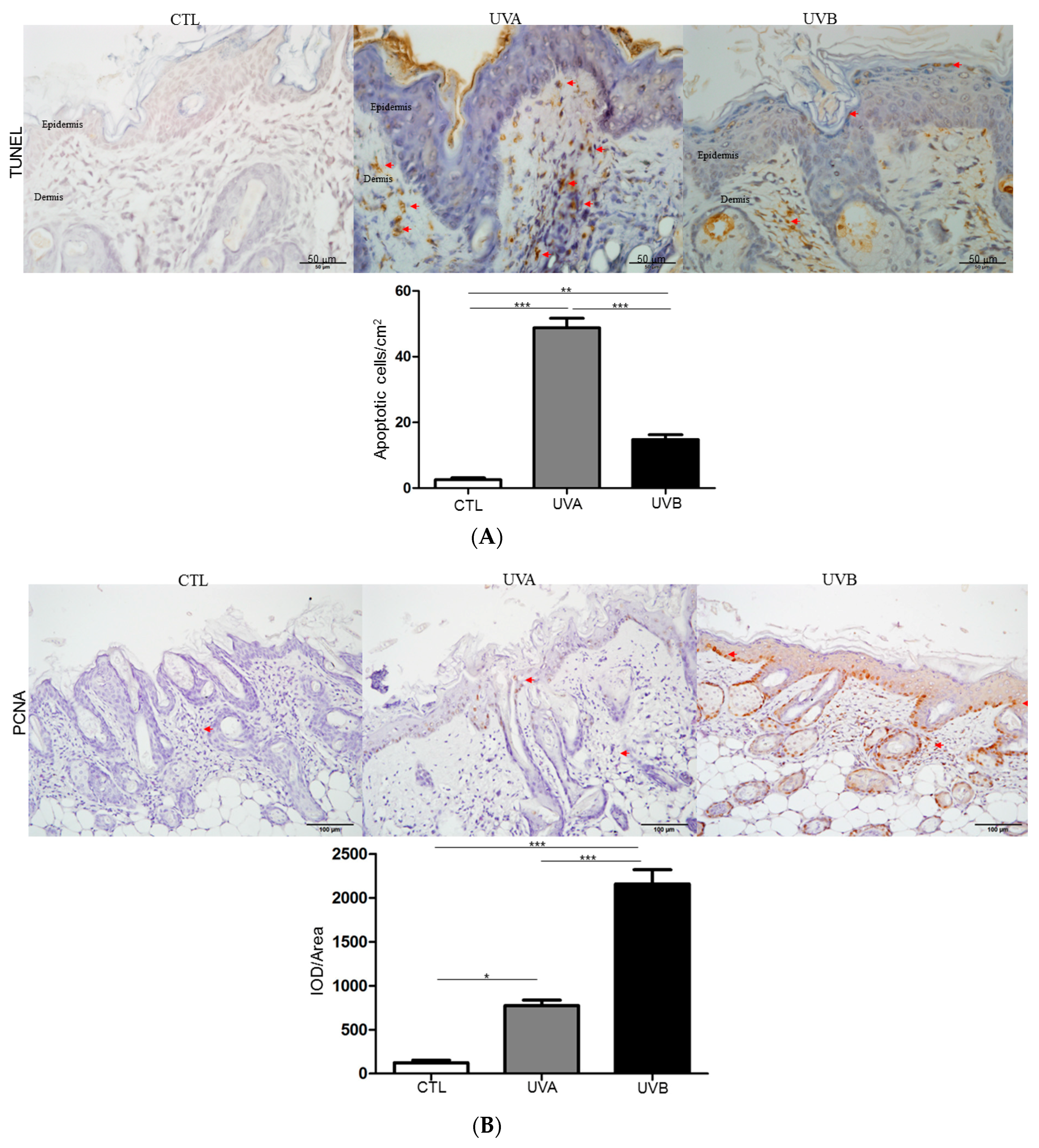
3.3. Effects of UVA and UVB Exposure in Cell Proliferation and Apoptosis
3.4. Exploring Target Proteins with Proteome Tools
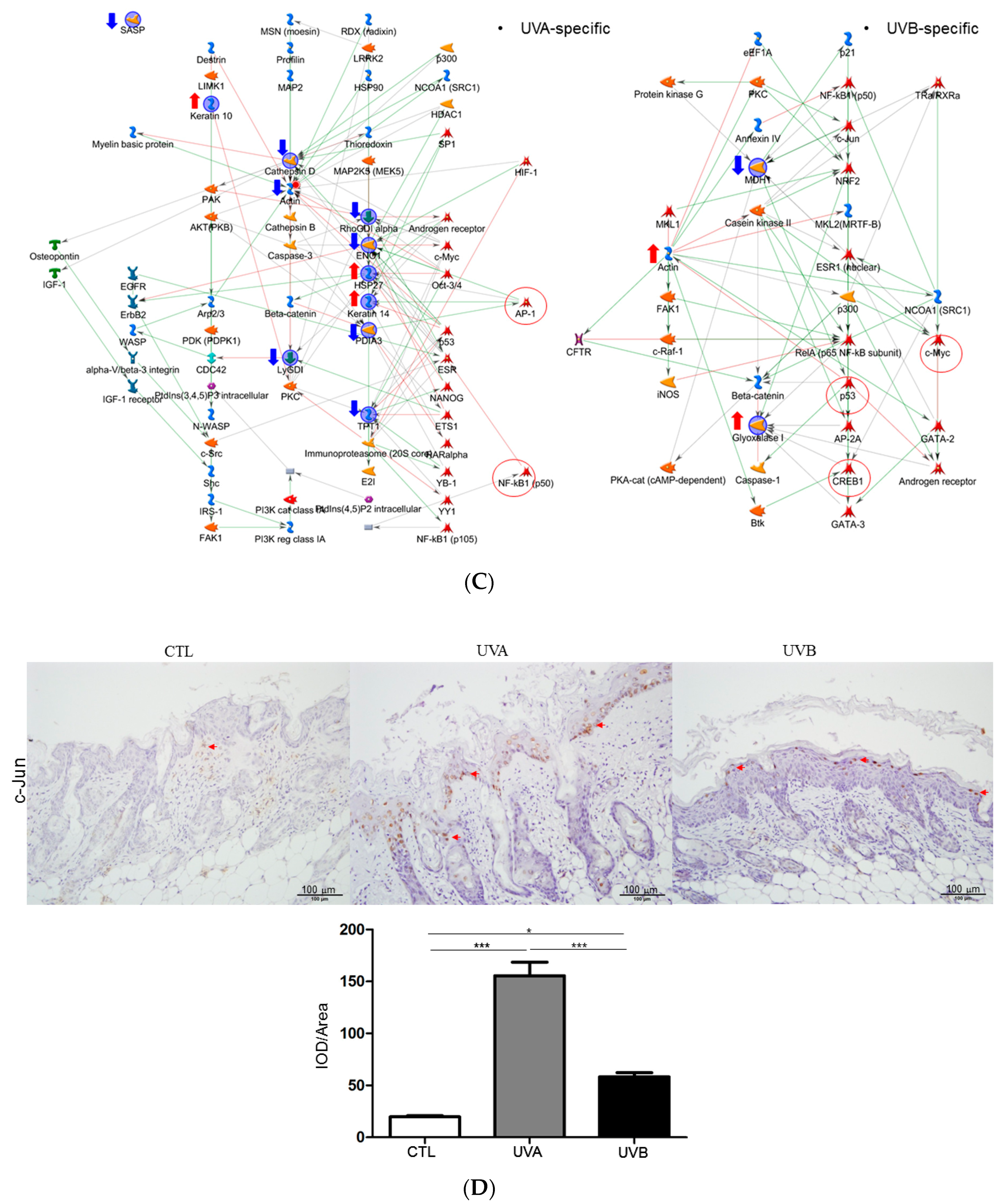
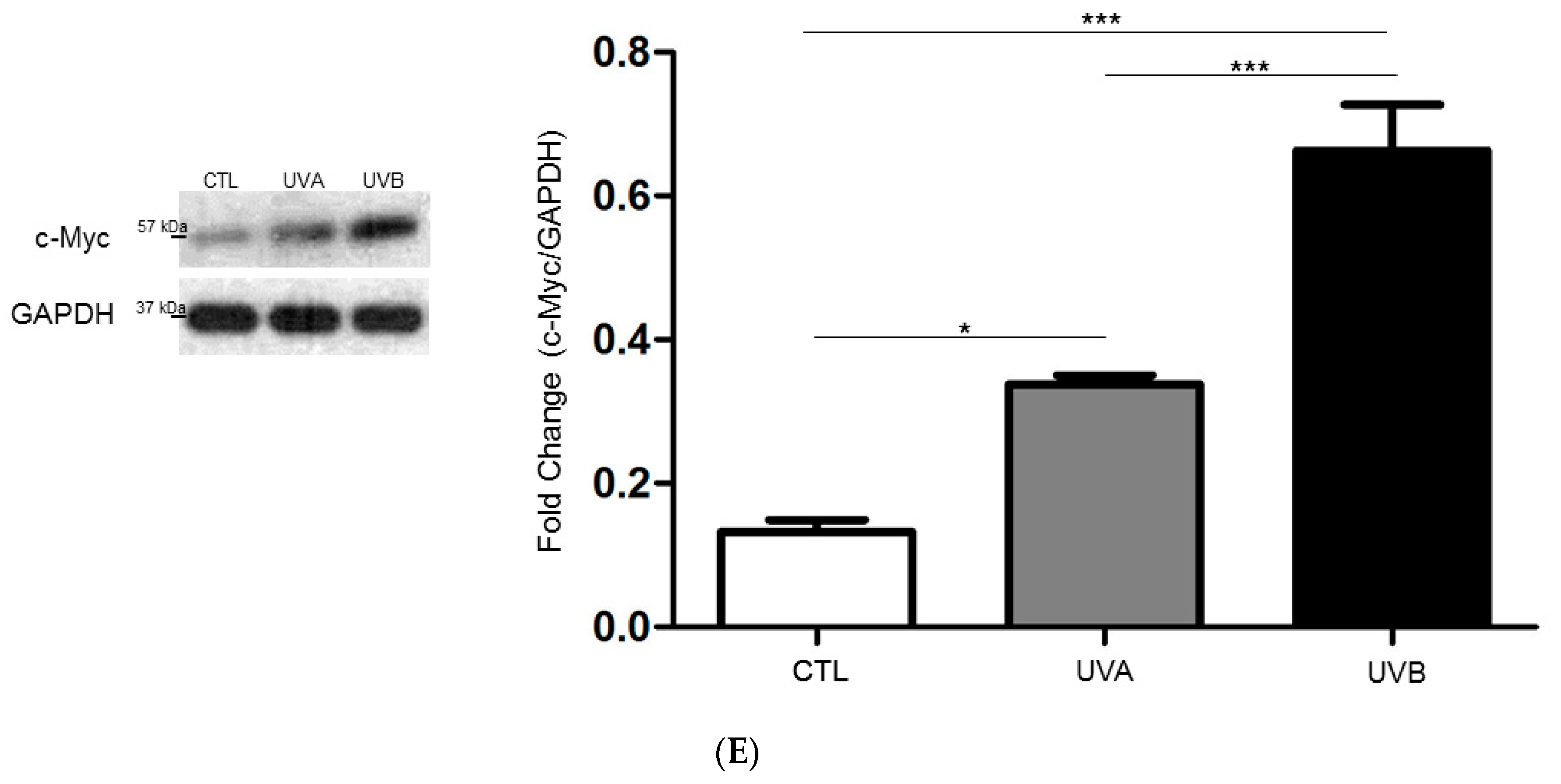
3.5. Network Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2-DE | two-dimensional electrophoresis |
| DNP | 2,4-dinitrophenylhydrazine |
| MALDI-TOF-MS | matrix-assisted laser desorption/ionization time of flight mass spectrometry |
| PMF | peptide mass fingerprinting |
| ROS | reactive oxygen species |
| IPG | immobilized pH gradient |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| IHC | immunohistochemistry |
| MAPK | mitogen-activated protein kinase |
| DTT | dithioerythritol |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| UV | ultraviolet |
| 8-OHdG | 8-Oxo-2’-deoxyguanosine |
| TBST | tris-buffered saline Tween-20 |
| PVDF | polyvinylidene difluoride |
| TUNEL | terminal deoxynucleotidyl transferase dUTP nick end labeling |
| ECM | extracellular matrix |
| UPR | unfolding protein response |
| MMP | matrix metallopeptidases |
| PCNA | proliferating cell nuclear antigen |
References
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef] [PubMed]
- Mohania, D.; Chandel, S.; Kumar, P.; Verma, V.; Digvijay, K.; Tripathi, D.; Choudhury, K.; Mitten, S.K.; Shah, D. Ultraviolet Radiations: Skin Defense-Damage Mechanism. Adv. Exp. Med. Biol. 2017, 996, 71–87. [Google Scholar] [PubMed]
- De Gruijl, F.R. Photocarcinogenesis: UVA vs. UVB. Methods Enzymol. 2000, 319, 359–366. [Google Scholar] [PubMed]
- Nishigori, C.; Hattori, Y.; Toyokuni, S. Role of reactive oxygen species in skin carcinogenesis. Antioxid. Redox Signal. 2004, 6, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Barolet, D.; Christiaens, F.; Hamblin, M.R. Infrared and skin: Friend or foe. J. Photochem. Photobiol. B 2016, 155, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Renert-Yuval, Y.; Guttman-Yassky, E. The ‘omics’ revolution: Redefining the understanding and treatment of allergic skin diseases. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Rice, R.H.; Durbin-Johnson, B.P.; Mann, S.M.; Salemi, M.; Urayama, S.; Rocke, D.M.; Phinney, B.S.; Sundberg, J.P. Corneocyte proteomics: Applications to skin biology and dermatology. Exp. Dermatol. 2018, 27, 931–938. [Google Scholar] [CrossRef]
- Villaseñor-Park, J.; Ortega-Loayza, A.G. Microarray technique, analysis, and applications in dermatology. J. Investig. Dermatol. 2013, 133, e7. [Google Scholar] [CrossRef]
- Jinnin, M. Recent progress in studies of miRNA and skin diseases. J. Dermatol. 2015, 42, 551–558. [Google Scholar] [CrossRef]
- Fang, J.Y.; Wang, P.W.; Huang, C.H.; Chen, M.H.; Wu, Y.R.; Pan, T.L. Skin aging caused by intrinsic or extrinsic processes characterized with functional proteomics. Proteomics 2016, 16, 2718–2731. [Google Scholar] [CrossRef]
- Hammers, C.M.; Tang, H.Y.; Chen, J.; Emtenani, S.; Zheng, Q.; Stanley, J.R. Research Techniques Made Simple: Mass Spectrometry for Analysis of Proteins in Dermatological Research. J. Investig. Dermatol. 2018, 138, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Lupu, M.; Caruntu, C.; Ghita, M.A.; Voiculescu, V.; Voiculescu, S.; Rosca, A.E.; Caruntu, A.; Moraru, L.; Popa, I.M.; Calenic, B.; et al. Gene Expression and Proteome Analysis as Sources of Biomarkers in Basal Cell Carcinoma. Dis. Markers 2016, 2016, 9831237. [Google Scholar] [CrossRef] [PubMed]
- De Jager, T.L.; Cockrell, A.E.; Du Plessis, S.S. Ultraviolet Light Induced Generation of Reactive Oxygen Species. Adv. Exp. Med. Biol. 2017, 996, 15–23. [Google Scholar] [PubMed]
- Natarajan, V.T.; Ganju, P.; Ramkumar, A.; Grover, R.; Gokhale, R.S. Multifaceted pathways protect human skin from UV radiation. Nat. Chem. Biol. 2014, 10, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, M.; Ueda, M.; Budiyanto, A.; Bito, T.; Oka, M.; Fukunaga, M.; Tsuru, K.; Horikawa, T. UV-induced skin damage. Toxicology 2003, 189, 21–39. [Google Scholar] [CrossRef]
- Radak, Z.; Zhao, Z.; Goto, S.; Koltai, E. Age-associated neurodegeneration and oxidative damage to lipids, proteins and DNA. Mol Aspects Med. 2011, 32, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Quintanilha, A.; Moradas-Ferreira, P. Protein oxidation, repair mechanisms and proteolysis in Saccharomyces cerevisiae. IUBMB Life 2007, 59, 293–298. [Google Scholar] [CrossRef]
- Kruk, J.; Duchnik, E. Oxidative stress and skin diseases: Possible role of physical activity. Asian Pac. J. Cancer Prev. 2014, 15, 561–568. [Google Scholar] [CrossRef]
- Silva, S.A.M.E.; Michniak-Kohn, B.; Leonardi, G.R. An overview about oxidation in clinical practice of skin aging. An. Bras. Dermatol. 2017, 92, 367–374. [Google Scholar] [CrossRef]
- Baraibar, M.A.; Friguet, B. Oxidative proteome modifications target specific cellular pathways during oxidative stress, cellular senescence and aging. Exp. Gerontol. 2013, 48, 620–625. [Google Scholar] [CrossRef]
- Hung, C.F.; Chen, W.Y.; Aljuffali, I.A.; Shih, H.C.; Fang, J.Y. The risk of hydroquinone and sunscreen over-absorption via photodamaged skin is not greater in senescent skin as compared to young skin: Nude mouse as an animal model. Int. J. Pharm. 2014, 471, 135–145. [Google Scholar] [CrossRef] [PubMed]
- McGinley, J.N.; Thompson, H.J. Quantitative assessment of mammary gland density in rodents using digital image analysis. Biol. Proced. Online 2011, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.L.; Wang, P.W.; Aljuffali, I.A.; Hung, Y.Y.; Lin, C.F.; Fang, J.Y. Dermal toxicity elicited by phthalates: Evaluation of skin absorption, immunohistology, and functional proteomics. Food Chem. Toxicol. 2014, 65, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.C.; Lin, S.M.; Chen, M.F.; Pan, T.L.; Wang, P.W.; Yeh, C.T. Evaluation of serum matrix metalloproteinase (MMP)-9 to MMP-2 ratio as a biomarker in hepatocellular carcinoma. Hepatogastroenterology 2010, 57, 98–102. [Google Scholar] [PubMed]
- Wang, P.W.; Cheng, Y.C.; Hung, Y.C.; Lee, C.H.; Fang, J.Y.; Li, W.T.; Wu, Y.R.; Pan, T.L. Red Raspberry Extract Protects the Skin against UVB-Induced Damage with Antioxidative and Anti-inflammatory Properties. Oxid. Med. Cell. Longev. 2019, 2019, 9529676. [Google Scholar] [CrossRef]
- Lin, C.H.; Wang, P.W.; Pan, T.L.; Bazylak, G.; Liu, E.K.; Wei, F.C. Proteomic profiling of oxidative stress in human victims of traffic-related injuries after lower limb revascularization and indication for secondary amputation. J. Pharm. Biomed. Anal. 2010, 51, 784–794. [Google Scholar] [CrossRef]
- Pan, T.L.; Wang, P.W.; Huang, C.C.; Yeh, C.T.; Hu, T.H.; Yu, J.S. Network analysis and proteomic identification of vimentin as a key regulator associated with invasion and metastasis in human hepatocellular carcinoma cells. J. Proteom. 2012, 75, 4676–4692. [Google Scholar] [CrossRef]
- Matsumura, Y.; Ananthaswamy, H.N. Toxic effects of ultraviolet radiation on the skin. Toxicol. Appl. Pharmacol. 2004, 195, 298–308. [Google Scholar] [CrossRef]
- Skotarczak, K.; Osmola-Mańkowska, A.; Lodyga, M.; Polańska, A.; Mazur, M.; Adamski, Z. Photoprotection: Facts and controversies. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 98–112. [Google Scholar]
- Awad, F.; Assrawi, E.; Louvrier, C.; Jumeau, C.; Giurgea, I.; Amselem, S.; Karabina, S.A. Photoaging and skin cancer: Is the inflammasome the missing link? Mech. Ageing Dev. 2018, 172, 131–137. [Google Scholar] [CrossRef]
- Polefka, T.G.; Meyer, T.A.; Agin, P.P.; Bianchini, R.J. Effects of solar radiation on the skin. J. Cosmet. Dermatol. 2012, 11, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Rittié, L.; Fisher, G.J. Natural and sun-induced aging of human skin. Cold Spring Harb. Perspect. Med. 2015, 5, a015370. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.; Qin, Z.; Xia, W.; Shao, Y.; Voorhees, J.J.; Fisher, G.J. Matrix-degrading metalloproteinases in photoaging. J. Investig. Dermatol. Symp. Proc. 2009, 14, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Hruza, L.L.; Pentland, A.P. Mechanisms of UV-induced inflammation. J. Investig. Dermatol. 1993, 100, 35S–41S. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Werner, S. Nrf2—A regulator of keratinocyte redox signaling. Free Radic. Biol. Med. 2015, 88, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Farrukh, M.R.; Nissar, U.A.; Afnan, Q.; Rafiq, R.A.; Sharma, L.; Amin, S.; Kaiser, P.; Sharma, P.R.; Tasduq, S.A. Oxidative stress mediated Ca(2+) release manifests endoplasmic reticulum stress leading to unfolded protein response in UV-B irradiated human skin cells. J. Dermatol. Sci. 2014, 75, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, H.; Simard-Bisson, C.; Larouche, D.; Tanguay, R.M.; Blouin, R.; Germain, L. The small heat-shock protein Hsp27 undergoes ERK-dependent phosphorylation and redistribution to the cytoskeleton in response to dual leucine zipper-bearing kinase expression. J. Investig. Dermatol. 2010, 130, 74–85. [Google Scholar] [CrossRef]
- Vashishta, A.; Saraswat Ohri, S.; Vetvickova, J.; Fusek, M.; Ulrichova, J.; Vetvicka, V. Procathepsin D secreted by HaCaT keratinocyte cells—A novel regulator of keratinocyte growth. Eur. J. Cell Biol. 2007, 86, 303–313. [Google Scholar] [CrossRef]
- Okada, R.; Wu, Z.; Zhu, A.; Ni, J.; Zhang, J.; Yoshimine, Y.; Peters, C.; Saftig, P.; Nakanishi, H. Cathepsin D deficiency induces oxidative damage in brain pericytes and impairs the blood-brain barrier. Mol. Cell. Neurosci. 2015, 64, 51–60. [Google Scholar] [CrossRef]
- Goy, C.; Czypiorski, P.; Altschmied, J.; Jakob, S.; Rabanter, L.L.; Brewer, A.C.; Ale-Agha, N.; Dyballa-Rukes, N.; Shah, A.M.; Haendeler, J. The imbalanced redox status in senescent endothelial cells is due to dysregulated Thioredoxin-1 and NADPH oxidase 4. Exp. Gerontol. 2014, 56, 45–52. [Google Scholar] [CrossRef]
- Bosch, R.; Philips, N.; Suárez-Pérez, J.A.; Juarranz, A.; Devmurari, A.; Chalensouk-Khaosaat, J.; González, S. Mechanisms of Photoaging and Cutaneous Photocarcinogenesis, and Photoprotective Strategies with Phytochemicals. Antioxidants 2015, 4, 248–268. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Suzuki, J.S.; Tanaka, M.; Ogiso, M.; Tohyama, C.; Kobayashi, S. Gene expression in skin tumors induced in hairless mice by chronic exposure to ultraviolet B irradiation. Photochem. Photobiol. 1997, 65, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Matsushima, K.; Miyamoto, K.; Oe, T. UV irradiation-induced methionine oxidation in human skin keratins: Mass spectrometry-based non-invasive proteomic analysis. J. Proteom. 2016, 133, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Thiele, J.J.; Hsieh, S.N.; Briviba, K.; Sies, H. Protein oxidation in human stratum corneum: Susceptibility of keratins to oxidation in vitro and presence of a keratin oxidation gradient in vivo. J. Investig. Dermatol. 1999, 113, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Gęgotek, A.; Domingues, P.; Skrzydlewska, E. Proteins involved in the antioxidant and inflammatory response in rutin-treated human skin fibroblasts exposed to UVA or UVB irradiation. J. Dermatol. Sci. 2018, 90, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Syed, D.N.; Afaq, F.; Mukhtar, H. Differential activation of signaling pathways by UVA and UVB radiation in normal human epidermal keratinocytes. Photochem. Photobiol. 2012, 88, 1184–1190. [Google Scholar] [CrossRef]
- López-Camarillo, C.; Ocampo, E.A.; Casamichana, M.L.; Pérez-Plasencia, C.; Alvarez-Sánchez, E.; Marchat, L.A. Protein kinases and transcription factors activation in response to UV-radiation of skin: Implications for carcinogenesis. Int. J. Mol. Sci. 2012, 13, 142–172. [Google Scholar] [CrossRef]
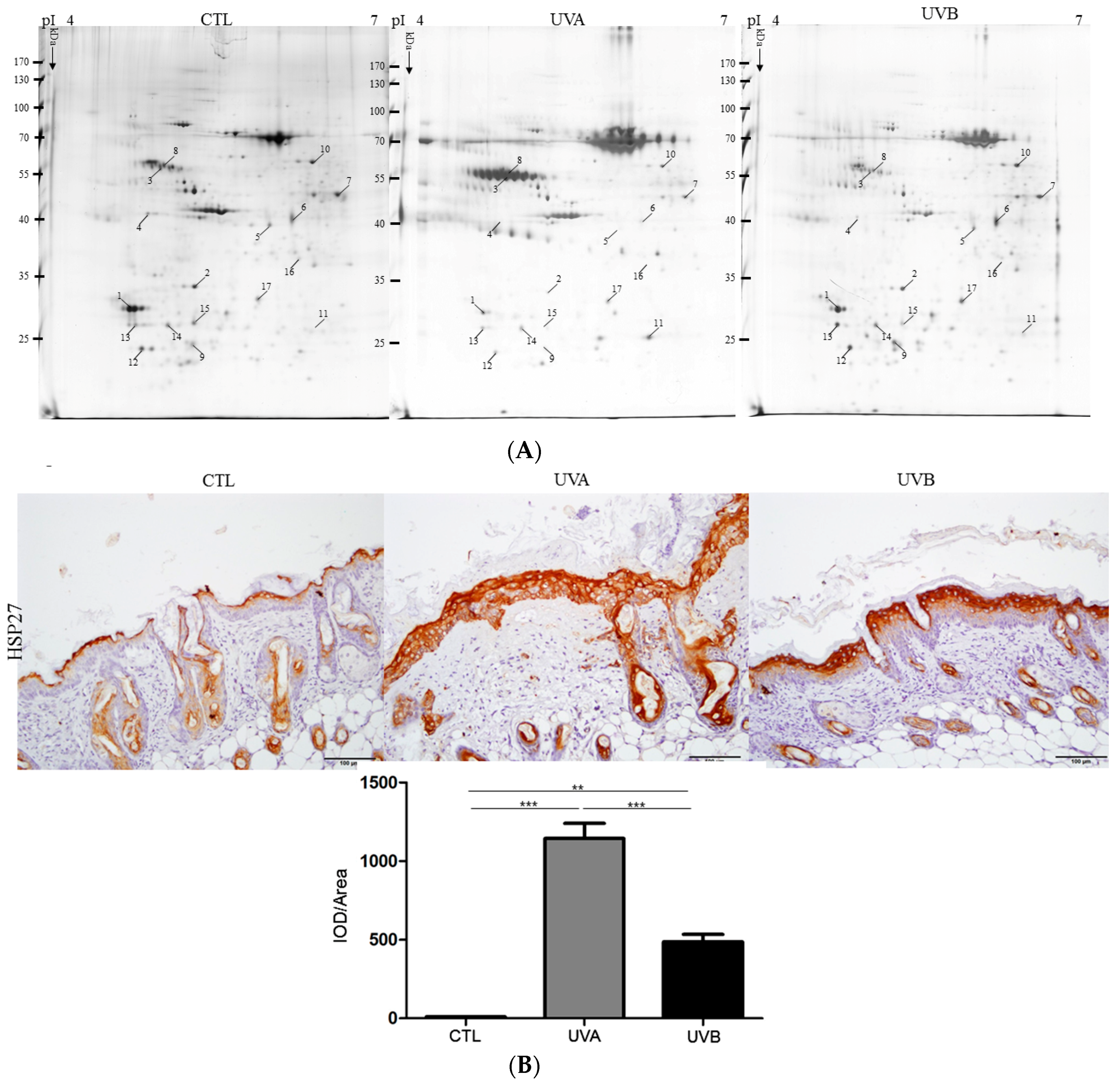
| Spot No. | Protein | Accession No. | Mw (kDa) | pI | Matched- Peptides | SCORE (a) (Sequence Coverage %) | Ratios (b) | Biological Function | ||
|---|---|---|---|---|---|---|---|---|---|---|
| UVA/CTL | UVB/CTL | p-Value (c) | ||||||||
| 1 | Stratifin (1433S) | O70456 | 27.803 | 4.72 | 12 | 101 (48%) | −6.62 ± 0.07 | −0.92 ± 0.04 | 0.002 | Involved in the regulation of both general and specialized signaling pathways. It also controls protein synthesis and epithelial cell growth via protein kinase B/mammalian target of rapamycin (Akt/mTOR) pathway. |
| 2 | Skin aspartic protease (SASP) | Q09PK2 | 33.637 | 5.07 | 11 | 65 (24%) | −5.84 ± 0.02 | −0.32 ± 0.06 | 0.037 | Majorly expresses in the epidermis and hair follicles. |
| 3 | Cytokeratin-14 (K1C14) | Q61781 | 53.176 | 5.10 | 37 | 329 (71%) | 7.67 ± 0.02 | −0.57 ± 0.02 | 0.005 | Enhances KRT5-KRT14 filaments to self-organize into large bundles and promotes the properties related to resilience of keratin intermediate filaments. |
| 4 | 40S Ribosomal protein SA (RSSA) | P14206 | 32.935 | 4.80 | 10 | 90 (34%) | −1.89 ± 0.02 | −0.57 ± 0.04 | 0.055 | Involved in cell adhesion to the basement membrane and activation of signaling transduction cascades. |
| 5 | Serpin B5/Maspin (SPB5) | P70124 | 42.484 | 5.55 | 18 | 195 (66%) | −2.96 ± 0.06 | −0.86 ± 0.01 | 0.037 | Inhibits the growth, invasion, and metastatic properties of mammary tumors. |
| 6 | Cathepsin D (CATD) | P18242 | 45.381 | 6.71 | 13 | 134 (31%) | −5.92 ± 0.01 | 0.55 ± 0.02 | 0.049 | Regulation in intracellular protein breakdown. |
| 7 | α-enolase (ENOA) | P17182 | 47.322 | 6.7 | 24 | 231 (58%) | −3.18 ± 0.04 | −1.53 ± 0.01 | 0.042 | Multifunctional enzyme to play a role in various processes including growth control, hypoxia tolerance, and allergic responses. |
| 8 | Cytokeratin-10 (K1C10) | P02535 | 57.178 | 5.00 | 12 | 71 (19%) | 6.52 ± 0.03 | −2.86 ± 0.02 | 0.008 | Establishment of the epidermal barrier on skin. |
| 9 | Lactoylglutathione lyase (LGUL) | Q9CPU0 | 20.967 | 5.24 | 13 | 139 (55%) | −1.59 ± 0.01 | 1.64 ± 0.02 | 0.003 | Regulation of tumor necrosis factor (TNF)-mediated activation of NF-kappa-B. |
| 10 | Protein disulfide-isomerase A3 (PDIA3) | P27773 | 57.099 | 5.88 | 27 | 260 (50%) | −2.23 ± 0.04 | −0.28 ± 0.02 | 0.084 | Induces the rearrangement of -S–S- bonds in proteins. |
| 11 | Heat shock 27 kDa protein (HSP27) | P14602 | 23.057 | 6.12 | 7 | 77 (35%) | 4.52 ± 0.03 | 1.86 ± 0.02 | 0.014 | Functions as a molecular chaperone to maintain denatured proteins in a folding-competent state. |
| 12 | Translationally-controlled tumor protein (TCTP) | P63028 | 19.592 | 4.72 | 12 | 103 (44%) | −2.52 ± 0.06 | 0.56 ± 0.02 | 0.038 | Calcium binding and microtubule stabilization. |
| 13 | Proteasome subunit alpha type-5 (PSA5) | Q9Z2U1 | 26.565 | 4.74 | 11 | 104 (56%) | −1.52 ± 0.03 | 2.86 ± 0.02 | 0.004 | Component of the 20S core proteasome complex linked to the degradation of intracellular proteins. |
| 14 | Rho GDP-dissociation inhibitor 2 (GDIR2) | Q61599 | 22.894 | 4.97 | 9 | 98 (69%) | −0.32 ± 0.13 | 0.86 ± 0.46 | 0.024 | Involved in reorganization of the actin cytoskeleton through Rho family members. |
| 15 | Rho GDP-dissociation inhibitor 1 (GDIR1) | Q99PT1 | 23.450 | 5.12 | 14 | 133 (59%) | −1.36 ± 0.08 | −0.56 ± 0.02 | 0.005 | Modulates Rho proteins homeostasis. |
| 16 | 60S acidic ribosomal protein P0 (RLA0) | P14869 | 34.366 | 5.91 | 8 | 79 (41%) | −0.38± 0.03 | −0.58±0.04 | 0.086 | Playing a pivotal role in the interaction of the ribosome with GTP-bound translation factors. |
| 17 | Beta-actin (ACTB; Frag.) | P60710 | 42.052 | 5.29 | 7 | 85 (47%) | −0.89 ± 0.01 | 1.64 ± 0.02 | 0.004 | Actin exists in both monomeric (G-actin) and polymeric (F-actin) forms to regulate functions including cell motility and contraction. It also localizes in the nucleus to control gene transcription, motility and repair of damaged DNA. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.-W.; Hung, Y.-C.; Lin, T.-Y.; Fang, J.-Y.; Yang, P.-M.; Chen, M.-H.; Pan, T.-L. Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry. Antioxidants 2019, 8, 569. https://doi.org/10.3390/antiox8120569
Wang P-W, Hung Y-C, Lin T-Y, Fang J-Y, Yang P-M, Chen M-H, Pan T-L. Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry. Antioxidants. 2019; 8(12):569. https://doi.org/10.3390/antiox8120569
Chicago/Turabian StyleWang, Pei-Wen, Yu-Chiang Hung, Tung-Yi Lin, Jia-You Fang, Pei-Ming Yang, Mu-Hong Chen, and Tai-Long Pan. 2019. "Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry" Antioxidants 8, no. 12: 569. https://doi.org/10.3390/antiox8120569
APA StyleWang, P.-W., Hung, Y.-C., Lin, T.-Y., Fang, J.-Y., Yang, P.-M., Chen, M.-H., & Pan, T.-L. (2019). Comparison of the Biological Impact of UVA and UVB upon the Skin with Functional Proteomics and Immunohistochemistry. Antioxidants, 8(12), 569. https://doi.org/10.3390/antiox8120569








