Abstract
Understanding growth mechanisms in brown algae is a current scientific and economic challenge that can benefit from the modeling of their metabolic networks. The sequencing of the genomes of Saccharina japonica and Cladosiphon okamuranus has provided the necessary data for the reconstruction of Genome–Scale Metabolic Networks (GSMNs). The same in silico method deployed for the GSMN reconstruction of Ectocarpus siliculosus to investigate the metabolic capabilities of these two algae, was used. Integrating metabolic profiling data from the literature, we provided functional GSMNs composed of an average of 2230 metabolites and 3370 reactions. Based on these GSMNs and previously published work, we propose a model for the biosynthetic pathways of the main carotenoids in these two algae. We highlight, on the one hand, the reactions and enzymes that have been preserved through evolution and, on the other hand, the specificities related to brown algae. Our data further indicate that, if abscisic acid is produced by Saccharina japonica, its biosynthesis pathway seems to be different in its final steps from that described in land plants. Thus, our work illustrates the potential of GSMNs reconstructions for formalizing hypotheses that can be further tested using targeted biochemical approaches.
1. Introduction
Saccharina japonica and Cladosiphon okamuranus are two brown algal species widely used in Asian aquaculture, known, respectively, as kombu and mozuku. S. japonica is the most important edible alga from an economic viewpoint. Its production was multiplied by 8 over the last 40 years: 0.8 million tons harvested in 1976 against 4.5 million tons in 2004 [1]. The annual production of C. okamuranus is estimated to be 20,000 tons [2,3]. Thus, studying the mechanisms of biomass production of those organisms through their Genome–Scale Metabolic Networks (GSMNs) may have a direct interest for algoculture [4]. Indeed, to produce molecules with high added value through biotechnological engineering, one first needs to understand their biosynthetic pathways. Brown algae produce specific carotenoids, some of them having potentially positive effects on human health, in particular, due to their antioxidant properties [5,6,7,8,9,10]. Some cleaved carotenoid derivatives are also signaling molecules and important phytohormones in land plants, like strigolactones or abscisic acid (ABA) [11,12,13].
Carotenoids are a ubiquitous class of molecules found in many organisms, such as plants, fungi, algae, and even metazoans. They are membrane–stabilizing hydrophobic molecules that also play a crucial role as photoprotective pigments in photosynthetic organisms [11,12,14,15]. Some carotenoids have been preserved through evolution, such as violaxanthin, which is detected in plants, green algae, and brown algae, among other organisms. This molecule is, on the one hand, the entry point for the biosynthesis of xanthophylls specific to brown algae, like fucoxanthin, which is the main marine carotenoid of this lineage [16,17,18] and, on the other hand, one of the precursors in the biosynthesis of ABA. Except for Laminariales [19], nothing is known about ABA in brown algae, including Ectocarpales like the model species Ectocarpus siliculosus or C. okamuranus. Given the potential importance of this signaling molecule in the biology of brown algae, one of our objectives when reconstructing the GSMNs of S. japonica and C. okamuranus was to clarify the possible contribution of land plant-like biosynthetic enzymes to the production of ABA in brown algae.
There have been extensive efforts to improve GSMN reconstructions from a computational perspective [20,21]. One current challenge is to integrate the knowledge of genome-based evidence and the knowledge coming from metabolome-based evidence, which is usually much more heterogeneous [22]. Following the protocol recommended in Palsson and Thiele [23] and in line with previous work on the model algae E. siliculosus [24], Tisochrysis luteae [21] and Chondrus crispus [25], the automated reconstruction methods and labor-intensive manual curation were combined to build a genome–scale metabolic model in the brown algae S. japonica and C. okamuranus, with a specific focus on the carotenoid biosynthesis pathway.
2. Materials and Methods
2.1. Data Sources and Cleaning
Genome and protein sequences of S. japonica and C. okamuranus are freely available, respectively, in references [26] and [3]. These algae are usually grown in non-axenic media, and the presence of contaminating sequences was demonstrated in the published genome of S. japonica [27]. Therefore, Taxoblast analysis (version 1.21beta) was carried out to discriminate prokaryote and eukaryote in order to filter out all prokaryotic sequences from the genome of S. japonica. Blast analyses were performed using diamond blastx (version 0.9.18) [28] against the nr database (downloaded on 13 August 2016) and the nodes.dmp (downloaded on 23 March 2018) from https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/. The cleaned version of the S. japonica genome is available under the following link: http://abims.sb-roscoff.fr/resources/genomic_resources.
2.2. Reconstruction of Genome–Scale Metabolic Networks
AuReMe (AUtomatic REconstruction of MEtabolic models—version 1.2.4) dedicated to “à la carte” reconstruction of GSMNs [21] was used to reconstruct the GSMNs of S. japonica and C. okamuranus. This workflow was installed locally from a docker image. As suggested for state-of-the-art GSMN reconstruction methods [23], this workflow allowed, through the encapsulation of several tools and the local installation of specialized tools in the docker container, to create a high-quality GSMNs based on genomic and metabolic profiling data. It combined 2 complementary approaches to extract information from the genome sequences. One results from the functional annotation of the genome (see Table S1 and Figure S1 of Supplementary Materials), and the other is derived from the comparison with GSMNs and protein sequences of organisms selected as templates. These intermediate networks were then merged, analyzed both qualitatively (topological analysis) and quantitatively (constraint-based analysis), refined by manual curation and enriched with metabolic profiling data extracted from the literature (see Table S2 and Table S3 of Supplementary Materials).
To reconstruct the intermediate annotation-based network, predicted coding regions in transcripts were functionally annotated using the Trinotate pipeline (version 3.0.1) [29]. An internal Trinotate script extracted the Gene Ontology Terms (GOT) [30,31]. The Kyoto Encyclopedia of Genes and Genomes identifiers (KEGG) [32] from the annotation file were used to retrieve the EC numbers associated with the genes. This step was performed via the KEGG–APi REST (REpresentational State Transfer) application programming interface (http://www.kegg.jp/kegg/rest/keggapi.html). The annotation concerning S. japonica was enriched by further analyses carried out with Kobas [33] and Blast2GO [34] with an e-value cutoff of 1e–05. A file created in the GenBank format, containing all the available data, was then generated using the following script: https://github.com/ArnaudBelcour/gbk_from_gff. This GenBank file was used as an input to the PathoLogic software from the Pathway Tools suite (version 20.5, default settings) [35]. The database containing the information from the annotation was then exported in attribute-value flat files, which were necessary for further analysis in the AuReMe workspace.
To reconstruct the intermediate orthology–supported network, templates from 3 model organisms were used: Arabidopsis thaliana [36], E. siliculosus [24], and Nannochloropsis salina [37]. Those templates were chosen according to the quality of their GSMNs or their phylogenetic proximity to the 2 studied algae. Orthology searches between the 2 studied algae, and these 3 templates were carried out using the OrthoMCL (version 1.4) [38] and Inparanoid (version 4.0) [39]. The results of the latter were combined using the pantograph (version 0.2) [37]. Since the S. japonica GSMN was the first to be generated, it was added to the 3 previous templates during the reconstruction of the C. okamuranus GSMN. As the A. thaliana and N. salina data refer to the KEGG [32] and BIGG [40] databases, respectively, a mapping operation, intrinsic to AuReMe, against the MetaCyc database (version 20.5) [41,42,43] was performed to standardize the identifiers with the MetaNetX dictionary [44].
Once the networks resulting from the annotation-based and orthology-based approaches were merged, an automatic gap-filling step was conducted using Meneco (version 1.5.0) [45]. This tool first tests the ability of the topological GSMNs to produce a set of metabolite targets defined by metabolite occurrences from the literature (Tables S2 and S3 of the Supplementary Materials) according to a Boolean abstraction of the metabolic network expansion [46]. We tested here whether or not metabolic paths existed between specific compounds known to be present in the studied algae. When one or more target(s) was (were) not reachable in the GSMNs, Meneco proposed a list of missing reactions to complete the GSMNs. For this qualitative analysis, it was necessary to provide 2 lists, one containing the target metabolites and the other containing a description of the culture medium, including the cofactors (seeds) essential for biochemical reactions (Table S4 of the Supplementary Materials).
One established criterion to consider the GSMN functional was the production of biomass with balanced growth. Flux Balance Analysis (FBA) is a widely used method to quantitatively model the behavior of the system under given conditions. This optimization problem was based on the principle of mass conservation and considered the steady state assumption (intracellular metabolites at equilibrium). Mathematically, this reaction was modeled by a linear function to be optimized. This point was tested using the quantitative FBA python scripts from the Padmet toolbox, based on the CobraPy package [47] and provided in AuReMe. To do this, the same production reactions, transport, and exchange of biomass used in the reconstruction of E. siliculosus GSMN [24] were added to our GSMNs (Table S5 of the Supplementary Materials). Biologically, the biomass reaction modeled the synthesis of essential amino acids, membrane lipids, and sugars by the organism. When the flux associated with a metabolite was nil, this implied the incompleteness of the GSMNs resulting either from missing reactions in the biosynthesis pathway or from an accumulation of one or more reaction products due to the absence of a degradation reaction for those metabolites. To overcome this issue, manual analysis was then carried out, either by adding outward transport reactions or by determining the reactions missing through the analysis of GSMNs of similar organisms. This manual analysis was guided by the predictions performed by the Fluto gap-filling tool [48].
The Venn diagrams presented in the results section illustrating the comparison between S. japonica, C. okamuranus, and E. siliculosus GSMNs were obtained using http://bioinformatics.psb.ugent.be/webtools/Venn/. The final versions of those GSMNs are available through their respective Wiki–websites: https://gem-aureme.genouest.org/sjapgem and https://gem-aureme.genouest.org/cokagem. They are designed to allow the visualization and specific search of the different components of a GSMN (pathways, metabolites, reactions, genes) [21]. The traceability of the reconstruction procedure was ensured by the display of the source(s) of each reaction: Annotations, orthology, and manual curation or gap-filling. These user-friendly wiki websites allow semantic searches according to the W3C standards, and they are designed to enable updates according to the expertise of the scientific community.
2.3. Exploration and Assessment of Carotenoid Biosynthesis Pathways in Brown Algae
The manual exploration of the carotenoid biosynthesis pathway (CAROTENOID–PWY) and the first cycle of xanthophylls (PWY–5945) was conducted starting from the pathways encoded in the MetaCyc database (version 22.6) [43] and completed by further literature searches. These 2 pathways encoded the activity of 10 enzymes whose protein sequences were selected, when identified, in E. siliculosus or, if not available, in A. thaliana (sometimes supplemented with other terrestrial plants). These protein sequences are accessible either via the Uniprot database (https://www.uniprot.org) or via the Online Resource for Community Annotation of Eukaryotes (ORCAE) website for E. siliculosus (https://bioinformatics.psb.ugent.be/orcae/overview/EctsiV2) [49]. Sequences homologous to these enzymes were then searched for in the proteomes of 3 green algae Chlamydomonas reinhardtii, Volvox carteri, Ulva mutabilis, 3 red algae Chondrus crispus, Porphyra umbilicalis, Galdieria sulphuraria and 6 stramenopiles Nannochloropsis gaditana, N. salina, Phaeodactylum tricornutum, C. okamuranus, S. japonica. With the exception of Ulva mutabilis and E. siliculosus, for which homology searches were carried out through the ORCAE portal [49], all proteomes are accessible in the Genome database (https://www.ncbi.nlm.nih.gov/genome) of the NCBI. All accession numbers used are available as Table S6 in Supplementary Materials. Proteome indexing was performed using formatdb (version 2.2.16), then homology searches were done with blastp (version 2.7.1+). Hits with an e-value of less than 1e–5 were selected for alignment after checking the organization of their protein domains (HmmerWeb [50] version 2.30.0—https://www.ebi.ac.uk/Tools/hmmer/search/phmmer). The sequence files previously obtained were aligned using the Muscle algorithm implemented in the Seaview software (version 4.4.2) [51]. The associated phylogenetic trees were generated in Seaview using maximum likelihood (PhyML) with the LG (Le et Gascuel) model, the BIONJ algorithm (BIO Neighbor–joining) for the starting tree, and 100 bootstrap replicates. The 5 trees presented in the supplementary data have been edited with Figtree (version 1.4.4). The modifications made to the GSMNs are detailed in Appendix A. Some of them are related to the second cycle of xanthophylls (PWY–7949), and fucoxanthin biosynthesis pathway (PWY–7950) encoded in MetaCyc.
3. Results
3.1. Genome–Scale Metabolic Network Reconstructions
Presented here are the functional GSMNs for S. japonica and C. okamuranus, both of which are of similar size. The S. japonica GSMN is composed of 3345 reactions and 2211 metabolites, and the C. okamuranus GSMN comprises of 3390 reactions, 2255 metabolites. Both GSMNs were expected to sustain biomass production since Flux Balance Analysis (FBA) analyses evidenced that their maximal growth rates were 3.67 mmol·gDW−1·h−1 and 3.56 mmol·gDW−1·h−1 (millimole per gram dry weight per hour), respectively. They constituted valuable tools for assessing and visualizing the currently available knowledge on the metabolism of these organisms.
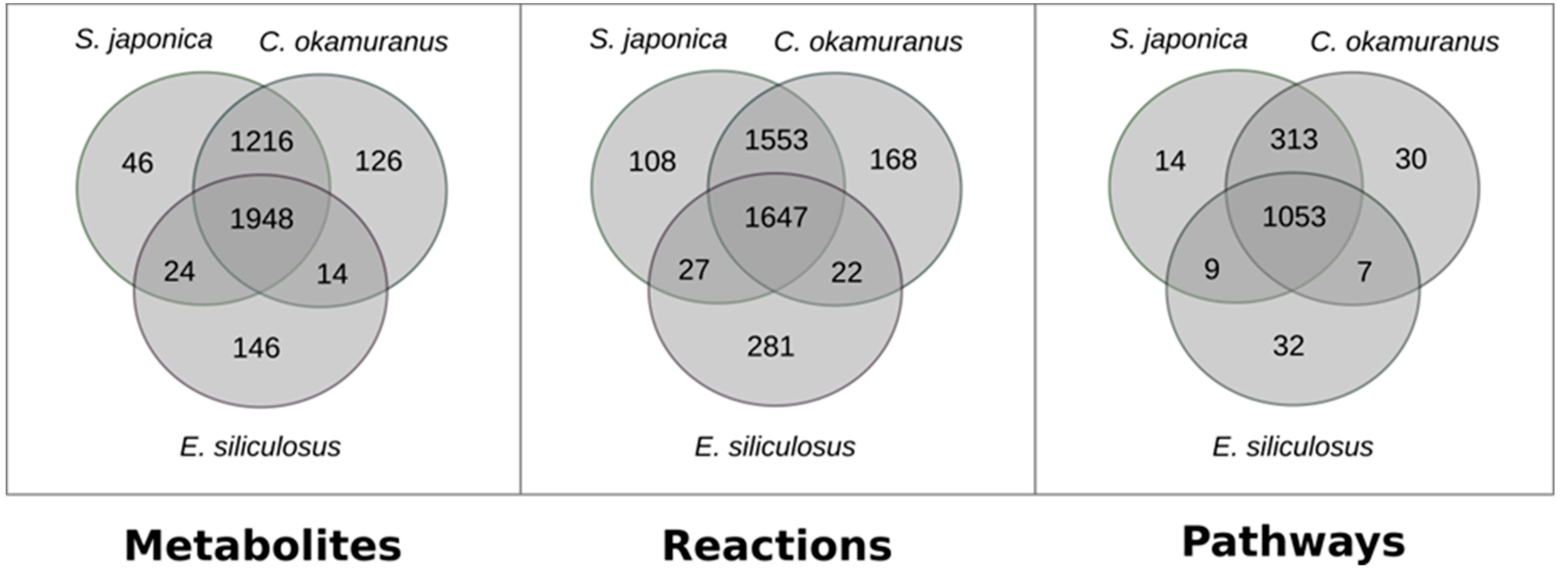
Both GSMNs were compared with the one from E. siliculosus [24], which was a brown algal model more closely related to C. okamuranus than to S. japonica. This was intended to test, at the global scale, if the differences between closely related species can be attributed to biological factors, or if they were merely due to technical differences in the reconstruction procedure. The results are shown in Figure 1.
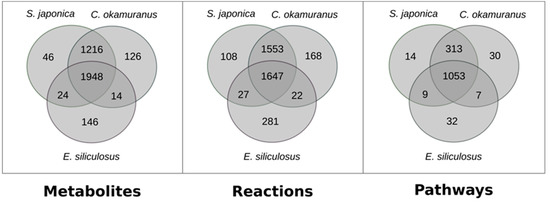
Figure 1.
Comparison of the content of genome–scale metabolic networks of the three brown algae S. japonica, C. okamuranus, and E. siliculosus.
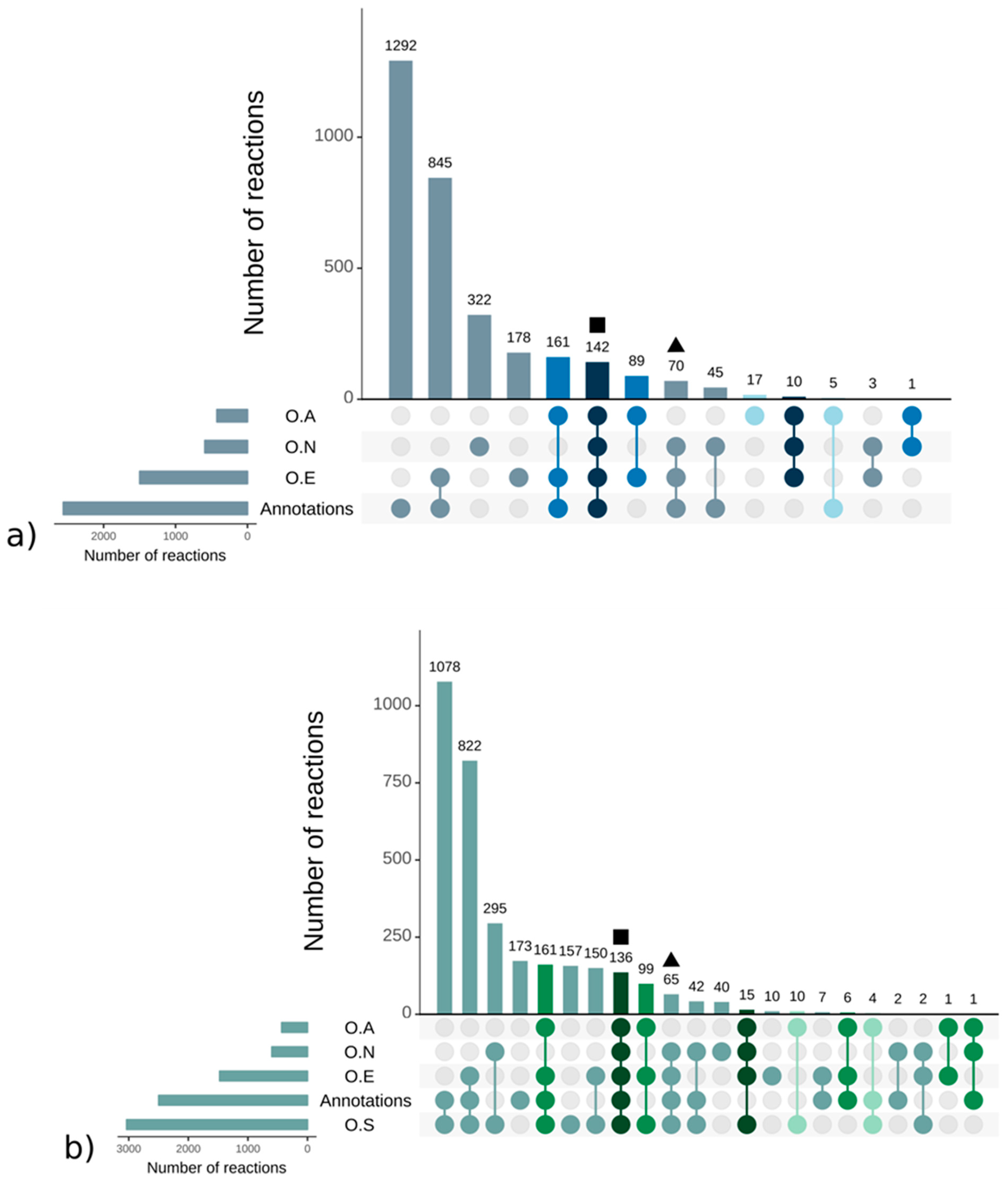
Strikingly, there were more reactions, metabolites, and pathways shared exclusively between S. japonica and C. okamuranus (1553 reactions) than exclusively between E. siliculosus and C. okamuranus (22 reactions), although the latter were phylogenetically closer. This was explained by examining the sources of reactions in the GSMNs (Figure 2). Of the reactions resulting from the annotations, 52% (S. japonica) and 44% (C. okamuranus) were not supported by the orthology with E. siliculosus. These differences were partly due to recent improvements in databases and annotation methods. The functional annotation of E. siliculosus that we used was done manually 10 years ago by an expert consortium [52], whereas we used Trinotate for S. japonica and C. okamuranus GSMNs. Therefore, the strong differences between E. siliculosus and the two other algae did not reflect biological differences, but different ways of annotation. One crucial point for the quality check was to build strong pathway-by-pathway expertise by scrutinizing them. AuReMe enabled the curation work to be stored and to reiterated on later versions of the GSMNs. To illustrate these points, intense manual curation centralized on the carotenoid biosynthesis pathway was performed.
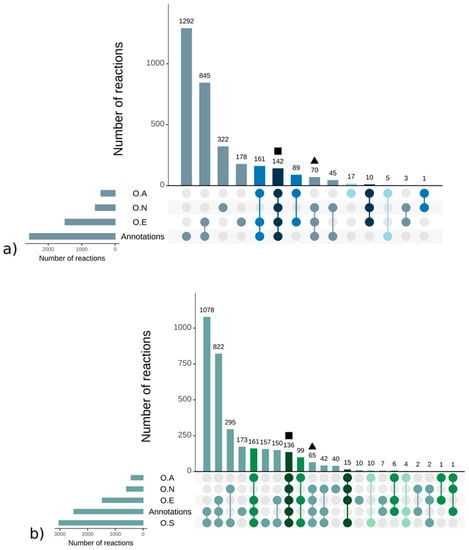
Figure 2.
Sources of reactions in the S. japonica (a) and C. okamuranus (b) genome—scale metabolic networks. Reactions supported by orthology with A. thaliana (O.A), E. siliculosus (O.E), N. salina (O.N), and S. japonica (O.S). The different colors refer to the core reactions (blue and green gradient, from darkest to lightest). The square and triangle shapes are the examples corresponding to the mevalonate pathway (MVA), methylerythritol phosphate pathway (MEP), and the geranylgeranyl diphosphate pathways presented in the main text.
3.2. Focused Exploration of GSMNs Regarding the Carotenoid Biosynthesis Pathway, Generalities, and Specificities
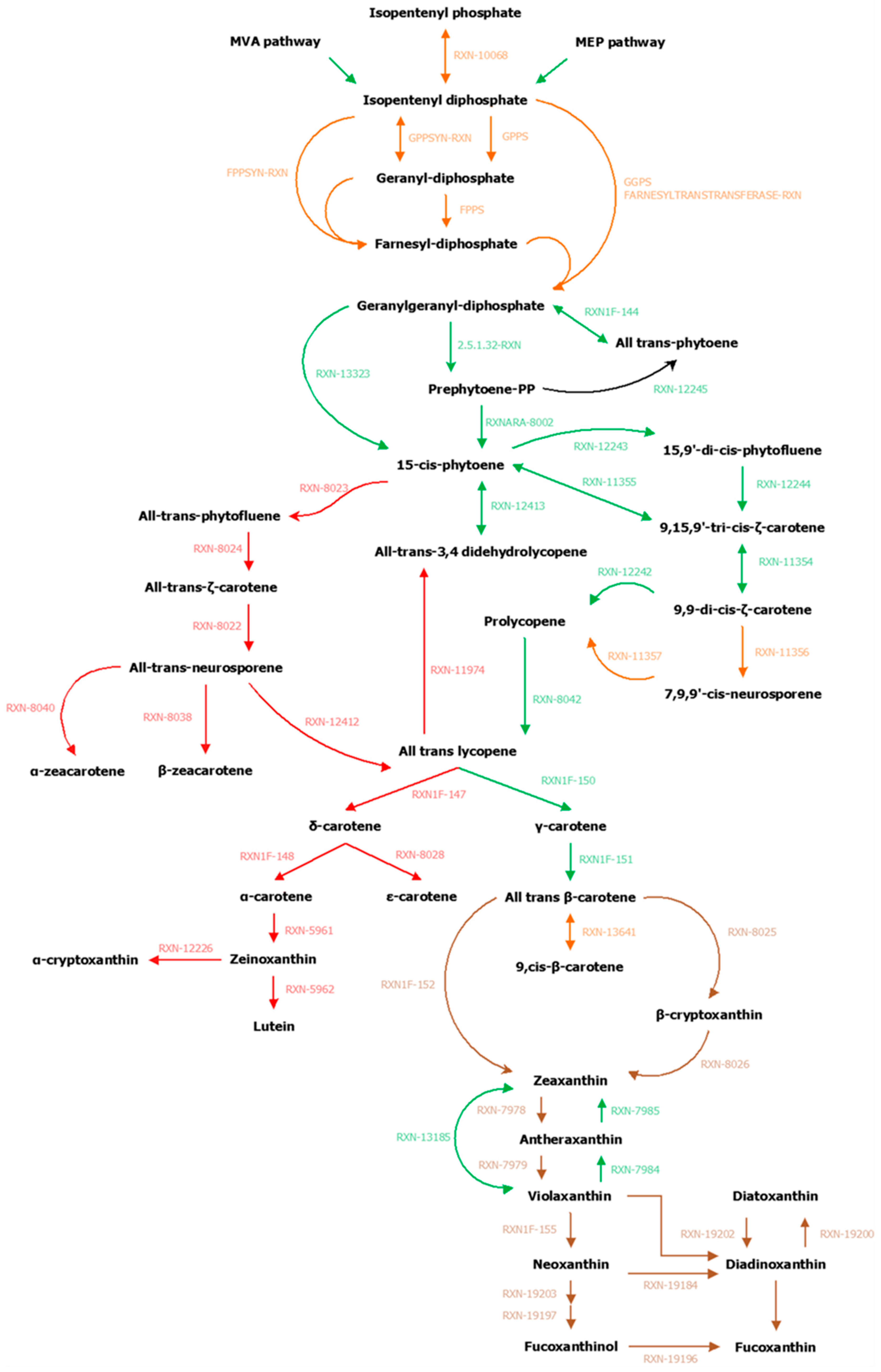
In order to facilitate the reading and understanding of the following sections, all the details related to the manual curation of GSMNs (names of enzymes or reactions, modifications of gene-reaction associations, adding or removing reactions, etc.) are available in Appendix A. Based on known pathways in terrestrial plants [53] and MetaCyc pathways (CAROTENOID-PWY), we conducted our exploration of the carotenoid pathway by starting with the transformation of geranylgeranyl diphosphate.
It should be noted, however, that this essential metabolic component was derived from isopentyl diphosphate (IPP). This fundamental and ubiquitous building block of the metabolism was itself obtained by the biosynthetic pathways methylerythritol phosphate pathway (MEP) and/or the mevalonate pathway (MVA) [54] (Figures S2 and S3 and Tables S7 and S8 of the Supplementary Materials). These pathways belonged to a set of reactions preserved through evolution that we will call core reactions. These reactions were easily and quickly identifiable within the GSMNs since they corresponded to those that have been inferred from the orthological search with a phylogenetically distant organism (here A. thaliana). In other words, these reactions were supported by all the components (orthology and annotations—square in Figure 2) that were used to build the GSMNs. Reactions associated with the enzyme, common to terrestrial plants and various algal phyla, which catalyzed the transformation of lycopene into β–carotene, also belonged to this set of core reactions.
From geranylgeranyl diphosphate, five conserved enzymes were involved in producing lycopene (Figure 3). The genes of these enzymes have been characterized in green, red, and brown algae [55]. Homology searches based on E. siliculosus proteins coupled with network annotations confirmed the accuracy of the enzymes associated with those reactions (triangle in Figure 2) in S. japonica and C. okamuranus.
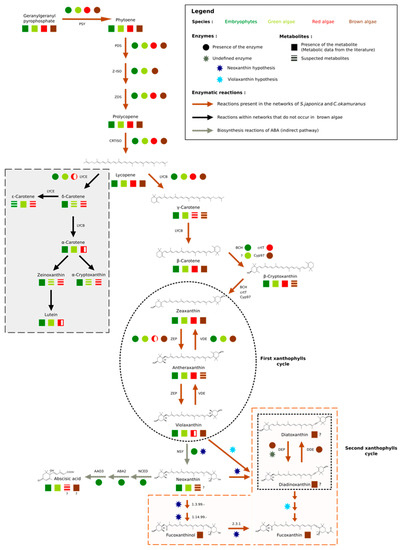
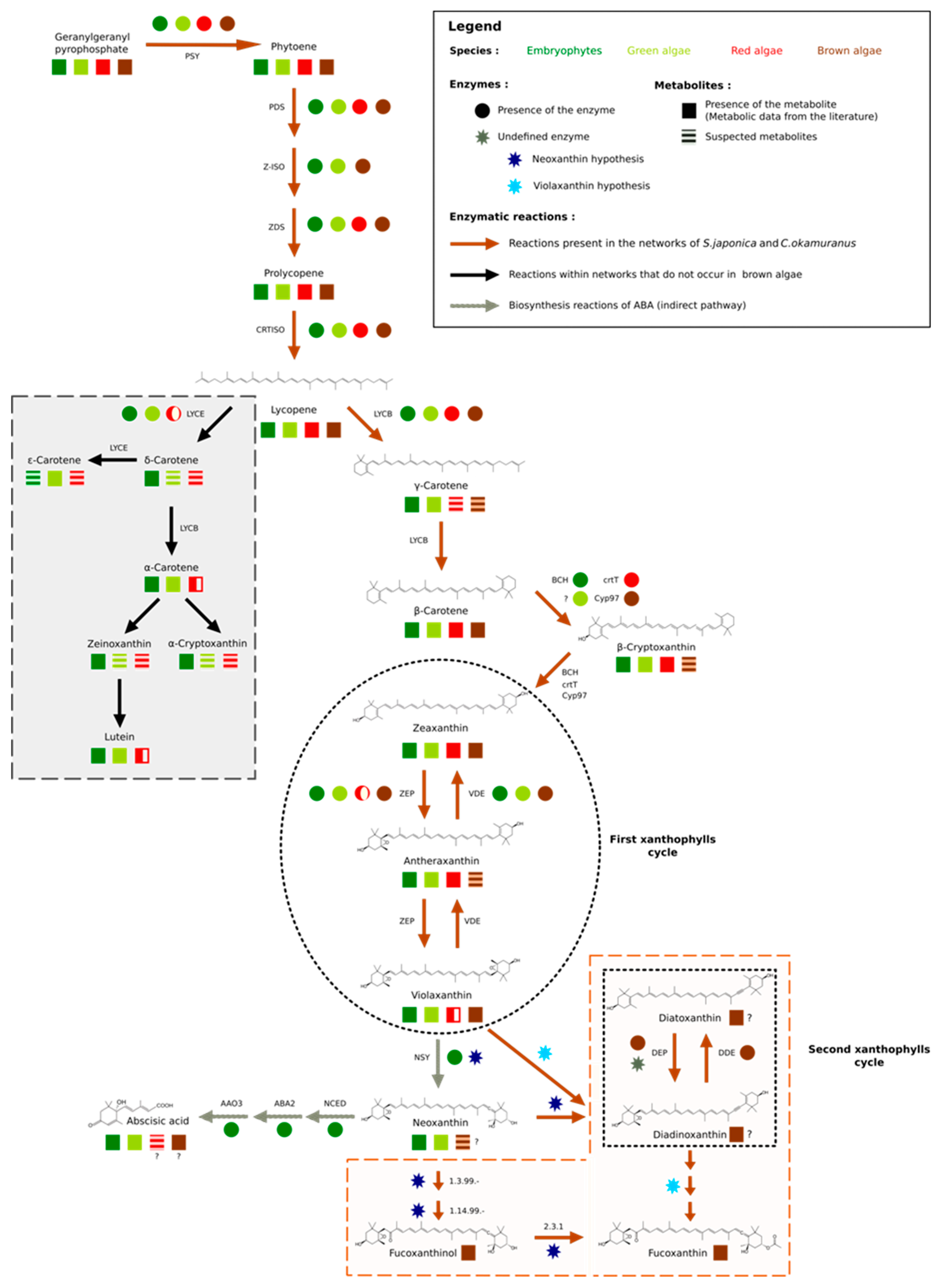
Figure 3.
Carotenoid biosynthesis. The mechanisms of carotenoid synthesis seem to be common to all algae and terrestrial plants. The first connection point is made from lycopene. The synthesis of δ–carotene and its derivatives is absent in red microalgae and brown algae (grey insert), while the synthesis of β–carotene is also common to all organisms. The second point of divergence appears during the first xanthophyll cycle (black circle) partially found in red algae. In terrestrial plants, violaxanthin is transformed into neoxanthin, the starting point for the synthesis of abscisic acid (ABA), among other metabolites. Nevertheless, if this step exists in various algal groups, it seems to be carried out by different enzymes. Finally, stramenopiles have a second xanthophyll cycle (black square), for which one of the supposed precursors is also neoxanthin and whose enzymes remain to be determined (brown insert). Since pathways have been hypothesized for diatoms, we extended this hypothesis to brown algae. The figure is based on our results and the following references [5,53,55,56,57,58,59,60,61,62].
Among the terrestrial plants, red, green, and brown algae, lycopene was the first point of connection in the synthesis of carotenoids, since it offered two possible routes: The one of α–carotene and the one of β–carotene. These pathways started respectively, with the action of lycopene ε–cyclase (LYCE) for α–carotene and lycopene β–cyclase (LYCB) for β–carotene [63]. β–carotene is a carotene present in the majority of photosynthetic organisms, while α–carotene seems to be absent in stramenopiles and red microalgae [56]. These results were confirmed manually by our homology search. This search also allowed us to identify a single sequence containing one lycopene cyclase domain (PF05834) within the proteomes of brown algae: The lycopene β–cyclase [55,56,64] (Figure S4 of the Supplementary Materials). Nevertheless, within the GSMNs of the two brown algae, all the reactions leading to the synthesis of α–carotene and its derivatives (the grey part of Figure 3) were wrongly predicted and supported by this lycopene β–cyclase. Manual curation, therefore, led us to correct and suppress the reactions associated with α–carotene synthesis within the GSMNs.
The topology of the GSMNs, and in particular the presence of gaps, also revealed the specificities of the lineages. For instance, within the initially reconstructed GSMNs, no reaction allowing β–carotene transformation into zeaxanthin was automatically inferred. However, we know that this transformation involved different enzymes in different lineages, and in stramenopile species. Recent studies have suggested that this modification was catalyzed by an enzyme belonging to the P450 monooxygenase family [58,64]. During a homology search within the proteomes of S. japonica and C. okamuranus, we found two CYP97 contiguous paralogs sequences that we associated with this transformation (Figure 3).
3.3. No Plant-Like Abscisic Acid Synthesis Pathway in Brown Algae
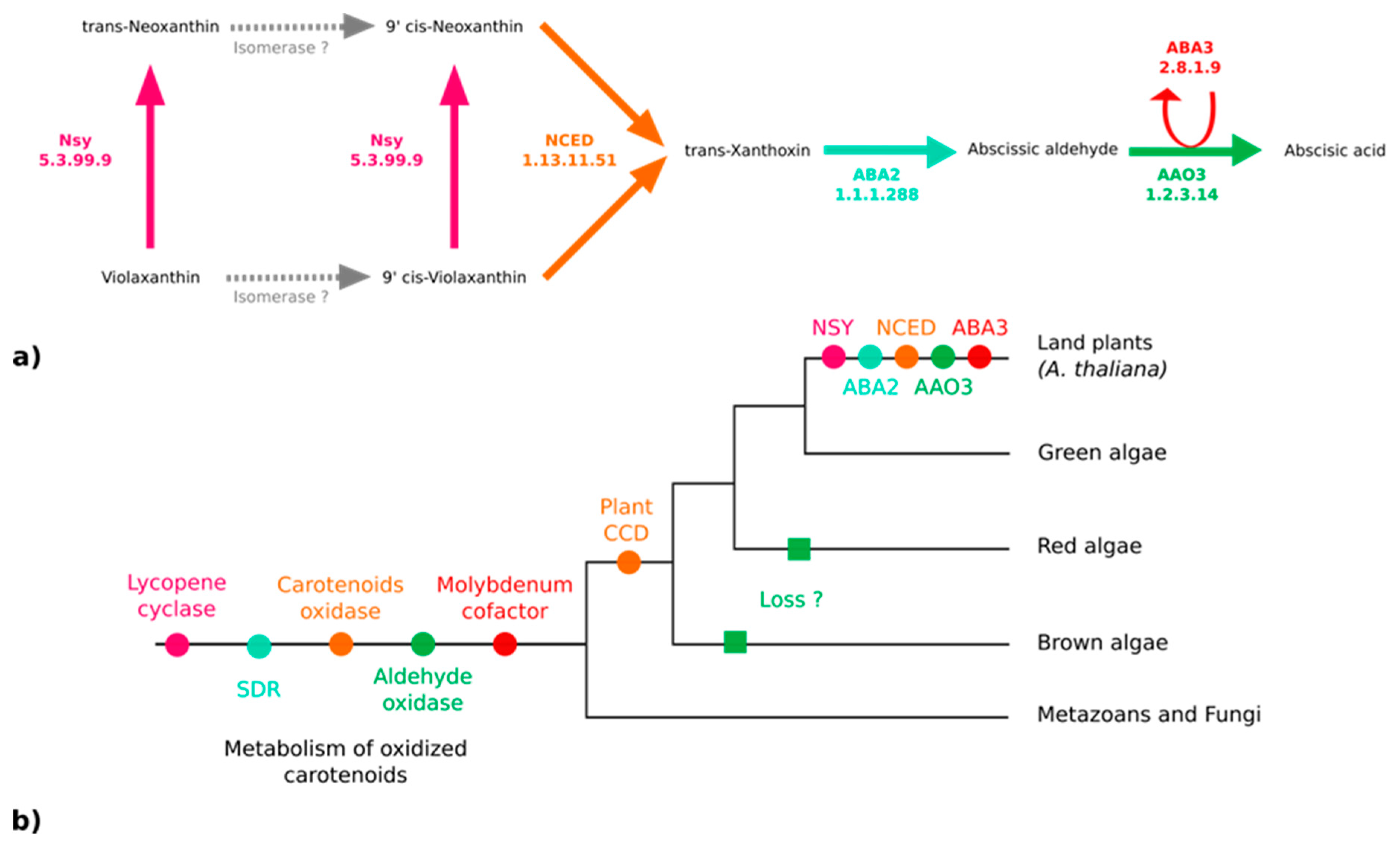
Because ABA has been reported in various kelp species [19] and associated with biological effects on kelp growth and maturation [65,66], we thought it would be important to clarify if both S. japonica and C. okamuranus can produce it. The enzymes responsible for the synthesis of ABA from violaxanthin have been identified and characterized in embryophytes [67]. According to the MetaCyc pathway PWY–695, five enzymes are essential for the biosynthesis of ABA [67,68,69,70] (Figure 4a). We searched for orthologs of these five proteins in the brown algal genomes, but only paralogs were found (Figure 4b). All of them are members of large multigenic families whose conserved domains are general and, therefore, prevent the inference of their precise catalytic activities (Figure 4b and Figures S4–S7, and Table S9 of the Supplementary Materials).
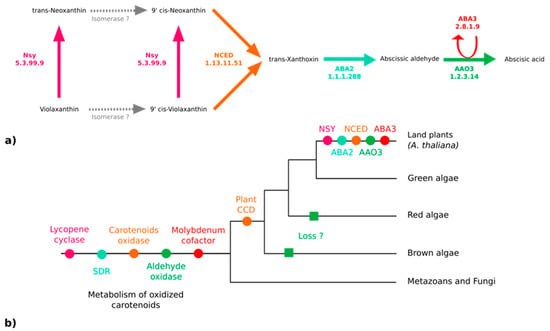
Figure 4.
Enzymes of the abscisic acid biosynthesis pathway. (a) The ABA biosynthesis pathway in embryophytes (A. thaliana) [67]. (b) Phylogenetic tree representing the putative apparition and loss of the biosynthetic enzymes. NSY: Neoxanthin synthase; NCED: Abscisic–aldehyde oxidase; ABA2: Xanthoxin dehydrogenase; ABA3: Molybdenum cofactor sulfurase; AAO3: Abscisic–aldehyde oxidase; SDR: Short-chain alcohol dehydrogenase/reductase; CCD: Carotenoid cleavage dioxygenase.
We also examined two reactions involved in the regulation of ABA availability through its conjugation with glucose by an ABA glucosyltransferase (AOG — RXN–8155). Glycosylated ABA is inactive and can be stored for regulatory purposes. The activation of glycosylated ABA pools was performed by glucosidase (BG1 or BG2 — RXN–11469) [71,72]. However, a homology search carried out on these enzymes in predicted brown algal proteomes did not reveal any homologous sequences either.
3.4. Extending the Fucoxanthin Biosynthesis Models from Diatoms to Brown Algae
With the exception of the interconversion of zeaxanthin, antheraxanthin, and violaxanthin, violaxanthin was not consumed and corresponds at the GSMN level, to a dead end [73]. This, in silico dead end does not represent a biological reality since we know that this molecule is the entry point for the biosynthesis of fucoxanthin, but also of diatoxanthin and diadinoxanthin, two xanthophylls strongly suspected of being present in brown algae [56,59]. The reconstruction of the GSMNs was based, among other things, on the functional annotation of their genome, but on average, 35% of the predicted coding sequences in the two algae were devoid of annotations (See Figure S1 of Supplementary Materials). Among the sequences without any type of annotation, 28% (1984 S. japonica sequences) and 22% (1015 C. okamuranus sequences) can be considered as taxonomically restricted or orphan genes (sequences from one species not homologous to the other species). They represent on average, 11% and 7% of all predicted coding sequences. The other sequences without annotation 5088 and 3640 sequences in S. japonica and C. okamuranus, respectively, represent conserved proteins of unknown function. These unannotated sequences constitute a protein reservoir with unknown biochemical functions, probably including these sought enzymes.
The number and nature of cycling xanthophylls, metabolites involved in photoprotection and which depend on light conditions, differ among the algal lineages (black insert Figure 3) [59,60,74,75]. Based on previous studies, two distinct cycles have been described in stramenopiles: The zeaxanthin to violaxanthin interconversion cycle [74,75] and the diatoxanthin to diadinoxanthin interconversion cycle [59]. The enzymes involved in the first cycle are zeaxanthin epoxidase (ZEP) and violaxanthin de-epoxidase (VDE). The sequences corresponding to those involved in the second cycle, diadinoxanthin de-epoxidase (DDE) and diatoxanthin–epoxidase (DEP), have not yet been identified. However, it would appear that these enzymes are very close to ZEP and VDE since they catalyze the same type of reaction (opening or epoxide formation on the cyclic ends of molecules) [5,18,60]. Indeed, it has previously been suggested that one copy of the VDE gene, identified as VDL, could correspond to the DDE enzyme [5,60]. Within the various algal proteomes queried, we found only one sequence associated with the ZEP and, as expected, three known out–paralogues of VDE: VDE, VDR-related, and VDL-like (Figure S8 of the Supplementary Materials). Consistent with the absence of a xanthophyll cycle, no copy of the VDE gene was found in red algal proteomes, and the presence of a VDL appeared to be a specificity of stramenopiles [5,61]. We decided to associate this sequence with the reaction catalyzed by the DDE within our reconstructions
Finally, a hypothetical pathway leading to the production of fucoxanthin has been proposed in P. tricornutum (diatoms), but the enzymes are not yet known biochemically [60]. Nevertheless, two production hypotheses exist: Either through neoxanthin, whose presence is discussed in the literature [5,56,60,76] or directly through violaxanthin [18,60,61]. However, the investigation of the proteomes of the two brown algae has shown that the conversion of violaxanthin to neoxanthin, if it takes place, cannot be performed by neoxanthin synthase (NSY), as in terrestrial plants [53,77]. Whatever the hypothesis considered, in view of molecular structures, these biosynthetic steps require the intervention of at least three enzymes. It is likely that the latter, if they exist, are in the set of non-annotated sequences. Awaiting biological confirmation, we propose these two pathways in our reconstructions (brown insert in Figure 3).
4. Discussion
The use of automatic or semi-automatic, GSMN reconstruction workflows allows efficient and rapid modeling of GSMNs, even for emerging model organisms, as is the case for S. japonica and C. okamuranus. The theoretical biomass reaction we have tested here is mainly composed of the list of amino acids and some essential sugars. All these compounds were predicted to be producible, and our GSMNs are considered functional. In fine, for instance, adding compounds such as fucoxanthin, a molecule with pharmaceutical interest [17], could make it possible to understand and improve their production in the coming years
The reconstruction of the GSMNs is partially based on the assumption that two orthologous sequences, resulting from a speciation event, share the same function. Such reactions, called core reactions, reflect an ancient evolutionary origin and indicate which metabolites are preserved through the various kingdoms of life. For the most part, they correspond to metabolites historically qualified as primary (amino acids, essential sugars, ATP, ADP) but also ubiquitous secondary metabolites such as lycopene. The preliminary steps to the IPP synthesis are highlighted by the orthology component of the reconstruction, suggesting and confirming their ancestral origin. In general, all the reactions present within the GSMNs that are supported by orthology with A. thaliana illustrate this phenomenon. However, the list proposed based on this criterion is not exhaustive since, on the one hand, the A. thaliana GSMN preferentially targets “primary metabolites” and, on the other hand, some reactions may have been lost during the mapping steps between the various databases. This point is illustrated by the reactions allowing the synthesis of lycopene from geranylgeranyl diphosphate (triangle in Figure 2). They belong to the core reaction set (similar enzymes in plants and various algal lineages) but are not supported by the A. thaliana component. In contrast, reactions supported only by annotation may indicate a specificity of the species or lineage.
A contrario, the presence of gaps, and a fortiori dead ends within GSMNs has a high predictive power by informing either on the computational limit and/or biological specificity related to one species. From a purely bioinformatic point of view, the presence of gaps is directly related to the quality and quantity of data present in the databases. Apart from encoding issues of some metabolites, there is currently little Phaeophyceae referenced data. As such, a gap in the GSMNs may be a direct reflection of the biological specificities of the studied lineages and open up to new questions and biological investigations (i.e., identification of fucoxanthin biosynthesis enzymes). The first source of answers is probably to be found in the set of unannotated sequences, which are not taken into account in the reconstruction of the GSMNs. Indeed, the reconstruction is based on the functional annotation of coding sequences. Among all sequences, about 35% of them do not have any annotation and are of sufficient size to encode functional enzymes (average of 525 amino acids — see Figure S9 of the Supplementary Materials). There is no doubt that these sequences constitute a reservoir/pool of candidates for the shadows that persist within the current knowledge of brown algae. Moreover, diversifying the tools used during the annotation step could be considered in order to compensate slightly for computational artifacts listed above. Indeed, using Kobas and Blast2GO on S. japonica data allow finding 3.2% additional GO terms and 6.0% Kegg identifiers, which are necessary to track EC numbers (Figure S10 of the Supplementary Materials) Among the unannotated sequences, some may be candidates to be assigned to a reaction inferred by gap-filling without any known associated enzymes. Correlating gene expression data, targeted chemical profiling can help to narrow down candidates for full biochemical characterization, as already done in some model bacteria [78].
The GSMNs have been tested and enriched with targeted metabolic profiling data from the literature. Few or none of these targets were reachable before the gap-filling stage. The effectiveness of this step, more than two-thirds of the targets are now achievable, allowing us to propose a better topology of the GSMNs that can mimic the biological behavior of the brown algae (see Appendix A). Nevertheless, despite the presence of a positive flux under constraint-based modeling, some of the target metabolites are topologically not structurally producible in the GSMNs. These problems of unproducible compounds within bacterial and eukaryotic GSMNs are known [22,25]. The first intuitive constatation is that if the targets are not found during the topological analysis, this implies that at least some genomic data (e.g., an enzyme) is missing and that there is a need for further development of functional approaches. However, among the 12 and 7 unreachable targets—in S. japonica and C. okamuranus GSMNs, respectively—we found mainly fatty acids, which are rarely found in their free forms. By focusing on conjugated forms, particularly with acetyl–CoA, we realize that some of these compounds are present in the GSMNs thanks to the addition of gap-filling reactions. To summarize, the non-producibility of these targets can also come from the encoding choice of the referenced metabolites, and in this case, the GSMNs already reflect the biochemical reality of fatty acid metabolism. An additional category of metabolites is those that could even not be incorporated as targets for gap filling because they are not yet connected to biochemical reaction models and thus not incorporated in the Metacyc database. For such molecules, specific tools have been developed to infer new reaction models using detailed comparisons of already known reactions and molecules substructures or to infer the precise structures of the unknown intermediates leading to known metabolites [25].
The curation of the carotenoid pathway has pointed out one of the limits of automatic GSMN reconstructions with the proposal of genes and reactions leading to the synthesis of α–carotene and its derivatives. This point highlights the need for manual curation steps since, in this specific case, only the user’s expertise could make it possible not to infer these reactions. Indeed, the in silico proposal of these pathways is not aberrant since the enzyme that catalyzes the transformation of lycopene into α–carotene, LYCE, is very similar to LYCB, which transforms the same substrate into β–carotene [57,77,79]. The approaches used here are based on the EC numbers of enzymes, and in this case, the proximity of their EC numbers (EC 5.5.1.18 and EC 5.5.1.19 respectively) explains why these undesired reactions were added and associated with the LYCB sequence.
In any case, as we have seen, these two in silico reconstructions of GSMNs provide a satisfactory model of the carotenoid biosynthesis pathway. We highlighted, on the one hand, the common skeleton (reactions and enzymes) that have been preserved through evolution and, on the other hand, the specificities related to brown algae. One of them corresponds to the presence of a second cycle of xanthophylls, and even if we are not able to propose a synthesis pathway with certainty, we propose a candidate for one of the two enzymes involved in the interconversion of diatoxanthin and diadinoxanthin. Another major point is the production of fucoxanthin. Here, we extend the biosynthetic hypotheses that were previously formulated in diatoms to brown algae.
On the contrary, there is not enough knowledge yet to identify the ABA synthesis pathway in filamentous brown algae from the kelp data for a number of reasons. In the European kelp species Laminaria hyperborea, Laminaria digitate, and Saccharina latissima, the presence of ABA in sporophytic tissues were reported about 20 years ago [19]. The characterization of this phytohormone has been performed by GC–MS (gas chromatography coupled with mass spectrometry). It has been reported that ABA concentration varies according to the seasons, with a maximum peak around November, and that a negative correlation exists between the increase in ABA concentration and vegetative growth of sporophytes [19]. Later, the presence of ABA was also detected in S. japonica by LC–MS/MS analysis (liquid chromatography coupled with two tandem mass spectrometers) [80]. In addition, it has been shown that the application of exogenous ABA may inhibit sporophyte growth and induce the accelerated formation of reproductive tissues called sori [65]. ABA has also been proposed to play a role in the control of elicitor-induced oxidative bursts [66].
Two pathways for the biosynthesis of ABA are known in the literature: The direct pathway and the indirect pathway. Chemically, two other pathways related to the regulation of ABA production have been proposed, but they are not supported by characterized enzymes thus far. ABA could be obtained by transformation of acid xanthoxin resulting from the oxidation of xanthoxin or by abscisic alcohol resulting from the oxidation of abscisic aldehyde [69]. In fungi, ABA is produced by the direct pathway within the cytoplasm from farnesyl diphosphate with ionylidene derivatives. In terrestrial plants, the existence of a pathway derived from farnesyl diphosphate is assumed but has not yet been identified [67]. The indirect pathway, the one explored here (see Figure 4a), is carried out in the plastids of the photosynthetic organisms following carotenoid cleavage [72,77,81]. Violaxanthin is transformed into neoxanthin by an intramolecular oxidoreductase, neoxanthin synthase (NSY). A series of structural modifications likely carried out by isomerases produces the 9-cis forms of the epoxycarotenoids (C40) violaxanthin or neoxanthin (main epoxycarotenoid) [67]. These two molecules undergo oxidative cleavage by a 9-cis-epoxycarotenoid dioxygenase belonging to the NCED family to form xanthoxin (C15). After the export of xanthoxin to the cytosol, an enzyme (ABA2) of the SDR family (short-chain alcohol dehydrogenase/reductase) oxidises this molecule. The opening of the epoxy forms the abscisic aldehyde. Finally, an abscisic aldehyde oxidase acid (AAO3) oxidises the abscisic aldehyde to ABA. This transformation is only achievable in the presence of a molybdenum cofactor sulfurase (MoCo - ABA3) that catalyzes the sulfarylation of a dioxo form of MoCo into a mono-oxo sulfide form necessary for the activation of abscisic aldehyde oxidase acid [82,83]. Homology searches did not allow us to find any orthologs of the corresponding genes in brown algae (see Figure 4b, Figures S4–S7, and Table S9 of Supplementary Materials). Thus, the paralogue sequences identified during the homology searches may be involved in the metabolism of carotenoids, but not specifically in the metabolism of plant-like intermediates in the ABA synthesis pathway from violaxanthin and this pathway, therefore, probably emerged in terrestrial plants.
To conclude, if brown algae are able to synthesize ABA, the corresponding pathway is either unknown at the moment or it could be close to that of fungi. Collecting metabolic data about biosynthetic intermediates would be key to discriminate between both hypotheses. Another possibility is that the metabolite reported as ABA is another structurally close oxidized carotenoid, like β–ionone, which could have an equivalent role [84]. Aside from ABA, there is a huge diversity of oxidized carotenoids involved in signaling processes [85], which gives ample room for possible structural variation in signaling molecules across lineages. Facing such diversity, integrative approaches through genome–scale metabolic models should be helpful tools to prioritize further experimental efforts, in a context where the discovery of drugs coming from natural products is experiencing a revival, fuelled by an increasing integration with genomics data [86].
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3921/8/11/564/s1. Table S1: Data from the annotation of algal protein–coding genes, Figure S1: Source of the S. japonica (a) and C. okamuranus (b) annotations, Table S2: Targets for S. japonica from metabolic data in the literature [19,87,88,89,90,91,92,93,94,95], Table S3: Targets for C. okamuranus from metabolic data in the literature [2,96,97,98,99], Table S4: List of cofactors (seeds) added to the S. japonica and C. okamuranus GSMNs, Table S5: List of biomass reactions added to the S. japonica and C. okamuranus GSMNs [24], Table S6: Accession numbers of enzymes and protein sequences used, Figure S2: Pathways of methylerythritol phosphate (MEP) within the S. japonica and C. okamuranus GSMNs [43,54], Table S7: List of reactions and genes associated with the MEP pathways [43,54], Figure S3: Pathways of mevalonate (MVA) within the S. japonica and C. okamuranus GSMNs [43,54], Table S8: List of reactions and genes associated with the MVA pathways [43,54], Figure S4: Maximum likelihood tree of the lycopene cyclase family [57,77,100], Figure S5: Maximum likelihood tree of the NCED family [12,14,101,102,103,104,105], Figure S6: Maximum likelihood tree of the xanthine oxidase family [106,107], Table S9: Type of SDR found in the genomes of S. japonica and C. okamuranus [108], Figure S7: Maximum likelihood tree of the Mocos family [109,110,111,112,113], Figure S8: Maximum likelihood tree of the VDE [5,18,60,61], Figure S9: Amino acid length of unannotated S. japonica and C. okamuranus coding sequences, Figure S10: Enrichment of the annotation in GO terms and KEGG identifiers: data fusion. The wiki–website is available under the following links: https://gem-aureme.genouest.org/sjapgem/index.php/Main_Page and https://gem-aureme.genouest.org/cokagem/index.php/Main_Page.
Author Contributions
Conceptualization, S.M.D., E.C. and G.V.M.; data curation, D.N., A.P.L., C.L., S.M.D., E.C., and G.V.M.; formal analysis, D.N., C.F., X.L., and A.P.L.; funding acquisition, C.L., A.S., S.M.D., and E.C.; investigation, D.N.; methodology, A.S.; resources, L.B.-G., P.B., O.G.; software, M.A., A.B., C.F., and A.S.; supervision, E.C. and G.V.M.; writing—original draft, D.N., E.C., and G.V.M.; Writing—review and editing, D.N., C.F., A.P.L., C.L., A.S., S.M.D., E.C., and G.V.M. All authors approved the submitted version and agreed to be personally accountable for the authors’ own contributions.
Funding
This research received funding from the French Government via the National Research Agency investment expenditure program IDEALG (ANR–10–BTBR–04) and from Région Bretagne via the grant « SAD 2016 – METALG (9673) ».
Acknowledgments
The authors thank Catherine Boyen, Jonas Collén, and Philippe Potin for their comments and suggestions. We acknowledge the GenOuest bioinformatics core facility (https://www.genouest.org) for providing the computing infrastructure.
Conflicts of Interest
The authors declare no conflict of interest
Appendix A
A.1. Qualitative Analysis of Genome–Scale Metabolic Networks and Gap-Filling
Lists of 61 and 29 metabolites were compiled for S. japonica and C. okamuranus (Tables S1 and S2 of Supplementary Materials) and used to test the topology of the GSMNs. According to topological criteria modeling the Boolean behavior of the system dynamics and before the gap-filling step conducted by Meneco, five targets (L–α–alanine, mannose–1–phosphate, GDP–mannose, D–mannitol 1–phosphate, GDP–mannuronate) were reachable in the S. japonica GSMN. The topological gap-filling allowed reaching 45 and 22 additional metabolites by adding 90 and 67 gap-filling reactions, respectively, in S. japonica and C. okamuranus GSMNs, including 46 common reactions to both algae. The targets that could not be reached were mainly fatty acids. From a dynamic point of view, the presence of a target was then verified and supported by the possibility of producing a flux. This means that all import, export, and production reactions of these compounds were present in the GSMNs. In our analyses, all targets had a positive flux.
A.2. Manual Curation Made to the S. japonica and C. okamuranus Genome–Scale Metabolic Networks
Carotenoids (xanthophylls and carotenes) are synthesized from geranylgeranyl diphosphate, which was transformed into phytoene and then into lycopene under the action of five enzymes, three of them being common to all photosynthetic organisms: Phytoene synthase (PSY — EC 2.5.1.32), phytoene desaturase (PDS — EC 1.3.5.5) and ζ–carotene desaturase (ZDS — EC 1.3.5.6). As the genes of these enzymes are known in algae [55], a homology search based on E. siliculosus proteins coupled with annotations from the GSMNs have made it possible to identify one candidate for PSY (SJ02885 and g11610), one candidate for PDS (SJ08891 and g10852), and one to two candidates for ZDS (SJ05680,SJ05681 and g16199) in S. japonica and C. okamuranus, respectively.
The two other steps in the synthesis of all-trans lycopene were either catalyzed by ζ–carotene isomerase (Z–iso — EC 5.2.1.12) and prolycopene isomerase (crtISO — EC 5.2.1.13) or compensated by photoisomerization [53,62,102]. Based on the results published in reference [55], annotations, and homology searches, it appeared that one copy of the Z–iso gene (SJ04715 and g9721) was found in the brown algae. However, two and three copies of crtISO were found in S. japonica and C. okamuranus, respectively. According to the evolutionary history of these sequences, the most likely hypothesis would be the loss of the copy annotated as amine oxidase in S. japonica after a successive triple duplication [55]. Therefore, we propose two potential candidates for this gene for each of the algae, one of which is annotated with the expected EC number (SJ05083 and g8850) and the other without EC annotation (SJ22161 and g4521).
All-trans lycopene is the first carotenoid from which the biosynthesis pathway diverges since it allows forming both β–carotene and α–carotene. The cyclization of lycopene is ensured by a lycopene cyclase, which adds one β–ring (lycopene β–cyclase — EC 5.5.1.19) or one ε–ring (lycopene ε–cyclase — EC 5.5.1.18) at the extremity of the molecule [63]. The α–carotene, composed of a ε–ring and a β–ring is the precursor of lutein while the β–carotene, composed of two β–rings is the entry point for xanthophylls biosynthesis. The search for proteins homologous to lycopene β–cyclase (LYCB) and lycopene ε-cyclase (LYCE) of A. thaliana coupled with the check for the presence of the lycopene cyclase protein domain PF05834 made it possible to identify a lycopene cyclase sequence for each of the studied algae (SJ04962 and g10898). The reconstruction of the phylogenetic tree (see Figure S4 of the Supplementary Materials for more details) of the previously selected sequences allows linking these sequences to the lycopene β–cyclase, thus confirming the absence of α–carotene and its derivatives in these two brown algae [55,56,57]. However, all these reactions were inferred from the GSMNs and were supported, among other things, by the gene identified as LYCB (EC 5.5.1.19). This point, therefore, led us to remove from the GSMNs reactions associated with the synthesis of δ–carotene (RXN1F–147), α–carotene (RXN1F–148), ε–carotene (RXN–8028), zeinoxanthin (RXN–5961), α–cryptoxanthin (RXN–12226—only for C. okamuranus GSMNs) and lutein (RXN–8962).
The addition of a hydroxyl group at each end of the β–carotene forms zeaxanthin. Within the reconstructed S. japonica and C. okamuranus GSMNs, no reaction allows this modification. However, there are two reactions (RXN–8025 and RXN–8026) within the MetaCyc database that ensure this transformation using an intermediate, β–cryptoxanthin. The enzyme responsible for this transformation in algae is probably a monooxygenase belonging to the P450 family and more particularly of the CYP97B type [58,64]. In each of the genomes, two genes (SJ07227 and SJ07228 for S. japonica; g4983 and g4984 for C. okamuranus) were identified as such and one of them could ensure the hydroxylation of β–carotene to zeaxanthin [58].
The first cycle of xanthophylls in brown algae involves two enzymes and allows, depending on light conditions, the interconversion of zeaxanthin to violaxanthin through antheraxanthin [59,74]. In poor light conditions, a zeaxanthin epoxidase (ZEP — EC 1.14.15.21) causes the opening of the cycles positioned at the ends of zeaxanthin and then antheraxanthin to form epoxides. Within the algal genomes, there is a copy of this enzyme (SJ19373 and g5910), but only one reaction, that corresponding to the transformation of antheraxanthin into violaxanthin (RXN–7979) appears within the GSMNs. As a result, the reaction allowing the transformation of zeaxanthin into antheraxanthin (RXN–7978) was added manually to the GSMNs. In excess light, violaxanthin de–epoxidase (VDE — EC 1.23.5.1) converts violaxanthin into antheraxanthin (RXN–7984), which is then further modified to create zeaxanthin (RXN–7985). Within the GSMNs obtained, there is also a reversible reaction (RXN–13185) allowing the direct conversion of violaxanthin to zeaxanthin. For each of the algae, three sequences are associated with these reactions and the analysis of their phylogeny makes it possible to identify the three known out–paralogs: VDE (SJ03764 and g11316), VDL (VDE–like SJ20456 and g16187), and VDR (VDE–related, SJ19927 and g4586). The second cycle of xanthophylls, known in diatoms [60,61], and suspected in brown algae [56,59] operates on the same principle as the first cycle. Thus, the reactions linked to the interconversion of diatoxanthin into diadinoxanthin (RXN–19200 and RXN–19202) were added to the GSMNs. VDL (SJ20456 and g16187) has been associated with the reaction that converts diadinoxanthin to diatoxanthin (RXN–19202) [5,61] (see Figure S8 of the Supplementary Materials for more details).
According to the two hypotheses reviewed in reference [18], we have added seven reactions related to the synthesis of fucoxanthin and diadinoxanthin. According to the violaxanthin hypothesis [59], we created two reactions to allow the transformation of violaxanthin into diadinoxanthin (opening of the epoxide and formation of a triple bond—probably related to the action of a single enzyme) and the transformation of diadinoxanthin into fucoxanthin (reactions probably carried out by three independent enzymes). According to the neoxanthin hypothesis, we have added the three reactions presented in reference [60] (RXN–19203, RXN–19197 and RXN–19196) and also chose to add the reaction related to the transformation of violaxanthin into neoxanthin (RXN1F–155), even if there is no NSY in the two brown algae. Finally, a reaction allowing the transformation of neoxanthin into diadinoxanthin having been proposed by reference [60], we also added it to the GSMNs.

Table A1.
List of manual modifications made to the GSMNs of S. japonica and C. okamuranus.
Table A1.
List of manual modifications made to the GSMNs of S. japonica and C. okamuranus.
| Enzymes | ID Reactions (MetaCyc) | Publication | S. japonica (Associated Genes) | C. okamuranus (Associated Genes) |
|---|---|---|---|---|
| Carotenoid biosynthesis and first xanthophyll cycle, well–known reactions whose genes are characterized in brown algae | ||||
| PSY (phytoene synthase) | RXN–13323 | [11,53,55,61,62,104] | SJ02885 | g11610 |
| 2.5.1.32–RXN | ||||
| RXNARA–8002 | ||||
| PDS (phytoene desaturase) | RXN–12243 | [11,53,55,61,62,104] | SJ08891 | g10852 |
| RXN–12244 | ||||
| RXN–11355 | ||||
| Z–iso (ζ–carotene isomerase) | RXN–11354 | [55,104] | SJ04715 | g9721 |
| ZDS (ζ–carotene desaturase) | RXN–12242 | [11,53,55,61,62,104] | SJ05680 SJ05681 | g16199 |
| RXN–11356 | ||||
| RXN–11357 | ||||
| crtISO (prolycopene isomerase) | RXN–8042 | [55,56,61,62,104] | SJ05083 | g8850 |
| LycB (lycopene β–cyclase) | RXN1F–150 | [11,53,55,56,57,61,62,63,104] | SJ04962 | g10898 |
| RXN1F–151 | ||||
| Cyp97B | RXN–8025 | [12,58,62,64,104] | SJ07227 SJ07228 | g4983 g4984 |
| RXN–8026 | ||||
| RXN1F–152 | ||||
| ZEP (zeaxanthin epoxidase) | RXN–7979 | [11,18,53,55,56,62,74,104] | SJ19373 | g5910 |
| RXN–7978 | ||||
| VDE (violaxanthin de–epoxidase) | RXN–7984 | [11,18,53,56] | SJ03764 SJ20456 VDL (VDE–like ~ DDE) SJ19927 VDR (VDE–related) | g11316 g16187 VDL (VDE–like ~ DDE) g4586 VDR (VDE–related) |
| RXN–7985 | ||||
| Reactions removed | ||||
| phytoene desaturase (fungi – al–1 or crtI) | RXN–12413 | SJ18358 (ANNOTATION) | g18971.t1 (ORTHOLOGY) | |
| RXN–11974 | ||||
| RXN–8023 | ||||
| RXN–8024 | ||||
| RXN–8022 | ||||
| RXN–12412 | ||||
| LycE (lycopene ε–cyclase) | RXN–8040 | 7 sequences (ORTHOLOGY x2) | 18 sequences (ORTHOLOGY x3) | |
| LycB (lycopene β–cyclase) | RXN–8038 | |||
| LycE (lycopene ε–cyclase) | RXN1F–147 | |||
| RXN–8028 | ||||
| LycB (lycopene β–cyclase) | RXN1F–148 | |||
| Carotene epsilon monooxygenase | RXN–5961 | 5 sequences (ORTHOLOGY) | 7 sequences (ORTHOLOGY x2) | |
| RXN–5962 | 6 sequences (ORTHOLOGY) | 8 sequences (ORTHOLOGY x2) | ||
| RXN–12226 | — | g13263.t1 (ANNOTATION) | ||
| Reactions added manually | ||||
| DDE (diadinoxanthin de–epoxidase) | RXN–19200 | [12,59,61] | SJ20456 (VDE–like ~ DDE) | g16187 (VDE–like ~ DDE) |
| DEP (diatoxanthin–epoxidase) | RXN–19202 | — | — | |
| 1.3.99. – | RXN–19203 | [5,18,60] | — | — |
| 1.14.99.M8 | RXN–19197 | — | — | |
| 2.3.1. – | RXN–19196 | — | — | |
| — | RXN–19184 | [60] | — | — |
| — | RXN1F–155 | — | — | |
| — | ConversionViolaxanthinToDiadinoxanthin | [18] | — | — |
| — | ConversionDiadinoxanthinToFucoxanthin | [18] | — | — |
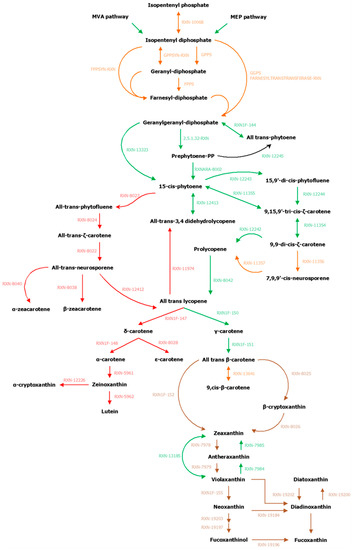
Figure A1.
Reconstruction and manual curation of the genome–scale metabolic networks of S. japonica and C. okamuranus. In green, the reactions present initially in the GSMNs, in red those that were deleted, in brown those added, and in orange those to be checked.
Figure A1.
Reconstruction and manual curation of the genome–scale metabolic networks of S. japonica and C. okamuranus. In green, the reactions present initially in the GSMNs, in red those that were deleted, in brown those added, and in orange those to be checked.
References
- Bartsch, I.; Wiencke, C.; Bischof, K.; Buchholz, C.M.; Buck, B.H.; Eggert, A.; Feuerpfeil, P.; Hanelt, D.; Jacobsen, S.; Karez, R.; et al. The genus Laminaria sensu lato: Recent insights and developments. Eur. J. Phycol. 2008, 43, 1–86. [Google Scholar] [CrossRef]
- Cheng, K.-C.; Kuo, P.-C.; Hung, H.-Y.; Yu, K.-H.; Hwang, T.-L.; Shieh, P.-C.; Chang, J.-S.; Wu, T.-S. Four new compounds from edible algae Cladosiphon okamuranus and Chlorella sorokiniana and their bioactivities. Phytochem. Lett. 2016, 18, 113–116. [Google Scholar] [CrossRef]
- Nishitsuji, K.; Arimoto, A.; Iwai, K.; Sudo, Y.; Hisata, K.; Fujie, M.; Arakaki, N.; Kushiro, T.; Konishi, T.; Shinzato, C.; et al. A draft genome of the brown alga, Cladosiphon okamuranus, S-strain: A platform for future studies of ‘mozuku’ biology. DNA Res. 2016, 23, 561–570. [Google Scholar] [CrossRef]
- Bleakley, S.; Hayes, M. Algal Proteins: Extraction, Application, and Challenges Concerning Production. Foods 2017, 6, 33. [Google Scholar] [CrossRef]
- Mikami, K.; Hosokawa, M. Biosynthetic Pathway and Health Benefits of Fucoxanthin, an Algae-Specific Xanthophyll in Brown Seaweeds. Int. J. Mol. Sci. 2013, 14, 13763–13781. [Google Scholar] [CrossRef]
- Christaki, E.; Bonos, E.; Giannenas, I.; Florou-Paneri, P. Functional properties of carotenoids originating from algae. J. Sci. Food Agric. 2013, 93, 5–11. [Google Scholar] [CrossRef]
- Álvarez, R.; Vaz, B.; Gronemeyer, H.; de Lera, Á.R. Functions, Therapeutic Applications, and Synthesis of Retinoids and Carotenoids. Chem. Rev. 2014, 114, 1–125. [Google Scholar] [CrossRef] [PubMed]
- Bohn, T. Carotenoids and Markers of Oxidative Stress in Human Observational Studies and Intervention Trials: Implications for Chronic Diseases. Antioxidants 2019, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J. Bioactive Properties of Carotenoids in Human Health. Nutrients 2019, 11, 2388. [Google Scholar] [CrossRef] [PubMed]
- Mounien, L.; Tourniaire, F.; Landrier, J.-F. Anti-Obesity Effect of Carotenoids: Direct Impact on Adipose Tissue and Adipose Tissue-Driven Indirect Effects. Nutrients 2019, 11, 1562. [Google Scholar] [CrossRef]
- Esteban, R.; Moran, J.F.; Becerril, J.M.; García-Plazaola, J.I. Versatility of carotenoids: An integrated view on diversity, evolution, functional roles and environmental interactions. Environ. Exp. Bot. 2015, 119, 63–75. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Antioxidant Protection from UV- and Light-Stress Related to Carotenoid Structures. Antioxidants 2019, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Kiser, P.D.; von Lintig, J.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. A Darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot. 2009, 60, 719–726. [Google Scholar] [CrossRef]
- Mise, T.; Ueda, M.; Yasumoto, T. Production of Fucoxanthin-Rich Powder from Cladosiphon okamuranus. Adv. J. Food Sci. Technol. 2011, 3, 73–76. [Google Scholar]
- Kanazawa, K.; Ozaki, Y.; Hashimoto, T.; Das, S.K.; Matsushita, S.; Hirano, M.; Okada, T.; Komoto, A.; Mori, N.; Nakatsuka, M. Commercial-Scale Preparation of Biofunctional Fucoxanthin from Waste Parts of Brown Sea Algae Laminaria japonica. FSTR 2008, 14, 573–582. [Google Scholar] [CrossRef]
- Kuczynska, P.; Jemiola-Rzeminska, M.; Strzalka, K. Photosynthetic Pigments in Diatoms. Mar. Drugs 2015, 13, 5847–5881. [Google Scholar] [CrossRef]
- Schaffelke, B. Abscisic Acid in Sporophytes of Three Laminaria Species (Phaeophyta). J. Plant Physiol. 1995, 146, 453–458. [Google Scholar] [CrossRef]
- Ebrahim, A.; Almaas, E.; Bauer, E.; Bordbar, A.; Burgard, A.P.; Chang, R.L.; Dräger, A.; Famili, I.; Feist, A.M.; Fleming, R.M.; et al. Do genome-scale models need exact solvers or clearer standards? Mol. Syst. Biol. 2015, 11, 831. [Google Scholar] [CrossRef]
- Aite, M.; Chevallier, M.; Frioux, C.; Trottier, C.; Got, J.; Cortés, M.P.; Mendoza, S.N.; Carrier, G.; Dameron, O.; Guillaudeux, N.; et al. Traceability, reproducibility and wiki-exploration for “à-la-carte” reconstructions of genome-scale metabolic models. PLoS Comput. Biol. 2018, 14, e1006146. [Google Scholar] [CrossRef] [PubMed]
- Frainay, C.; Schymanski, E.; Neumann, S.; Merlet, B.; Salek, R.; Jourdan, F.; Yanes, O. Mind the Gap: Mapping Mass Spectral Databases in Genome-Scale Metabolic Networks Reveals Poorly Covered Areas. Metabolites 2018, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [PubMed]
- Prigent, S.; Collet, G.; Dittami, S.M.; Delage, L.; Ethis de Corny, F.; Dameron, O.; Eveillard, D.; Thiele, S.; Cambefort, J.; Boyen, C.; et al. The genome-scale metabolic network of Ectocarpus siliculosus (EctoGEM): A resource to study brown algal physiology and beyond. Plant J. 2014, 80, 367–381. [Google Scholar] [CrossRef]
- Belcour, A.; Girard, J.; Aite, M.; Delage, L.; Trottier, C.; Marteau, C.; Leroux, C.; Dittami, S.M.; Sauleau, P.; Corre, E.; et al. Inferring biochemical reactions and metabolite structures to cope with metabolic pathway drift. bioRxiv 2018, 462556. [Google Scholar] [CrossRef]
- Ye, N.; Zhang, X.; Miao, M.; Fan, X.; Zheng, Y.; Xu, D.; Wang, J.; Zhou, L.; Wang, D.; Gao, Y.; et al. Saccharina genomes provide novel insight into kelp biology. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Dittami, S.M.; Corre, E. Detection of bacterial contaminants and hybrid sequences in the genome of the kelp Saccharina japonica using Taxoblast. PeerJ 2017, 5, e4073. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Karp, P.D.; Paley, S.; Romero, P. The Pathway Tools Software. Bioinformatics 2002, 18, S225–S232. [Google Scholar] [CrossRef]
- De Oliveira Dal’Molin, C.G.; Quek, L.-E.; Palfreyman, R.W.; Brumbley, S.M.; Nielsen, L.K. AraGEM, a Genome-Scale Reconstruction of the Primary Metabolic Network in Arabidopsis. Plant Physiol. 2010, 152, 579–589. [Google Scholar] [CrossRef]
- Loira, N.; Mendoza, S.; Paz Cortés, M.; Rojas, N.; Travisany, D.; Genova, A.D.; Gajardo, N.; Ehrenfeld, N.; Maass, A. Reconstruction of the microalga Nannochloropsis salina genome-scale metabolic model with applications to lipid production. BMC Syst. Biol. 2017, 11, 66. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- O’Brien, K.P.; Remm, M.; Sonnhammer, E.L.L. Inparanoid: A comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005, 33, D476–D480. [Google Scholar] [CrossRef]
- King, Z.A.; Lu, J.; Dräger, A.; Miller, P.; Federowicz, S.; Lerman, J.A.; Ebrahim, A.; Palsson, B.O.; Lewis, N.E. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016, 44, D515–D522. [Google Scholar] [CrossRef]
- Caspi, R.; Foerster, H.; Fulcher, C.A.; Kaipa, P.; Krummenacker, M.; Latendresse, M.; Paley, S.; Rhee, S.Y.; Shearer, A.G.; Tissier, C.; et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008, 36, D623–D631. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Dreher, K.; Karp, P.D. The challenge of constructing, classifying and representing metabolic pathways. FEMS Microbiol. Lett. 2013, 345, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Martin, O.; Van Du Tran, T.; Bridge, A.; Morgat, A.; Pagni, M. MetaNetX/MNXref—Reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016, 44, D523–D526. [Google Scholar] [CrossRef] [PubMed]
- Prigent, S.; Frioux, C.; Dittami, S.M.; Thiele, S.; Larhlimi, A.; Collet, G.; Gutknecht, F.; Got, J.; Eveillard, D.; Bourdon, J.; et al. Meneco, a Topology-Based Gap-Filling Tool Applicable to Degraded Genome-Wide Metabolic Networks. PLoS Comput. Biol. 2017, 13, e1005276. [Google Scholar] [CrossRef]
- Ebenhöh, O.; Handorf, T.; Heinrich, R. Structural analysis of expanding metabolic networks. Genome Inform. 2004, 15, 35–45. [Google Scholar]
- Ebrahim, A.; Lerman, J.A.; Palsson, B.O.; Hyduke, D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013, 7, 74. [Google Scholar] [CrossRef]
- Frioux, C.; Schaub, T.; Schellhorn, S.; Siegel, A.; Wanko, P. Hybrid metabolic network completion. Theory Pract. Log. Program. 2019, 19, 83–108. [Google Scholar] [CrossRef]
- Sterck, L.; Billiau, K.; Abeel, T.; Rouzé, P.; Van de Peer, Y. ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods 2012, 9, 1041. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; Sterck, L.; Rouzé, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.-M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Zhao, L.; Chang, W.; Xiao, Y.; Liu, H.; Liu, P. Methylerythritol Phosphate Pathway of Isoprenoid Biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, L.; Chi, S.; Wang, G.; Wang, X.; Liu, T.; Tang, X. Phylogenetic analyses of the genes involved in carotenoid biosynthesis in algae. Acta Oceanol. Sin. 2018, 37, 89–101. [Google Scholar] [CrossRef]
- Takaichi, S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef]
- Cui, H.; Wang, Y.; Qin, S. Molecular Evolution of Lycopene Cyclases Involved in the Formation of Carotenoids in Eukaryotic Algae. Plant Mol. Biol. Rep. 2011, 29, 1013–1020. [Google Scholar] [CrossRef]
- Teng, L.; Fan, X.; Nelson, D.R.; Han, W.; Zhang, X.; Xu, D.; Renault, H.; Markov, G.V.; Ye, N. Diversity and evolution of cytochromes P450 in stramenopiles. Planta 2019, 249, 647–661. [Google Scholar] [CrossRef]
- Lohr, M.; Wilhelm, C. Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. USA 1999, 96, 8784–8789. [Google Scholar] [CrossRef]
- Dambek, M.; Eilers, U.; Breitenbach, J.; Steiger, S.; Büchel, C.; Sandmann, G. Biosynthesis of fucoxanthin and diadinoxanthin and function of initial pathway genes in Phaeodactylum tricornutum. J. Exp. Bot. 2012, 63, 5607–5612. [Google Scholar] [CrossRef]
- Coesel, S.; Oborník, M.; Varela, J.; Falciatore, A.; Bowler, C. Evolutionary Origins and Functions of the Carotenoid Biosynthetic Pathway in Marine Diatoms. PLoS ONE 2008, 3, e2896. [Google Scholar] [CrossRef] [PubMed]
- Frommolt, R.; Werner, S.; Paulsen, H.; Goss, R.; Wilhelm, C.; Zauner, S.; Maier, U.G.; Grossman, A.R.; Bhattacharya, D.; Lohr, M. Ancient Recruitment by Chromists of Green Algal Genes Encoding Enzymes for Carotenoid Biosynthesis. Mol. Biol. Evol. 2008, 25, 2653–2667. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X.; Pogson, B.; Sun, Z.; McDonald, K.A.; DellaPenna, D.; Gantt, E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 1996, 8, 1613–1626. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Yu, X.; Wang, Y.; Cui, Y.; Li, X.; Liu, Z.; Qin, S. Evolutionary origins, molecular cloning and expression of carotenoid hydroxylases in eukaryotic photosynthetic algae. BMC Genom. 2013, 14, 457. [Google Scholar] [CrossRef]
- Nimura, K.; Mizuta, H. Inducible effects of abscisic acid on sporophyte discs from Laminaria japonica Areschoug (Laminariales, Phaeophyceae). J. Appl. Phycol. 2002, 14, 159–163. [Google Scholar] [CrossRef]
- Shimizu, K.; Uji, T.; Yasui, H.; Mizuta, H. Control of elicitor-induced oxidative burst by abscisic acid associated with growth of Saccharina japonica (Phaeophyta, Laminariales) sporophytes. J. Appl. Phycol. 2018, 30, 1371–1379. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.-K. Regulation of Abscisic Acid Biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Rock, C.D. Abscisic Acid Biosynthesis and Response. Arab. Book 2002, 1, e0058. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [PubMed]
- Mackie, A.; Keseler, I.M.; Nolan, L.; Karp, P.D.; Paulsen, I.T. Dead End Metabolites—Defining the Known Unknowns of the E. coli Metabolic Network. PLoS ONE 2013, 8, e75210. [Google Scholar] [CrossRef] [PubMed]
- Sajilata, M.G.; Singhal, R.S.; Kamat, M.Y. The Carotenoid Pigment Zeaxanthin—A Review. Compr. Rev. Food Sci. Food Saf. 2008, 7, 29–49. [Google Scholar] [CrossRef]
- Jahns, P.; Latowski, D.; Strzalka, K. Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim. Biophys. Acta (BBA)-Bioenerg. 2009, 1787, 3–14. [Google Scholar] [CrossRef]
- Haugan, J.A. Algal carotenoids 54. Carotenoids of brown algae (Phaeophyceae). Biochem. Syst. Ecol. 1994, 22, 31–41. [Google Scholar] [CrossRef]
- Bouvier, F.; D’Harlingue, A.; Backhaus, R.A.; Kumagai, M.H.; Camara, B. Identification of neoxanthin synthase as a carotenoid cyclase paralog: Plastid neoxanthin synthase. Eur. J. Biochem. 2000, 267, 6346–6352. [Google Scholar] [CrossRef]
- Pan, S.; Reed, J.L. Advances in gap-filling genome-scale metabolic models and model-driven experiments lead to novel metabolic discoveries. Curr. Opin. Biotechnol. 2018, 51, 103–108. [Google Scholar] [CrossRef]
- Cunningham, F.X.; Gantt, E. One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases. Proc. Natl. Acad. Sci. USA 2001, 98, 2905–2910. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, P.; Liu, X.; Chen, J.; Xu, J.; Chen, H.; Yan, X. Quantitative profiling method for phytohormones and betaines in algae by liquid chromatography electrospray ionization tandem mass spectrometry: Determination of phytohormones and betaines in algae by LC-MS/MS. Biomed. Chromatogr. 2014, 28, 275–280. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the Indirect Pathway of Abscisic Acid Biosynthesis by Mutants, Genes, and Enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 Is a Molybdenum Cofactor Sulfurase Required for Activation of Aldehyde Oxidase and Xanthine Dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef] [PubMed]
- Kaufholdt, D.; Baillie, C.-K.; Meinen, R.; Mendel, R.R.; Hänsch, R. The Molybdenum Cofactor Biosynthesis Network: In Vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 2017, 8, 1946. [Google Scholar] [CrossRef] [PubMed]
- Salt, S.D.; Tuzun, S.; Kuć, J. Effects of β-ionone and abscisic acid on the growth of tobacco and resistance to blue mold. Mimicry of effects of stem infection by Peronospora tabacina Adam. Physiol. Mol. Plant Pathol. 1986, 28, 287–297. [Google Scholar] [CrossRef]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Chi, S.; Liu, T.; Wang, X.; Wang, R.; Wang, S.; Wang, G.; Shan, G.; Liu, C. Functional genomics analysis reveals the biosynthesis pathways of important cellular components (alginate and fucoidan) of Saccharina. Curr. Genet. 2018, 64, 259–273. [Google Scholar] [CrossRef]
- Park, J.-N.; Ali-Nehari, A.; Woo, H.-C.; Chun, B.-S. Thermal stabilities of polyphenols and fatty acids in Laminaria japonica hydrolysates produced using subcritical water. Korean J. Chem. Eng. 2012, 29, 1604–1609. [Google Scholar] [CrossRef]
- Getachew, P.; Kang, J.-Y.; Choi, J.-S.; Hong, Y.-K. Does bryozoan colonization alter the biochemical composition of Saccharina japonica affecting food safety and quality? Bot. Mar. 2015, 58, 267–274. [Google Scholar] [CrossRef]
- Patterson, G.W. Sterols of Laminaria. Comp. Biochem. Physiol. 1968, 24, 501–505. [Google Scholar] [CrossRef]
- Zhang, P.; Shao, Z.; Jin, W.; Duan, D. Comparative Characterization of Two GDP-Mannose Dehydrogenase Genes from Saccharina japonica (Laminariales, Phaeophyceae). BMC Plant Biol. 2016, 16, 62. Available online: https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-016-0750-3 (accessed on 9 January 2018). [CrossRef] [PubMed]
- Duan, D.; Liu, X.; Pan, F.; Liu, H.; Chen, N.; Fei, X. Extraction and Identification of Cytokinin from Laminaria japonica Aresch. Bot. Mar. 1995, 38, 409–412. [Google Scholar] [CrossRef]
- Honya, M.; Kinoshita, T.; Ishikawa, M.; Mori, H.; Nisizawa, K. Seasonal variation in the lipid content of cultured Laminaria japonica: Fatty acids, sterols, β-carotene and tocopherol. J. Appl. Phycol. 1994, 6, 25–29. [Google Scholar] [CrossRef]
- Hwang, J.-H.; Kim, N.-G.; Woo, H.-C.; Rha, S.-J.; Kim, S.-J.; Shin, T.-S. Variation in the chemical composition of Saccharina japonica with harvest area and culture period. J. Aquac. Res. Dev. 2014, 5, 286. [Google Scholar]
- Groisillier, A.; Shao, Z.; Michel, G.; Goulitquer, S.; Bonin, P.; Krahulec, S.; Nidetzky, B.; Duan, D.; Boyen, C.; Tonon, T. Mannitol metabolism in brown algae involves a new phosphatase family. J. Exp. Bot. 2014, 65, 559–570. [Google Scholar] [CrossRef]
- Saito, H.; Xue, C.; Yamashiro, R.; Moromizato, S.; Itabashi, Y. High Polyunsaturated Fatty Acid Levels in Two Subtropical Macroalgae, Cladosiphon Okamuranus and Caulerpa Lentillifera. J. Phycol. 2010, 46, 665–673. [Google Scholar] [CrossRef]
- Tako, M.; Yoza, E.; Tohma, S. Chemical Characterization of Acetyl Fucoidan and Alginate from Commercially Cultured Cladosiphon okamuranus. Bot. Mar. 2005, 43, 393–398. [Google Scholar] [CrossRef]
- Lim, S.J.; Wan Aida, W.M.; Schiehser, S.; Rosenau, T.; Böhmdorfer, S. Structural elucidation of fucoidan from Cladosiphon okamuranus (Okinawa mozuku). Food Chem. 2019, 272, 222–226. [Google Scholar] [CrossRef]
- Kakisawa, H.; Asari, F.; Kusumi, T.; Toma, T.; Sakurai, T.; Oohusa, T.; Hara, Y.; Chiharai, M. An allelopathic fatty acid from the brown alga Cladosiphon okamuranus. Phytochemistry 1988, 27, 731–735. [Google Scholar] [CrossRef]
- Al-Babili, S.; Hugueney, P.; Schledz, M.; Welsch, R.; Frohnmeyer, H.; Laule, O.; Beyer, P. Identification of a novel gene coding for neoxanthin synthase from Solanum tuberosum. FEBS Lett. 2000, 485, 168–172. [Google Scholar] [CrossRef]
- Tan, B.-C.; Joseph, L.M.; Deng, W.-T.; Liu, L.; Li, Q.-B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.; Siva, R. Phylogenetic analysis and evolutionary studies of plant carotenoid cleavage dioxygenase gene. Gene 2014, 548, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef]
- Harrison, P.J.; Bugg, T.D.H. Enzymology of the carotenoid cleavage dioxygenases: Reaction mechanisms, inhibition and biochemical roles. Arch. Biochem. Biophys. 2014, 544, 105–111. [Google Scholar] [CrossRef]
- Seo, M.; Aoki, H.; Koiwai, H.; Kamiya, Y.; Nambara, E.; Koshiba, T. Comparative Studies on the Arabidopsis Aldehyde Oxidase (AAO) Gene Family Revealed a Major Role of AAO3 in ABA Biosynthesis in Seeds. Plant Cell Physiol. 2004, 45, 1694–1703. [Google Scholar] [CrossRef]
- Rodríguez-Trelles, F.; Tarrío, R.; Ayala, F.J. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc. Natl. Acad. Sci. USA 2003, 100, 13413–13417. [Google Scholar] [CrossRef]
- Moummou, H.; Kallberg, Y.; Tonfack, L.B.; Persson, B.; van der Rest, B. The Plant Short-Chain Dehydrogenase (SDR) superfamily: Genome-wide inventory and diversification patterns. BMC Plant Biol. 2012, 12, 219. [Google Scholar] [CrossRef]
- Peng, T.; Xu, Y.; Zhang, Y. Comparative genomics of molybdenum utilization in prokaryotes and eukaryotes. BMC Genom. 2018, 19, 691. [Google Scholar] [CrossRef]
- Filiz, E.; Distelfeld, A.; Fahima, T.; Metin, Ö.K.; Nevo, E.; Weining, S.; Uncuoğlu, A.A. Barley molybdenum cofactor sulfurase (MCSU): Sequencing, modeling, and its comparison to other higher plants. Turk. J. Agric. For. 2015, 39, 786–796. [Google Scholar] [CrossRef]
- Mendel, R.R. The Molybdenum Cofactor. J. Biol. Chem. 2013, 288, 13165–13172. [Google Scholar] [CrossRef] [PubMed]
- Hille, R.; Nishino, T.; Bittner, F. Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 2011, 255, 1179–1205. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Hänsch, R. Molybdoenzymes and molybdenum cofactor in plants. J. Exp. Bot. 2002, 53, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).