Exploring the Impact of Bioactive Compounds Found in Extra Virgin Olive Oil on NRF2 Modulation in Alzheimer’s Disease
Abstract
1. Introduction
2. Methods
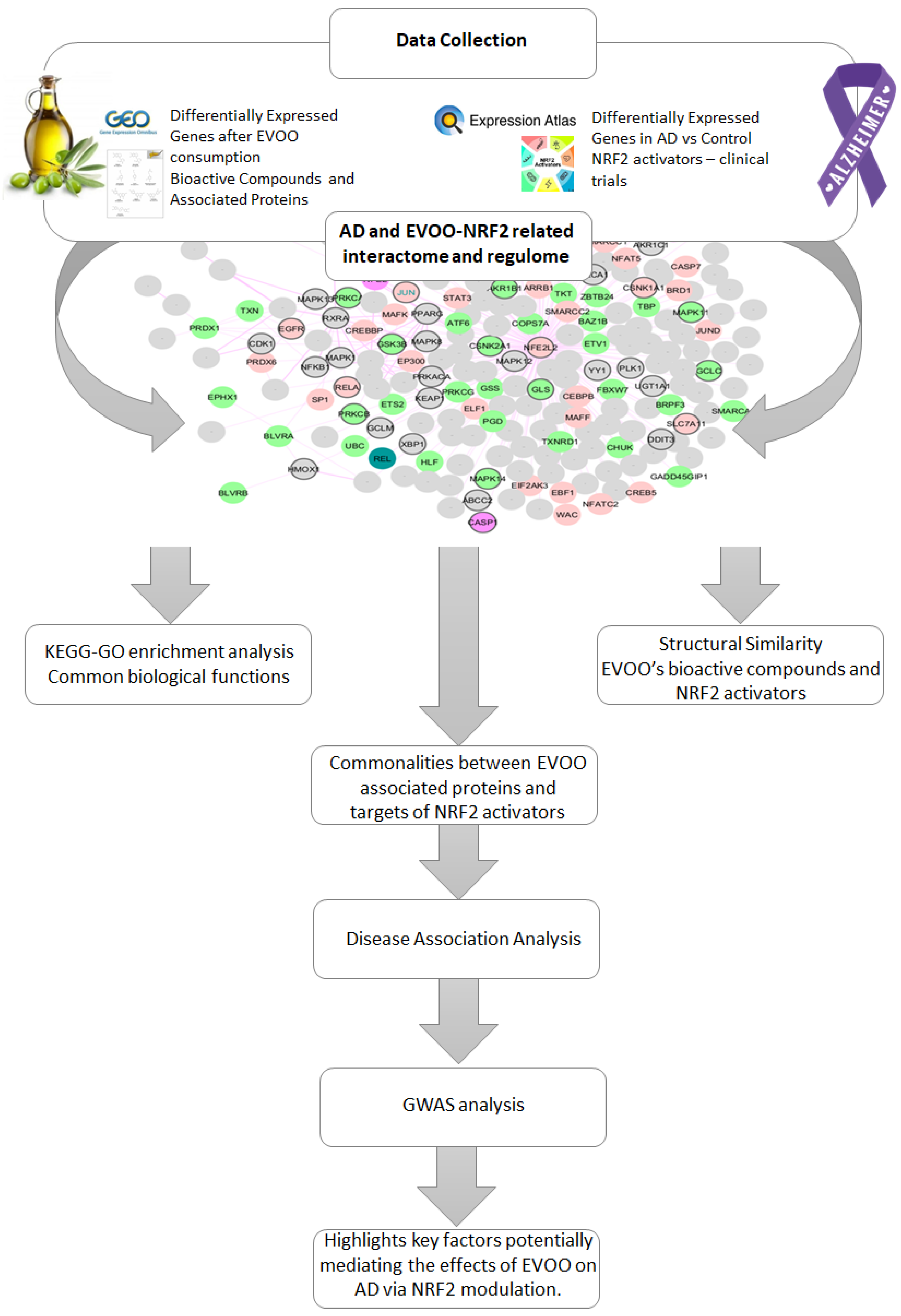
2.1. Methodology Overview
2.2. AD-Related NRF2 Interactome and Regulome
2.3. EVOO’s Data Collection
2.4. NRF2 Activator Collection
2.5. Structural Similarity
2.6. Genetic Investigation
3. Results
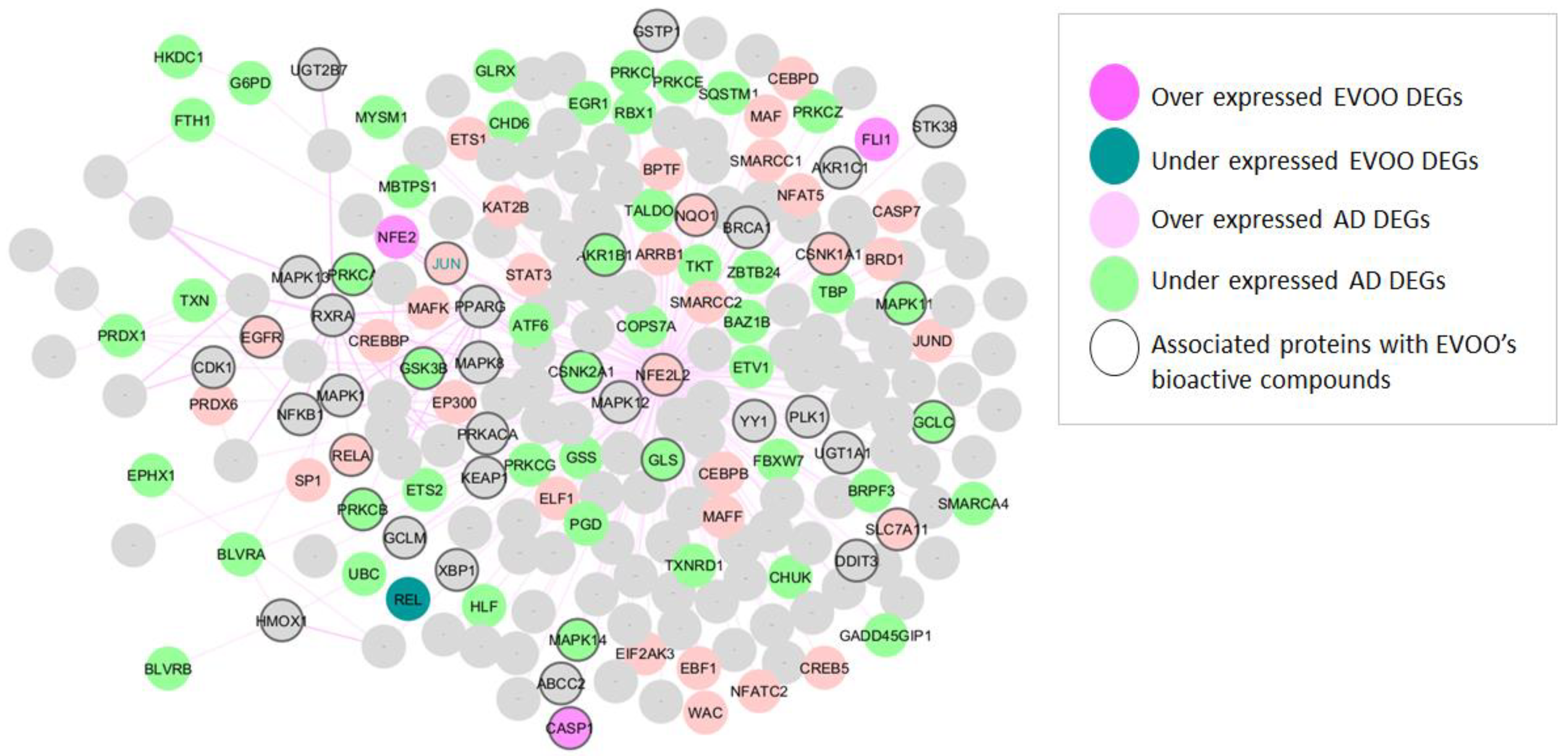
3.1. NRF2-Related Genes Found in AD and EVOO
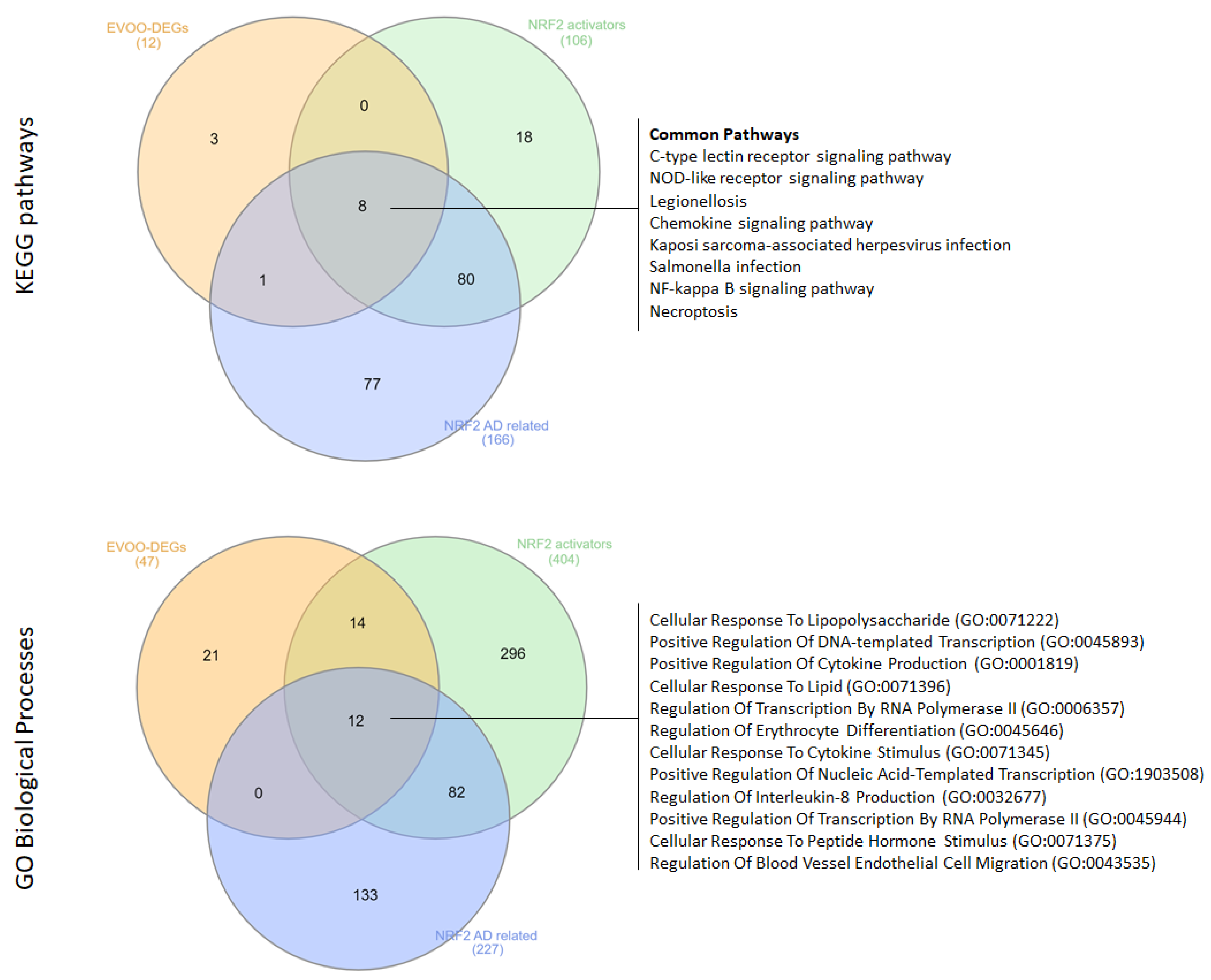
3.2. Computational Insights into the Functional Landscape of the Protective Role of EVOO Consumption in AD
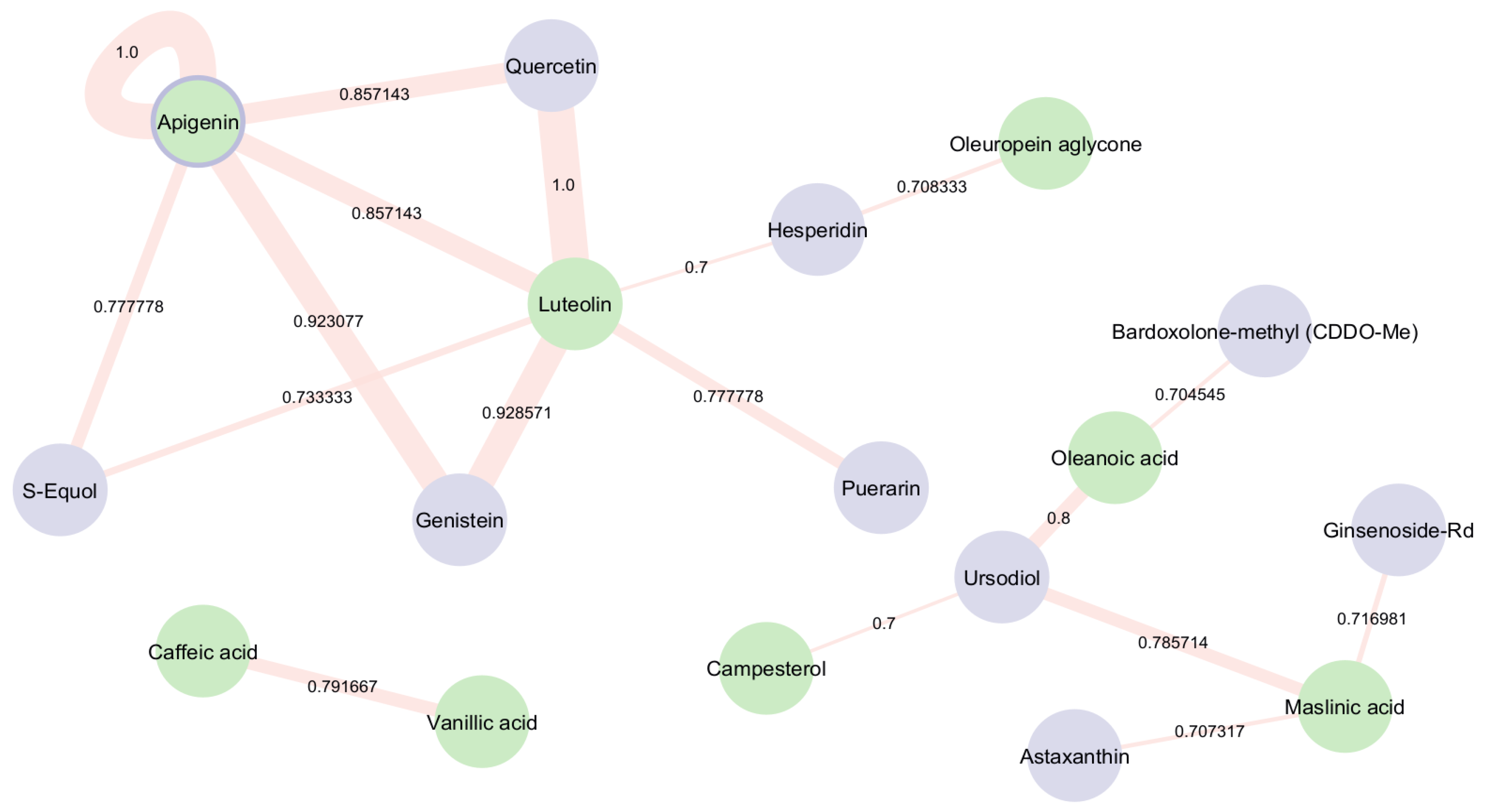
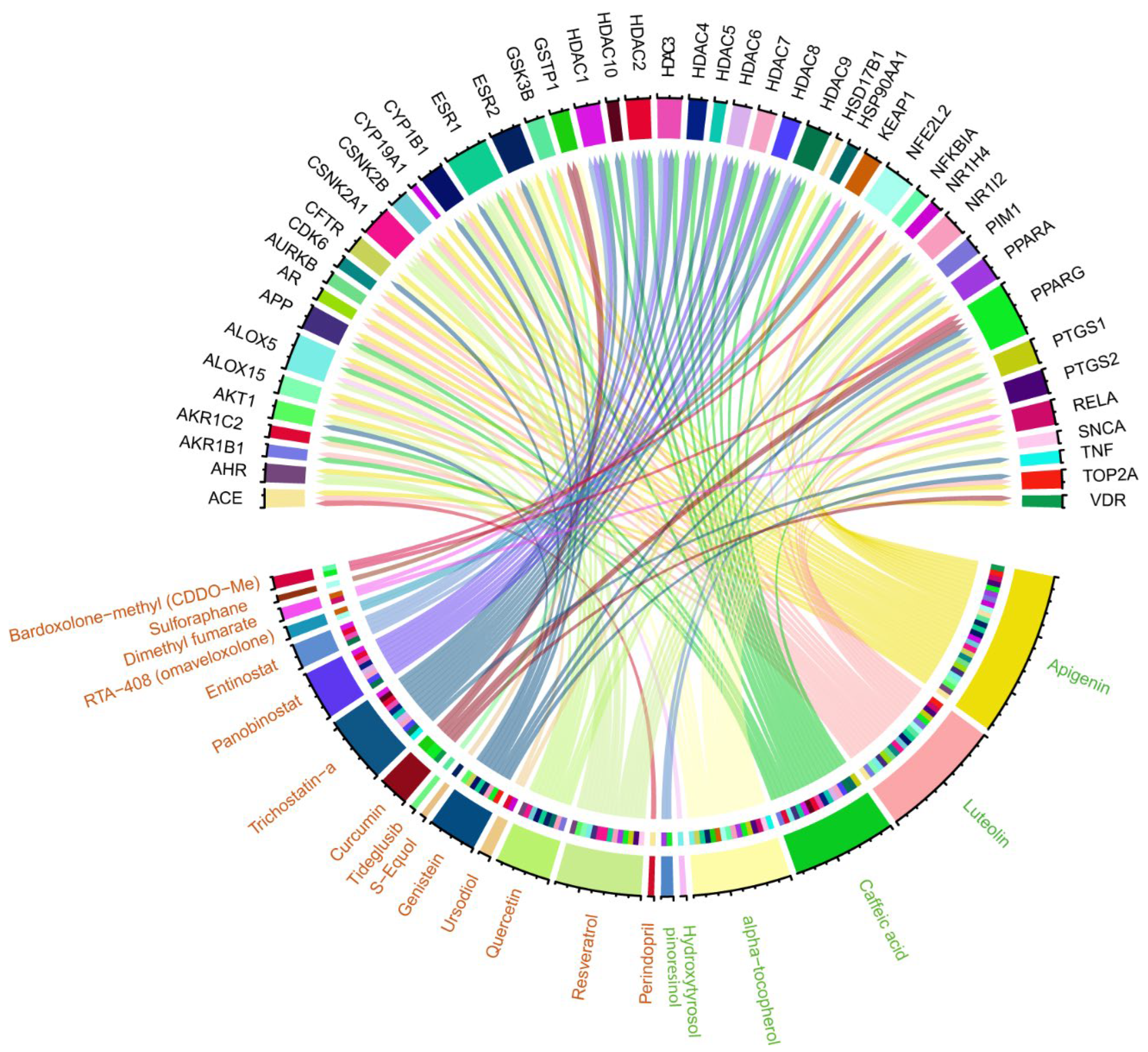
3.3. Structural Investigation of EVOO’s Bioactive Compounds and Drugs with NRF2 Activation Ability Under Ongoing Clinical Trial Investigation
3.4. Identifying Commonalities Between Proteins Associated with EVOO Bioactive Compounds and Targets of NRF2-Activating Drugs
3.5. Genetic Associations of the Common Genes Between the Associated Proteins of EVOO and Targets of NRF2 Activators in Ongoing Clinical Trials with Alzheimer’s Disease
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajmohan, R.; Reddy, P.H. Amyloid-Beta and Phosphorylated Tau Accumulations Cause Abnormalities at Synapses of Alzheimer’s disease Neurons. J. Alzheimer’s Dis. 2017, 57, 975–999. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, B. Oxidative stress and the pathogenesis of Alzheimer’s disease. Oxid. Med. Cell. Longev. 2013, 2013, 316523. [Google Scholar] [CrossRef]
- Guo, C.; Sun, L.; Chen, X.; Zhang, D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen Res. 2013, 8, 2003–2014. [Google Scholar]
- Bhatt, S.; Puli, L.; Patil, C.R. Role of reactive oxygen species in the progression of Alzheimer’s disease. Drug Discov. Today 2021, 26, 794–803. [Google Scholar] [CrossRef]
- Smith, M.A.; Rottkamp, C.A.; Nunomura, A.; Raina, A.K.; Perry, G. Oxidative stress in Alzheimer’s disease. Biochim. Biophys. Acta 2000, 1502, 139–144. [Google Scholar] [CrossRef]
- Ngo, V.; Duennwald, M.L. Nrf2 and Oxidative Stress: A General Overview of Mechanisms and Implications in Human Disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef]
- Kim, J.; Cha, Y.N.; Surh, Y.J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat. Res. 2010, 690, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Suzen, S.; Tucci, P.; Profumo, E.; Buttari, B.; Saso, L. A Pivotal Role of Nrf2 in Neurodegenerative Disorders: A New Way for Therapeutic Strategies. Pharmaceuticals 2022, 15, 692. [Google Scholar] [CrossRef]
- De Plano, L.M.; Calabrese, G.; Rizzo, M.G.; Oddo, S.; Caccamo, A. The Role of the Transcription Factor Nrf2 in Alzheimer’s Disease: Therapeutic Opportunities. Biomolecules 2023, 13, 549. [Google Scholar] [CrossRef] [PubMed]
- Petersson, S.D.; Philippou, E. Mediterranean Diet, Cognitive Function, and Dementia: A Systematic Review of the Evidence. Adv. Nutr. 2016, 7, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Alkhalifa, A.E.; Al-Ghraiybah, N.F.; Kaddoumi, A. Extra-Virgin Olive Oil in Alzheimer’s Disease: A Comprehensive Review of Cellular, Animal, and Clinical Studies. Int. J. Mol. Sci. 2024, 25, 1914. [Google Scholar] [CrossRef] [PubMed]
- Sidiropoulou, G.A.; Metaxas, A.; Kourti, M. Natural antioxidants that act against Alzheimer’s disease through modulation of the NRF2 pathway: A focus on their molecular mechanisms of action. Front. Endocrinol. 2023, 14, 1217730. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Huelamo, M.; Rodriguez-Morato, J.; Boronat, A.; de la Torre, R. Modulation of Nrf2 by Olive Oil and Wine Polyphenols and Neuroprotection. Antioxidants 2017, 6, 73. [Google Scholar] [CrossRef]
- Petryszak, R.; Keays, M.; Tang, Y.A.; Fonseca, N.A.; Barrera, E.; Burdett, T.; Fullgrabe, A.; Fuentes, A.M.; Jupp, S.; Koskinen, S.; et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016, 44, D746–D752. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Lee, J.-Y.; Huang, S.-H.; Hsu, Y.-C.; Hsu, N.-Y.; Yang, J.-M. FooDisNET: A database of food-compound-protein-disease associations. In Proceedings of the IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 190–195. [Google Scholar]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Bourdakou, M.M.; Fernandez-Gines, R.; Cuadrado, A.; Spyrou, G.M. Drug repurposing on Alzheimer’s disease through modulation of NRF2 neighborhood. Redox Biol. 2023, 67, 102881. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; Leon, R.; Lopez, M.G.; Oliva, B.; et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev 2018, 70, 348–383. [Google Scholar] [CrossRef]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Walker, D.G.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genom. 2007, 28, 311–322. [Google Scholar] [CrossRef]
- D’Amore, S.; Vacca, M.; Cariello, M.; Graziano, G.; D’Orazio, A.; Salvia, R.; Sasso, R.C.; Sabba, C.; Palasciano, G.; Moschetta, A. Genes and miRNA expression signatures in peripheral blood mononuclear cells in healthy subjects and patients with metabolic syndrome after acute intake of extra virgin olive oil. Biochim. Biophys. Acta 2016, 1861, 1671–1680. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Bourdakou, M.M.; Melliou, E.; Magiatis, P.; Spyrou, G.M. Computational investigation of the functional landscape of the protective role that extra virgin olive oil consumption may have on chronic lymphocytic leukemia. J. Transl. Med. 2024, 22, 869. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Osama, A.; Zhang, J.; Yao, J.; Yao, X.; Fang, J. Nrf2: A dark horse in Alzheimer’s disease treatment. Ageing Res. Rev. 2020, 64, 101206. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Anton, N.; Fernandez-Gines, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Cao, D.S.; Xiao, N.; Xu, Q.S.; Chen, A.F. Rcpi: R/Bioconductor package to generate various descriptors of proteins, compounds and their interactions. Bioinformatics 2015, 31, 279–281. [Google Scholar] [CrossRef]
- Sieberts, S.K.; Perumal, T.M.; Carrasquillo, M.M.; Allen, M.; Reddy, J.S.; Hoffman, G.E.; Dang, K.K.; Calley, J.; Ebert, P.J.; Eddy, J.; et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci. Data 2020, 7, 340. [Google Scholar] [CrossRef]
- Tesi, N.; van der Lee, S.; Hulsman, M.; van Schoor, N.M.; Huisman, M.; Pijnenburg, Y.; van der Flier, W.M.; Reinders, M.; Holstege, H. Cognitively healthy centenarians are genetically protected against Alzheimer’s disease. Alzheimer’s Dement. 2024, 20, 3864–3875. [Google Scholar] [CrossRef]
- Amin, F.U.; Shah, S.A.; Kim, M.O. Vanillic acid attenuates Abeta(1-42)-induced oxidative stress and cognitive impairment in mice. Sci. Rep. 2017, 7, 40753. [Google Scholar] [CrossRef]
- Pojero, F.; Aiello, A.; Gervasi, F.; Caruso, C.; Ligotti, M.E.; Calabro, A.; Procopio, A.; Candore, G.; Accardi, G.; Allegra, M. Effects of Oleuropein and Hydroxytyrosol on Inflammatory Mediators: Consequences on Inflammaging. Int. J. Mol. Sci. 2022, 24, 380. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.H.; Lee, H.G.; Smith, M.A.; Shah, K. Direct and indirect roles of cyclin-dependent kinase 5 as an upstream regulator in the c-Jun NH2-terminal kinase cascade: Relevance to neurotoxic insults in Alzheimer’s disease. Mol. Biol. Cell. 2009, 20, 4611–4619. [Google Scholar] [CrossRef]
- Yadav, E.; Yadav, P.; Khan, M.M.U.; Singh, H.; Verma, A. Resveratrol: A potential therapeutic natural polyphenol for neurodegenerative diseases associated with mitochondrial dysfunction. Front. Pharmacol. 2022, 13, 922232. [Google Scholar] [CrossRef] [PubMed]
- Banji, O.J.F.; Banji, D.; Makeen, H.A.; Alqahtani, S.S.; Alshahrani, S. Neuroinflammation: The Role of Anthocyanins as Neuroprotectants. Curr. Neuropharmacol. 2022, 20, 2156–2174. [Google Scholar] [CrossRef]
- Kumju Youn, C.-T.H. Mira Jun, Multifaceted neuroprotective effects of (-)-epigallocatechin-3-gallate (EGCG) in Alzheimer’s disease: An overview of pre-clinical studies focused on β-amyloid peptide. Food Sci. Hum. Wellness 2022, 11, 483–493. [Google Scholar] [CrossRef]
- Sano, M.; Ernesto, C.; Thomas, R.G.; Klauber, M.R.; Schafer, K.; Grundman, M.; Woodbury, P.; Growdon, J.; Cotman, C.W.; Pfeiffer, E.; et al. A controlled trial of selegiline, alpha-tocopherol, or both as treatment for Alzheimer’s disease. The Alzheimer’s Disease Cooperative Study. N. Engl. J. Med. 1997, 336, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Gugliandolo, A.; Chiricosta, L.; Silvestro, S.; Bramanti, P.; Mazzon, E. alpha-Tocopherol Modulates Non-Amyloidogenic Pathway and Autophagy in an In Vitro Model of Alzheimer’s Disease: A Transcriptional Study. Brain Sci. 2019, 9, 196. [Google Scholar] [CrossRef]
- Garcia-Garcia, V.A.; Alameda, J.P.; Page, A.; Casanova, M.L. Role of NF-kappaB in Ageing and Age-Related Diseases: Lessons from Genetically Modified Mouse Models. Cells 2021, 10, 1906. [Google Scholar] [CrossRef]
- Serreli, G.; Deiana, M. Extra Virgin Olive Oil Polyphenols: Modulation of Cellular Pathways Related to Oxidant Species and Inflammation in Aging. Cells 2020, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, X.; Zhang, R.; Wang, Z.; Gan, J.; Gao, Q.; Yang, L.; Xu, P.; Jiang, X. Inflammatory signaling pathways in the treatment of Alzheimer’s disease with inhibitors, natural products and metabolites (Review). Int. J. Mol. Med. 2023, 52, 111. [Google Scholar] [CrossRef]
- Reis e Sousa, C.; Yamasaki, S.; Brown, G.D. Myeloid C-type lectin receptors in innate immune recognition. Immunity 2024, 57, 700–717. [Google Scholar] [CrossRef]
- Almeida-da-Silva, C.L.C.; Savio, L.E.B.; Coutinho-Silva, R.; Ojcius, D.M. The role of NOD-like receptors in innate immunity. Front. Immunol. 2023, 14, 1122586. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Murray, P.J. Cytokine signaling modules in inflammatory responses. Immunity 2008, 28, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- Chen, Z.; Balachandran, Y.L.; Chong, W.P.; Chan, K.W.Y. Roles of Cytokines in Alzheimer’s Disease. Int. J. Mol. Sci. 2024, 25, 5803. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. Chemokines and chemokine receptors: Standing at the crossroads of immunobiology and neurobiology. Immunity 2009, 31, 711–721. [Google Scholar] [CrossRef]
- Wang, H.; Zong, Y.; Zhu, L.; Wang, W.; Han, Y. Chemokines in patients with Alzheimer’s disease: A meta-analysis. Front. Aging Neurosci. 2023, 15, 1047810. [Google Scholar] [CrossRef]
- Linzer, N.; Trumbull, A.; Nar, R.; Gibbons, M.D.; Yu, D.T.; Strouboulis, J.; Bungert, J. Regulation of RNA Polymerase II Transcription Initiation and Elongation by Transcription Factor TFII-I. Front. Mol. Biosci. 2021, 8, 681550. [Google Scholar] [CrossRef]
- Balez, R.; Steiner, N.; Engel, M.; Munoz, S.S.; Lum, J.S.; Wu, Y.; Wang, D.; Vallotton, P.; Sachdev, P.; O’Connor, M.; et al. Neuroprotective effects of apigenin against inflammation, neuronal excitability and apoptosis in an induced pluripotent stem cell model of Alzheimer’s disease. Sci. Rep. 2016, 6, 31450. [Google Scholar] [CrossRef]
- Rita, L.; Neumann, N.R.; Laponogov, I.; Gonzalez, G.; Veselkov, D.; Pratico, D.; Aalizadeh, R.; Thomaidis, N.S.; Thompson, D.C.; Vasiliou, V.; et al. Alzheimer’s disease: Using gene/protein network machine learning for molecule discovery in olive oil. Hum. Genom. 2023, 17, 57. [Google Scholar] [CrossRef]
- Ansari, M.A.; Abdul, H.M.; Joshi, G.; Opii, W.O.; Butterfield, D.A. Protective effect of quercetin in primary neurons against Abeta(1-42): Relevance to Alzheimer’s disease. J. Nutr. Biochem. 2009, 20, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Zaplatic, E.; Bule, M.; Shah, S.Z.A.; Uddin, M.S.; Niaz, K. Molecular mechanisms underlying protective role of quercetin in attenuating Alzheimer’s disease. Life Sci. 2019, 224, 109–119. [Google Scholar] [CrossRef]
- Ahmad, S.; Jo, M.H.; Ikram, M.; Khan, A.; Kim, M.O. Deciphering the Potential Neuroprotective Effects of Luteolin against Abeta(1)-(42)-Induced Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 9583. [Google Scholar] [CrossRef] [PubMed]
- Servili, M.; Sordini, B.; Esposto, S.; Urbani, S.; Veneziani, G.; Di Maio, I.; Selvaggini, R.; Taticchi, A. Biological Activities of Phenolic Compounds of Extra Virgin Olive Oil. Antioxidants 2013, 3, 1–23. [Google Scholar] [CrossRef]
- Manach, C.; Scalbert, A.; Morand, C.; Remesy, C.; Jimenez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, C.; Carpena, M.; Lourenco-Lopes, C.; Gallardo-Gomez, M.; Lorenzo, J.M.; Barba, F.J.; Prieto, M.A.; Simal-Gandara, J. Bioactive Compounds and Quality of Extra Virgin Olive Oil. Foods 2020, 9, 1014. [Google Scholar] [CrossRef]
- Sayed-Ahmad, B.U.M.; Hijazi, A.; Saad, Z.; Cerny, M.; Evon, P.; Talou, T.; Merah, O. Amaranth Oilseed Composition and Cosmetic Applications. Separations 2022, 9, 181. [Google Scholar] [CrossRef]
- Ryan, E.; Galvin, K.; O’Connor, T.P.; Maguire, A.R.; O’Brien, N.M. Phytosterol, squalene, tocopherol content and fatty acid profile of selected seeds, grains, and legumes. Plant Foods Hum. Nutr. 2007, 62, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Karadeniz, F. Biological importance and applications of squalene and squalane. Adv. Food Nutr. Res. 2012, 65, 223–233. [Google Scholar]
- Zhao, L.; Wang, J.L.; Liu, R.; Li, X.X.; Li, J.F.; Zhang, L. Neuroprotective, anti-amyloidogenic and neurotrophic effects of apigenin in an Alzheimer’s disease mouse model. Molecules 2013, 18, 9949–9965. [Google Scholar] [CrossRef]
- Ali Mohammadkhanizadeh, M.S. Soroush Taherkhani, Davood Nourabadi, Seyed Mahdi Mohamadi-Zarch, Farnaz Nikbakht, Yaser Azizi, Protective effects of apigenin in neurodegeneration: An update on the potential mechanisms. Brain Disord. 2025, 17, 100189. [Google Scholar] [CrossRef]
- He, Z.; Li, X.; Wang, Z.; Cao, Y.; Han, S.; Li, N.; Cai, J.; Cheng, S.; Liu, Q. Protective effects of luteolin against amyloid beta-induced oxidative stress and mitochondrial impairments through peroxisome proliferator-activated receptor gamma-dependent mechanism in Alzheimer’s disease. Redox Biol. 2023, 66, 102848. [Google Scholar] [CrossRef]
- Kabir, E.R.; Chowdhury, N.M.; Yasmin, H.; Kabir, M.T.; Akter, R.; Perveen, A.; Ashraf, G.M.; Akter, S.; Rahman, M.H.; Sweilam, S.H. Unveiling the Potential of Polyphenols as Anti-Amyloid Molecules in Alzheimer’s Disease. Curr. Neuropharmacol. 2023, 21, 787–807. [Google Scholar] [CrossRef]
- Chang, W.; Huang, D.; Lo, Y.M.; Tee, Q.; Kuo, P.; Wu, J.S.; Huang, W.; Shen, S. Protective Effect of Caffeic Acid against Alzheimer’s Disease Pathogenesis via Modulating Cerebral Insulin Signaling, beta-Amyloid Accumulation, and Synaptic Plasticity in Hyperinsulinemic Rats. J. Agric. Food Chem. 2019, 67, 7684–7693. [Google Scholar] [CrossRef] [PubMed]
- Azargoonjahromi, A.; Abutalebian, F. Unraveling the therapeutic efficacy of resveratrol in Alzheimer’s disease: An umbrella review of systematic evidence. Nutr. Metab. 2024, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Ramassamy, C. Emerging role of polyphenolic compounds in the treatment of neurodegenerative diseases: A review of their intracellular targets. Eur. J. Pharmacol. 2006, 545, 51–64. [Google Scholar] [CrossRef]
- Spencer, J.P. The interactions of flavonoids within neuronal signalling pathways. Genes. Nutr. 2007, 2, 257–273. [Google Scholar] [CrossRef]
- Chen, Z.; Zhong, C. Oxidative stress in Alzheimer’s disease. Neurosci. Bull. 2014, 30, 271–281. [Google Scholar] [CrossRef]
- Ganguly, G.; Chakrabarti, S.; Chatterjee, U.; Saso, L. Proteinopathy, oxidative stress and mitochondrial dysfunction: Cross talk in Alzheimer’s disease and Parkinson’s disease. Drug Des. Devel. Ther. 2017, 11, 797–810. [Google Scholar] [CrossRef]
- Zhou, J.; Chao, G.; Li, Y.; Wu, M.; Zhong, S.; Feng, Z. Activation of NRF2/ARE by isosilybin alleviates Abeta25-35-induced oxidative stress injury in HT-22 cells. Neurosci. Lett. 2016, 632, 92–97. [Google Scholar] [CrossRef]
- Ramsey, C.P.; Glass, C.A.; Montgomery, M.B.; Lindl, K.A.; Ritson, G.P.; Chia, L.A.; Hamilton, R.L.; Chu, C.T.; Jordan-Sciutto, K.L. Expression of Nrf2 in neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 2007, 66, 75–85. [Google Scholar] [CrossRef]
- Manach, C.; Williamson, G.; Morand, C.; Scalbert, A.; Remesy, C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005, 81 (Suppl. S1), 230S–242S. [Google Scholar] [CrossRef]
- Scalbert, A.; Manach, C.; Morand, C.; Remesy, C.; Jimenez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 2005, 45, 287–306. [Google Scholar] [CrossRef] [PubMed]
- Kamatham, P.T.; Shukla, R.; Khatri, D.K.; Vora, L.K. Pathogenesis, diagnostics, and therapeutics for Alzheimer’s disease: Breaking the memory barrier. Ageing Res. Rev. 2024, 101, 102481. [Google Scholar] [CrossRef] [PubMed]
| Bioactive Compounds | Structures | Associated Proteins |
|---|---|---|
| Caffeic acid | C1=CC(=C(C=C1C=CC(=O)O)O)O | SULT1A3, SULT1C2, TP53, TYR, UBE2C, UGT1A1, UGT1A10, UGT1A3, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT2B15, UGT2B17, UGT2B4, UGT2B7, MIF, KDM4E, MEN1, HDAC3, CA12, AKR1B10, KDM4A, SNUPN, ALDH1A1, EGFR, F2, CA1, CA2, LMNA, TTR, ESR1, MMP1, APP, GLA, POLB, CA3, HSP90AA1, HSP90AB1, MMP2, CYP3A4, ALOX5, GAA, MAPT, THRB, MMP9, AKR1B1, ARSA, HPGD, AKR1C4, PTPN1, NFKB1, ACHE, CA4, PTGS1, CA6, GPT, DPP4, APEX1, MPG, CA5A, PTPN7, AKR1C3, CA7, XDH, DUSP3, AKR1C2, HDAC4, ANTXR2, PMP22, KMT2A, AKR1C1, PTPN11, HYAL1, KCNH2, PLA2G7, HDAC1, KPNB1, CA9, HCAR2, HDAC7, HDAC2, HDAC10, HDAC11, EHMT2, HSD17B10, HDAC8, EGLN1, SLCO1B3, TDP1, HDAC6, POLK, BAZ2B, HDAC9, CA14, HDAC5, CA5B, SLCO1B1, P4HA1, PNLIP |
| Hydroxytyrosol | C1=CC(=C(C=C1CCO)O)O | ALOX5 |
| Tyrosol | C1=CC(=CC=C1CCO)O | |
| Oleuropein aglycone | CC=C1C(C(=COC1O)C(=O)OC)CC(=O)OCCC2=CC(=C(C=C2)O)O | |
| Oleacein | CC=C(C=O)C(CC=O)CC(=O)OCCC1=CC(=C(C=C1)O)O | |
| Oleocanthal | CC=C(C=O)C(CC=O)CC(=O)OCCC1=CC=C(C=C1)O | |
| 1-acetoxypinoresinol | CC(=O)OC12COC(C1COC2C3=CC(=C(C=C3)O)OC)C4=CC(=C(C=C4)O)OC | |
| Pinoresinol | COC1=C(C=CC(=C1)C2C3COC(C3CO2)C4=CC(=C(C=C4)O)OC)O | PPARG, PPARD, PPARA, ATAD5, RACGAP1 |
| Apigenin | C1=CC(=CC=C1C2=CC(=O)C3=C(C=C(C=C3O2)O)O)O | TTR, DAPK1, CSNK2A1, TNKS2, KDM4E, SGK1, MEN1, PLK4, STK25, GABRP, CHEK1, GABRD, RGS12, NPC1, MAPK13, PDPK1, DAPK3, AKR1B10, ROCK2, GMNN, RPS6KA5, USP2, STK16, IDH1, PDE5A, USP1, STK10, GLS, CCNB2, TNKS, OXSR1, PAK4, CHEK2, ALDH1A1, EGFR, LMNA, ESR1, GBA, NR3C1, TP53, CYP1A1, CYBB, APP, MPO, CYP1A2, INSR, LCK, BCHE, CDK1, LYN, ABCB1, ELANE, CYP3A4, FGR, PARP1, ALOX5, ADORA3, CXCL8, AR, CYP2D6, MAPT, THRB, HSPA5, PIM1, TOP2A, VDR, CYP19A1, CYP2C9, CFTR, HSD17B1, PKM, CCNB1, GABRA1, ARSA, HPGD, ALOX15, TSHR, PRKCA, PRKACA, GABRG2, CSNK2A2, RXRA, NFKB1, MAOA, UGT1A1, UGT1A4, PRKACG, PRKACB, RPS6KB1, JAK1, CDK2, PON1, MAOB, MAPK3, DPP4, APEX1, GABRB3, MAPK1, AKT1, CYP2C19, ABCC1, PTGS2, OPRM1, UGT1A3, AHR, MAP2K2, FLT3, HSD17B2, PPARG, BRCA1, FEN1, THPO, OPRD1, OPRK1, CSK, SYK, MAPK8, MAPK9, GABRB2, XDH, CSNK1A1, CSNK1D, MAPKAPK2, CSNK1E, CLK1, CLK2, CLK3, RGS4, GSK3A, GSK3B, RORC, RPS6KA3, NEK2, MAP2K6, PLK1, MAPK12, MAPK10, PRKAA2, UGT2B15, BACE1, GNAI1, CSNK2B, HBB, GABRE, CSNK1G2, SMAD3, CDKL1, CDK6, CDK5, CDK16, NFKB2, RUNX1, PMP22, MAP2K1, TOP2B, KMT2A, PPARD, PTH1R, RELA, PPARA, DMPK, KCNMA1, STK4, PIN1, CAMK2B, CAMK2G, CAMK2D, DYRK1A, CBFB, CDK5R1, PDK1, STK38, RPS6KA1, MAPK11, NFE2L2, MAPK14, CAMK4, SMN1, MAPK6, HIF1A, CYP1B1, QPCT, NPSR1, PIM3, VRK2, CAMK1D, VRK3, MAPKAPK5, CCNB3, ESR2, ABCC2, PBK, CAMK1G, ATAD5, NR1H4, CAMKK2, MPHOSPH8, MAP3K5, ATXN2, HSD17B10, MYOC, VRK1, RIOK2, SLK, UGT1A10, UGT1A8, NEK6, CSNK1G1, SLCO1B3, NOX4, PAK6, TDP1, PIM2, PAK5, STK26, POLK, STK17A, TNIK, POLI, ABCG2, CAMK2A, POLH, NEU2, MAP4K5, SLCO1B1, CSNK1G3, HSD17B3, AURKB, DPP3, PTPN1, ACHE, CA1, CA2, CA3, CA4, ACE |
| Luteolin | C1=CC(=C(C=C1C2=CC(=O)C3=C(C=C(C=C3O2)O)O)O)O | TTR, PPARG, DAPK1, GALNT2, PBRM1, IPMK, TNKS2, KDM4E, MEN1, TNFRSF10B, ALOX15B, CA12, AKR1B10, KDM4A, GMNN, IDH1, USP1, GLS, CCNB2, SNUPN, TNKS, RAPGEF3, ALDH1A1, PLG, CA1, CA2, LMNA, ESR1, MMP1, GBA, NR3C1, TP53, CYP1A1, CYBB, APP, CYP1A2, LCK, BCHE, GLA, CDK1, POLB, ABCB1, ELANE, MMP2, MMP3, CYP3A4, PARP1, ALOX5, GAA, AR, BCL2, CYP2D6, MAPT, THRB, HSPA5, PIM1, TOP1, TOP2A, CYP2C9, CCNB1, TYR, MMP9, HPGD, ALOX15, TSHR, ALOX12, TFE3, CSNK2A2, NFKB1, MAOA, CA4, PTGS1, CDK2, PON1, MAOB, DPP4, APEX1, CD38, CYP2C19, PTGS2, FLT3, BRCA1, FEN1, MMP12, CA7, SYK, MMP13, RECQL, XDH, FASN, GSK3A, GSK3B, GALK1, BLM, RAN, CSNK2B, CSNK2A1, SMAD3, CDK6, CDK5, NFKB2, RUNX1, KMT2A, RELA, GLO1, BCL2L1, PIN1, CBFB, WRN, KPNB1, CDK5R1, PDK1, NFE2L2, SMN1, HIF1A, CYP1B1, CCNB3, EHMT2, ATAD5, ATXN2, HSD17B10, GPR35, NOX4, MBNL1, TDP1, POLK, BAZ2B, POLI, ABCG2, POLH, NEU2, AMY1A, MAPK14, MAPK10, PSIP1, AURKB, DPP3, CA3, ACE, AKR1B1, ACHE |
| Beta-sitosterol | CCC(CCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C)C(C)C | |
| Campesterol | CC(C)C(C)CCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C | |
| Squalene | CC(=CCCC(=CCCC(=CCCC=C(C)CCC=C(C)CCC=C(C)C)C)C)C | |
| Alpha-tocopherol | CC1=C(C2=C(CCC(O2)(C)CCCC(C)CCCC(C)CCCC(C)C)C(=C1O)C)C | CFTR, CLCN3, CPT1A, CRABP2, CRP, CXCL8, CYP2E1, CYP3A4, CYP3A5, CYP4F2, DDIT3, EIF2A, FN1, FYN, GCLC, GCLM, GPX1, GPX4, HBG2, HMOX1, HP, ICAM1, IKBKB, IL10, IL6, JUN, KCNH2, KEAP1, LCK, LDHA, LGALS3BP, LYN, MAPK1, MAPK14, MAPK3, MAPK8, MAPK9, MBP, MUC4, NFE2L2, NFKB1, NFKBIA, NOS2, NOS3, NQO1, NR1I2, PARK7, PARP1, PGK1, PLG, PPARA, PPARG, PTGS2, RELA, RPS6, RXRA, SELE, SLC1A2, SLC7A11, SLPI, SMAD4, SNCA, SOD1, TERT, TF, TNF, TNFRSF10B, TP53, VCAM1, XBP1, YY1, PRKCB, ALOX5, PRKCA, DGKA, PPP2CB, PPP2CA, SEC14L4, SEC14L3, SEC14L2, TTPA, AGTR2, EGFR, CYP1A2, DRD3, SLC6A3, CHRM1, OPRK1, ADRA2C, ADORA1, ADORA2A, ADRB3, ELANE, CCR4, HRH1, CTSG, PTGS1, CXCR2, TBXAS1, ADORA3, MMP9, PPP3CA, CYP2C9, CYP2D6, MC3R, NPY2R, MMP1, EDNRA, ESR2, UGT2B7, HTR6, TACR1, ESR1, HMGCR, CHRM4, OPRM1, CYP2C19, BDKRB2, SLC6A2, CALCR, AVPR1A, ADRA2B, DRD1, CASP1, CCKAR, CNR1, PTAFR, CHRM3, CA2, ADRA1D, ERBB2, LMNA, ADRB2, KMT2A, MEN1, CYP2A6, TACR2, VIPR1, SLC6A4, NR3C1, CCR2, PTPRC, HTR2A, USP1, HTR2B, HRH2, OPRD1, ADRB1, RAPGEF3, MC5R, CHRM5, DRD2, DRD4, NPY1R, MAPT, CCR5, ADRA2A, CHRM2, CYSLTR1, HTR2C, SIGMAR1, PDE5A, SNUPN, KPNB1, RAN, CXCR1, FLT1, MAOA, MC4R, PTPN1, GSTP1, NOX4 |
| Oleanolic acid | CC1(CCC2(CCC3(C(=CCC4C3(CCC5C4(CCC(C5(C)C)O)C)C)C2C1)C)C(=O)O)C | |
| Maslinic acid | CC1(CCC2(CCC3(C(=CCC4C3(CCC5C4(CC(C(C5(C)C)O)O)C)C)C2C1)C)C(=O)O)C |
| Source | Compound Name | Smiles | Gene Targets |
|---|---|---|---|
| Clinical trials investigating drugs with NRF2 activation ability used in other diseases and demonstrating their positive effects on AD in vivo models | Dimethyl fumarate | COC(=O)C=CC(=O)OC | |
| Artemether | CC1CCC2C(C(OC3C24C1CCC(O3)(OO4)C)OC)C | ATP1A1 | |
| Ginsenoside-Rd | CC(=CCCC(C)(C1CCC2(C1C(CC3C2(CC(C4C3(CCC(C4(C)C)O)C)OC5C(C(C(C(O5)CO)O)O)OC6C(C(C(C(O6)CO)O)O)O)C)O)C)OC7C(C(C(C(O7)CO)O)O)O)C | ||
| Astaxanthin | CC1=C(C(CC(C1=O)O)(C)C)C=CC(=CC=CC(=CC=CC=C(C)C=CC=C(C)C=CC2=C(C(=O)C(CC2(C)C)O)C)C)C | NFKBIA | |
| Apigenin | C1=CC(=CC=C1C2=CC(=O)C3=C(C=C(C=C3O2)O)O)O | AKR1B1, AR, CDK6, CFTR, CYP19A1, CYP1B1, HSD17B1 | |
| Hesperidin | CC1C(C(C(C(O1)OCC2C(C(C(C(O2)OC3=CC(=C4C(=O)CC(OC4=C3)C5=CC(=C(C=C5)OC)O)O)O)O)O)O)O)O | AURKB, CACNA1B | |
| Ebselen | C1=CC=C(C=C1)N2C(=O)C3=CC=CC=C3[Se]2 | EPHX2, ALB | |
| Puerarin | C1=CC(=CC=C1C2=COC3=C(C2=O)C=CC(=C3C4C(C(C(C(O4)CO)O)O)O)O)O | ||
| Antroquinonol | CC1C(C(C(=C(C1=O)OC)OC)O)CC=C(C)CCC=C(C)CCC=C(C)C | ||
| Vanillic acid | COC1=C(C=CC(=C1)C(=O)O)O | ||
| Allicin | C=CCSS(=O)CC=C | TRPA1, TRPV1 | |
| Drugs with NRF2 activation ability demonstrated in AD clinical trials | DL-3-n-butylphthalide | O=C1C2=CC=CC=C2C(CCCC)O1 | |
| Resveratrol | C1=CC(=CC=C1C=CC2=CC(=CC(=C2)O)O)O | NQO2, CSNK2A1, PTGS1, PTGS2, ALOX15, ALOX5, AHR, PI4K2B, ITGA5, ITGB3, APP, SNCA, SIRT1, ESR1, MTNR1A, MTNR1B, CLEC14A, NR1I2, NR1I3, SLC2A1, CBR1, PPARA, PPARG, AKT1, KHSRP, YARS, CSNK2A1 | |
| Curcumin | COC1=C(C=CC(=C1)C=CC(=O)CC(=O)C=CC2=CC(=C(C=C2)O)OC)O | PPARG, VDR, ABCC5, CBR1, GSTP1 | |
| Sulforaphane | CS(=O)CCCCN=C=S | NFE2L2 | |
| Lipoic Acid | C1CSSC1CCCCC(=O)O | LIPT1, SLC5A6, LIAS | |
| Perindopril | CCCC(C(=O)OCC)NC(C)C(=O)N1C2CCCCC2CC1C(=O)O | ACE, SFRP4 | |
| S-Equol | C1C(COC2=C1C=CC(=C2)O)C3=CC=C(C=C3)O | ESR2, SHBG | |
| Doxycycline | CC1C2C(C3C(C(=O)C(=C(C3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O | MMP8 | |
| Quercetin | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O)O | PIK3CG, UGT3A1, ATP5A1, ATP5B, ATP5C1, PIM1, HIBCH, STK17B, ESR1, ESR2, NQO2, AHR, CYP1B1, ACTB, CSNK2A1, CSNK2B, EIF3F, HSP90AA1, HSPA2, RUVBL2, SF3B3, UBA1, SHBG, CBR1, CEBPB, NR1I2, HCK | |
| Genistein | C1=CC(=CC=C1C2=COC3=CC(=CC(=C3C2=O)O)O)O | NCOA1, ESR1, NCOA2, ESRRB, ESRRA, NR1I2, AKT1, GPER1, CYP1B1, SHBG, ESR2, TOP2A, PTK2B, CFTR, ESRRG, PPARG, TRPC5 | |
| Tideglusib | C1=CC=C(C=C1)CN2C(=O)N(SC2=O)C3=CC=CC4=CC=CC=C43 | GSK3B | |
| Pyridoxine | CC1=NC=C(C(=C1O)CO)CO | PDXK | |
| Benfotiamine | CC1=NC=C(C(=N1)N)CN(C=O)C(=C(CCOP(=O)(O)O)SC(=O)C2=CC=CC=C2)C | AGER | |
| Selected electrophilic activators of NRF2 under clinical development | Bardoxolone-methyl (CDDO-Me) | CC1(CCC2(CCC3(C(C2C1)C(=O)C=C4C3(CCC5C4(C=C(C(=O)C5(C)C)C#N)C)C)C)C(=O)OC)C | NFKBIA, PPARG, STAT3 |
| RTA-408 (omaveloxolone) | CC1(CCC2(CCC3(C(C2C1)C(=O)C=C4C3(CCC5C4(C=C(C(=O)C5(C)C)C#N)C)C)C)NC(=O)C(C)(F)F)C | KEAP1, NFE2L2 | |
| Dimethyl fumarate | COC(=O)C=CC(=O)OC | KEAP1, RELA | |
| ALKS-8700 | COC(=O)C=CC(=O)OCCN1C(=O)CCC1=O | CHRNA10 | |
| Oltipraz | CC1=C(SSC1=S)C2=NC=CN=C2 | ANG | |
| Ursodiol | CC(CCC(=O)O)C1CCC2C1(CCC3C2C(CC4C3(CCC(C4)O)C)O)C | AKR1C2, NR1H4 | |
| Sulforaphane | CS(=O)CCCCN=C=S | NFE2L2 | |
| Curcumin | COC1=C(C=CC(=C1)C=CC(=O)CC(=O)C=CC2=CC(=C(C=C2)O)OC)O | PPARG, VDR, ABCC5, CBR1, GSTP1 | |
| Resveratrol | C1=CC(=CC=C1C=CC2=CC(=CC(=C2)O)O)O | NQO2, CSNK2A1, PTGS1, PTGS2, ALOX15, ALOX5, AHR, PI4K2B, ITGA5, ITGB3, APP, SNCA, SIRT1, ESR1, MTNR1A, MTNR1B, CLEC14A, NR1I2, NR1I3, SLC2A1, CBR1, PPARA, PPARG, AKT1, KHSRP, YARS, CSNK2A1 | |
| CXA-10 | CCCCCCCCC(=CCCCCCCCC(=O)O)[N+](=O)[O−] | ||
| Candidate repurposed drugs for AD with NRF2 activation ability | Curcumin | COC1=C(C=CC(=C1)C=CC(=O)CC(=O)C=CC2=CC(=C(C=C2)O)OC)O | PPARG, VDR, ABCC5, CBR1, GSTP1 |
| Trichostatin-a | CC(C=C(C)C=CC(=O)NO)C(=O)C1=CC=C(C=C1)N(C)C | HDAC1, HDAC10, HDAC2, HDAC3, HDAC4, HDAC5, HDAC6, HDAC7, HDAC8, HDAC9, CASP8, TNF | |
| Panobinostat | CC1=C(C2=CC=CC=C2N1)CCNCC3=CC=C(C=C3)C=CC(=O)NO | HDAC1, HDAC2, HDAC3, HDAC4, HDAC6, HDAC7, HDAC8, HDAC9 | |
| Entinostat | C1=CC=C(C(=C1)N)NC(=O)C2=CC=C(C=C2)CNC(=O)OCC3=CN=CC=C3 | HDAC1, HDAC2, HDAC3, HDAC9 | |
| Parthenolide | CC1=CCCC2(C(O2)C3C(CC1)C(=C)C(=O)O3)C |
| Disease | p-Value | q-Value | Genes |
|---|---|---|---|
| dementia | 2.64 × 10−6 | 5.27 × 10−5 | [APP, TNF, SNCA] |
| migraine | 0.000635 | 0.004234 | [TNF, ESR1] |
| myocardial infarction | 0.000635 | 0.004234 | [ACE, ESR1] |
| asthma | 0.001325 | 0.00655 | [ALOX5, TNF] |
| Alzheimer’s disease | 0.001695 | 0.00655 | [APP, ACE] |
| breast cancer | 0.001965 | 0.00655 | [AKT1, ESR1] |
| obesity | 0.002407 | 0.006876 | [AKR1C2, PPARG] |
| diabetes | 0.013451 | 0.033626 | [ACE, PPARG] |
| osteoporosis | 0.025555 | 0.049865 | [VDR] |
| ectodermal dysplasia | 0.025555 | 0.049865 | [NFKBIA] |
| fibrosis | 0.027846 | 0.049865 | [APP, TNF, SNCA] |
| ovarian cancer | 0.030132 | 0.049865 | [TNF, ESR1] |
| malaria | 0.032412 | 0.049865 | [ACE, ESR1] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourdakou, M.M.; Loizidou, E.M.; Spyrou, G.M. Exploring the Impact of Bioactive Compounds Found in Extra Virgin Olive Oil on NRF2 Modulation in Alzheimer’s Disease. Antioxidants 2025, 14, 952. https://doi.org/10.3390/antiox14080952
Bourdakou MM, Loizidou EM, Spyrou GM. Exploring the Impact of Bioactive Compounds Found in Extra Virgin Olive Oil on NRF2 Modulation in Alzheimer’s Disease. Antioxidants. 2025; 14(8):952. https://doi.org/10.3390/antiox14080952
Chicago/Turabian StyleBourdakou, Marilena M., Eleni M. Loizidou, and George M. Spyrou. 2025. "Exploring the Impact of Bioactive Compounds Found in Extra Virgin Olive Oil on NRF2 Modulation in Alzheimer’s Disease" Antioxidants 14, no. 8: 952. https://doi.org/10.3390/antiox14080952
APA StyleBourdakou, M. M., Loizidou, E. M., & Spyrou, G. M. (2025). Exploring the Impact of Bioactive Compounds Found in Extra Virgin Olive Oil on NRF2 Modulation in Alzheimer’s Disease. Antioxidants, 14(8), 952. https://doi.org/10.3390/antiox14080952









