Dynamic Transcriptomic and Cellular Remodeling Underlie Cuprizone-Induced Demyelination and Endogenous Repair in the CNS
Abstract
1. Introduction
2. Methods
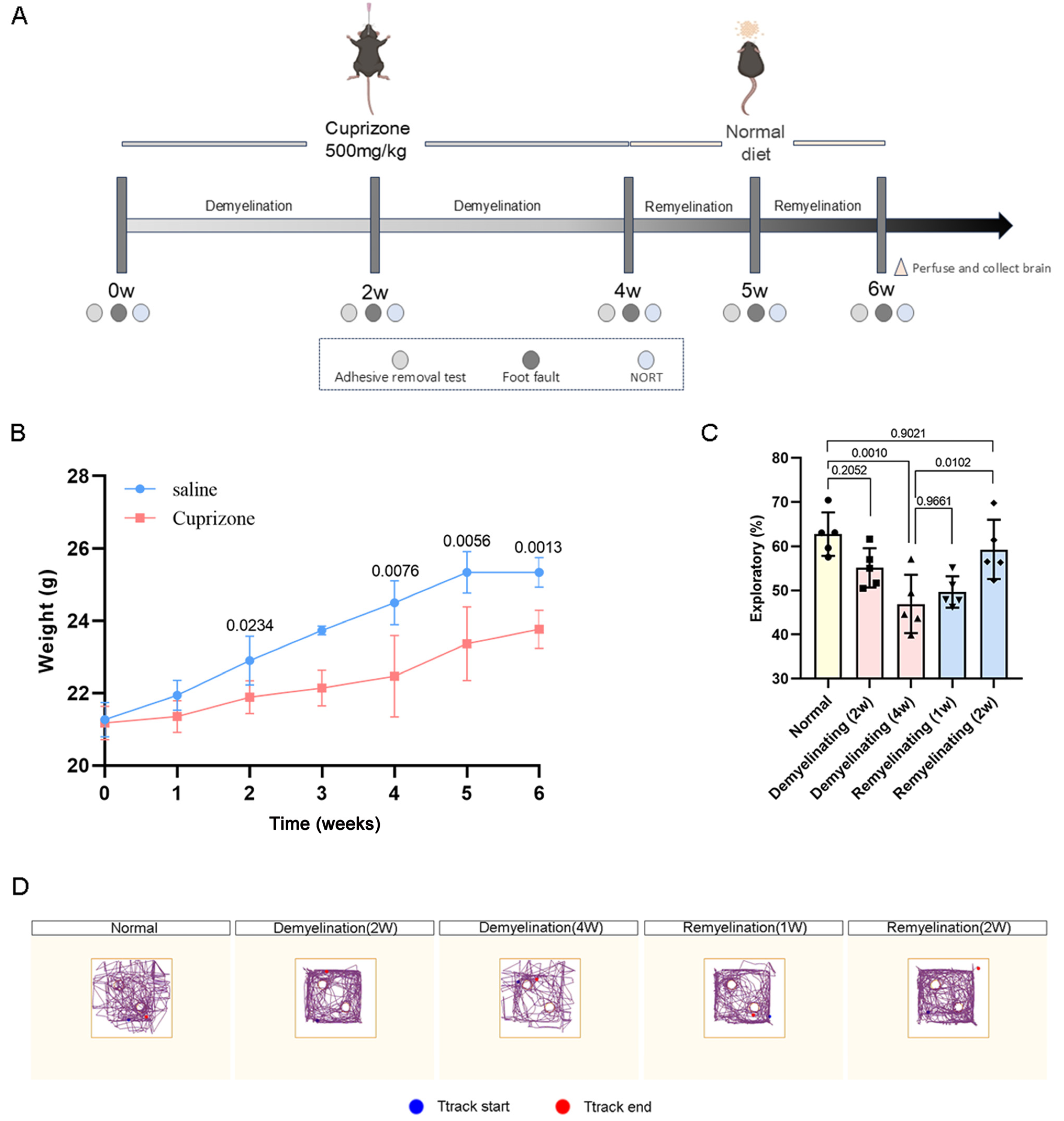
2.1. Mice and Experimental Model
2.2. Neurobehavioral Tests
2.3. Luxol Fast Blue Staining
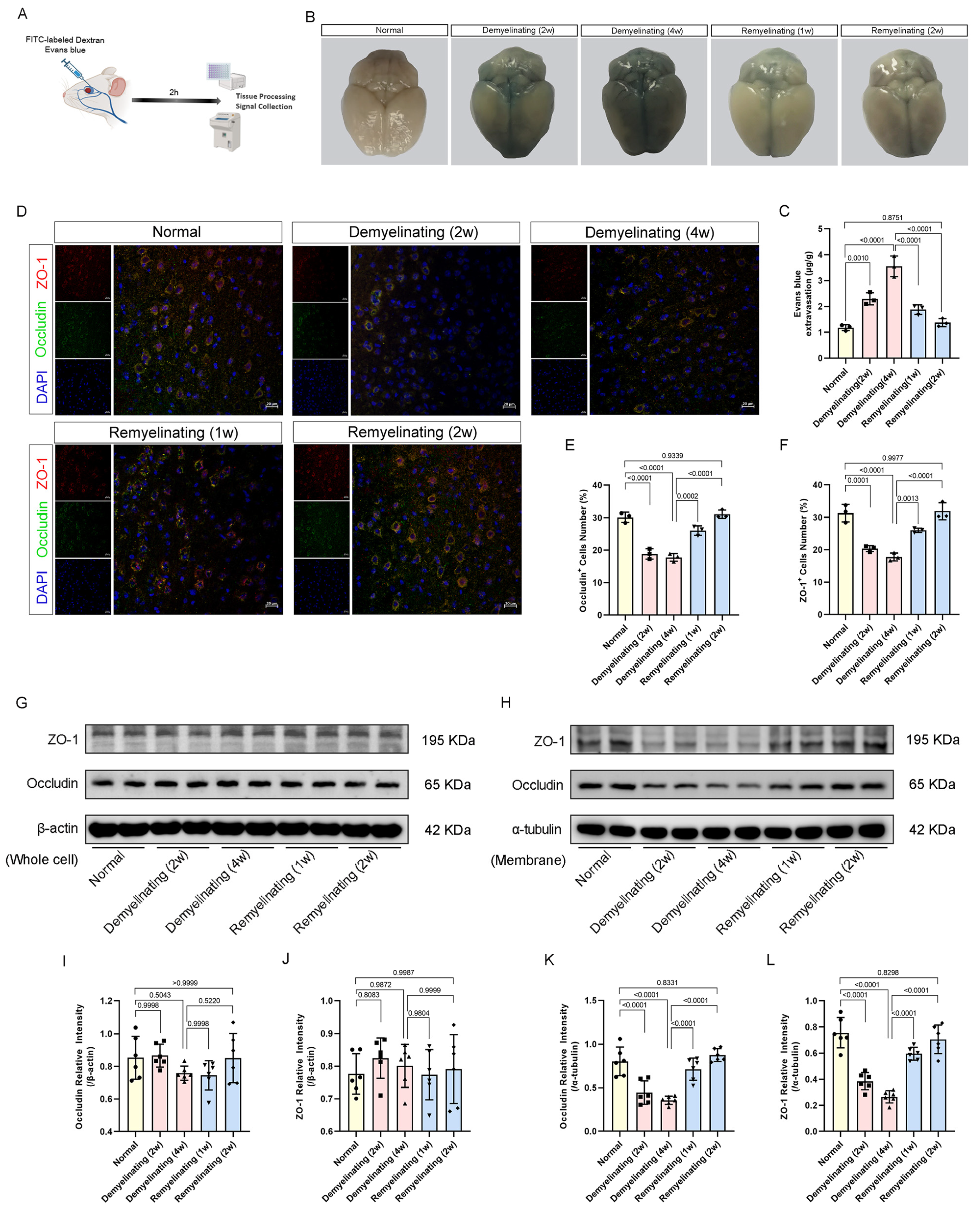
2.4. Evans Blue Permeability Assay
2.5. FITC–Dextran Tracer Assay
2.6. Immunofluorescence Staining
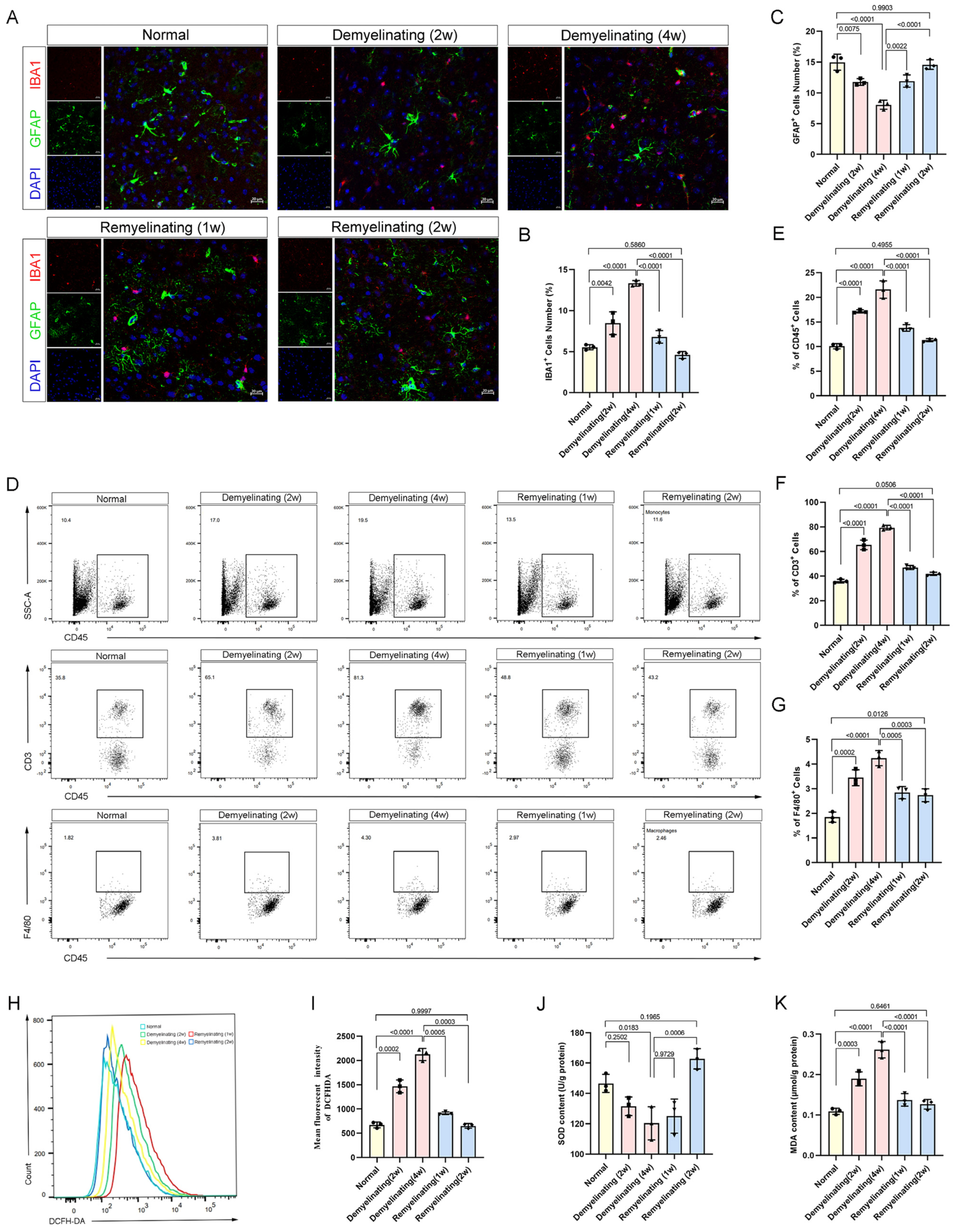
2.7. Flow Cytometry
2.8. Western Blot
2.9. Quantitative Real-Time PCR (qRT-PCR)
2.10. Oxidative Stress-Related Factor Detection
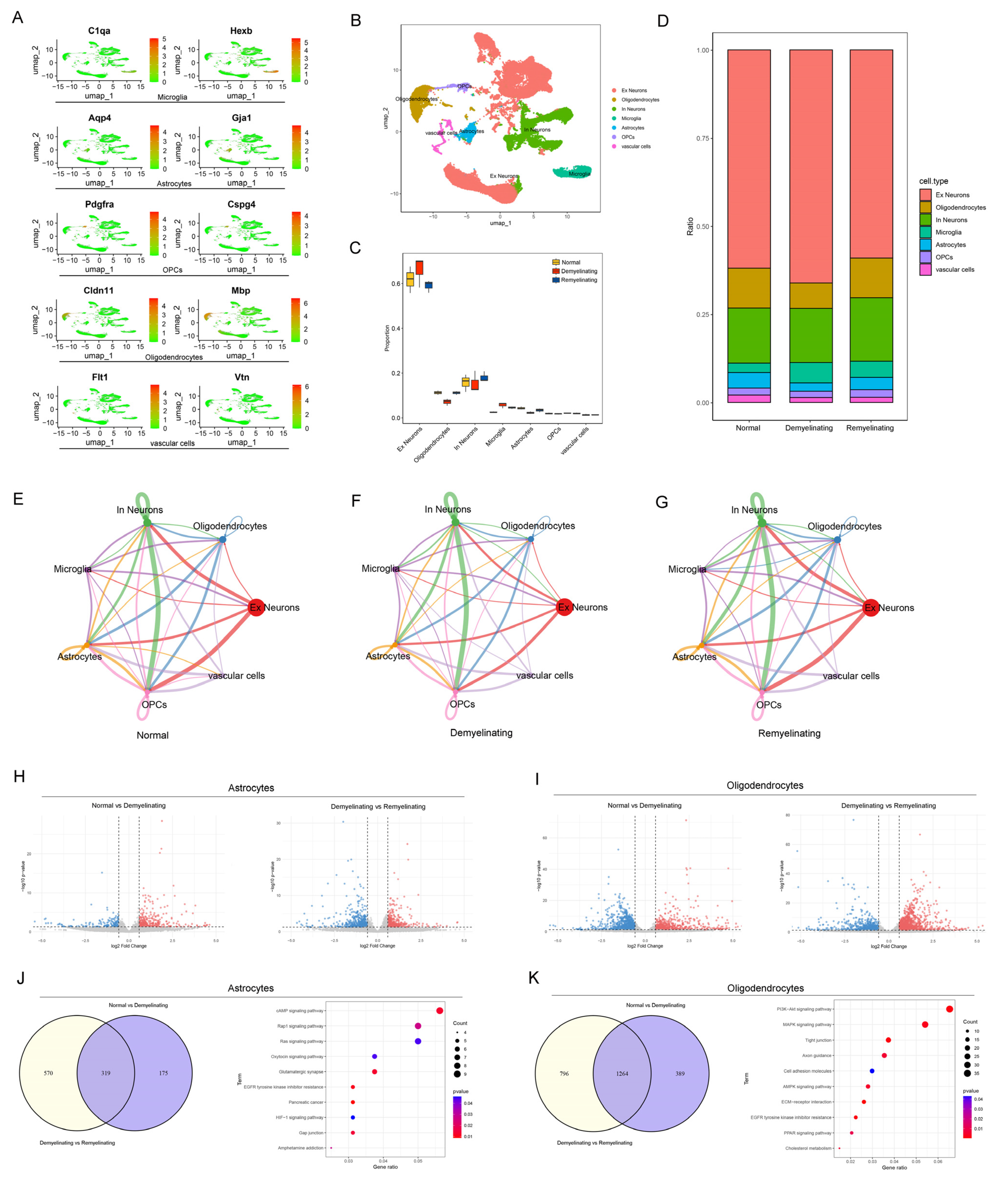
2.11. scRNA-seq and Data Mining
2.12. Bulk Transcriptome Analysis
2.13. Statistical Analysis
3. Results
3.1. Neurological and Motor Functions in Cuprizone-Induced Mice
3.2. Dynamic Reconstruction of Oligodendrocytes During Cuprizone-Induced Demyelination and Endogenous Remyelination
3.3. BBB Permeability Dynamics During Cuprizone-Induced Demyelination and Remyelination
3.4. A Single-Cell Atlas Revealed Dynamic Cellular Remodeling and Signaling Reprogramming During Demyelination and Remyelination
3.5. Neuroimmune Activation and Oxidative Stress Dynamics During the Demyelination-Remyelination Cycle
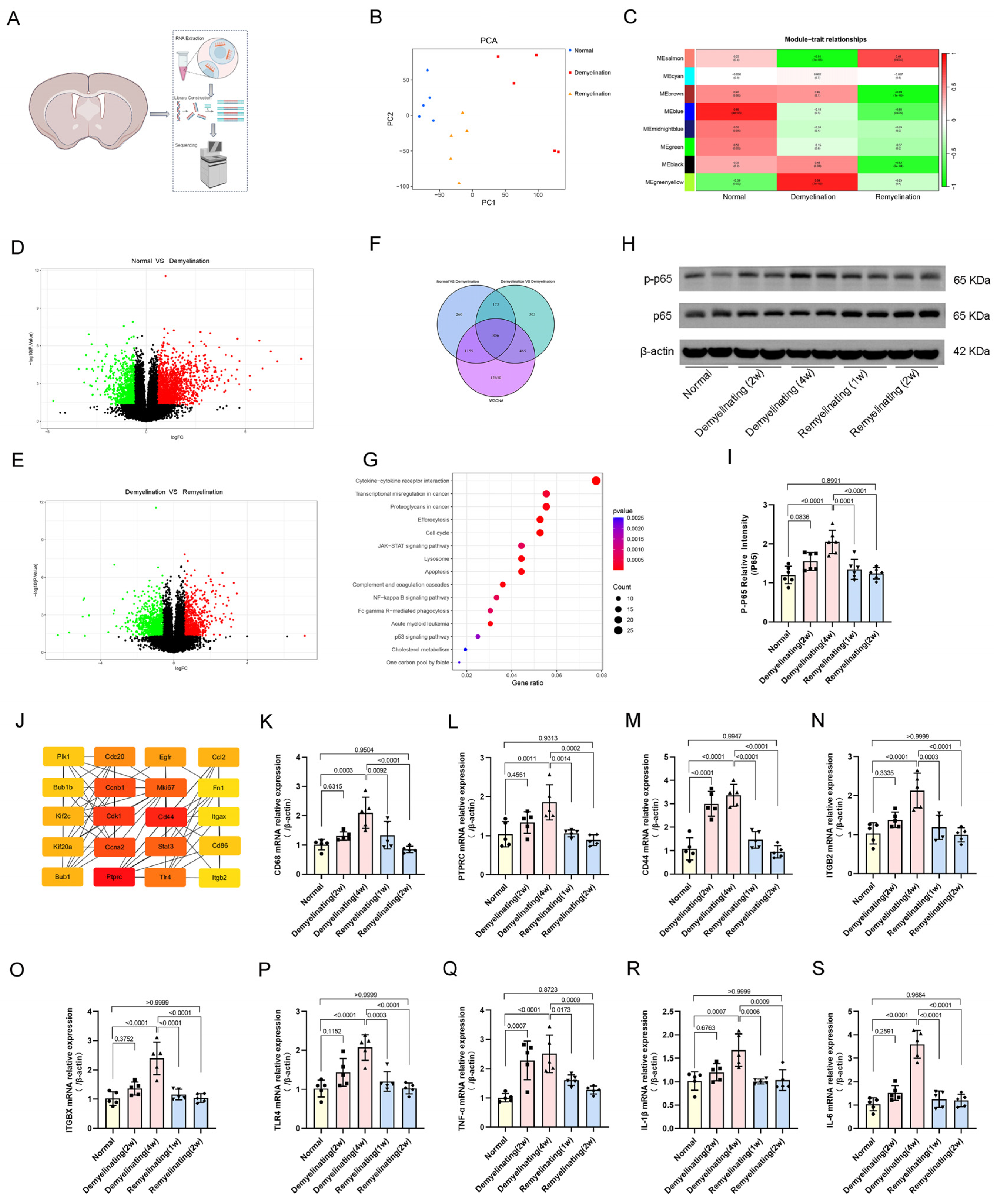
3.6. Transcriptomic Analysis and Inflammatory Regulatory Networks in Cuprizone-Induced Demyelination and Remyelination
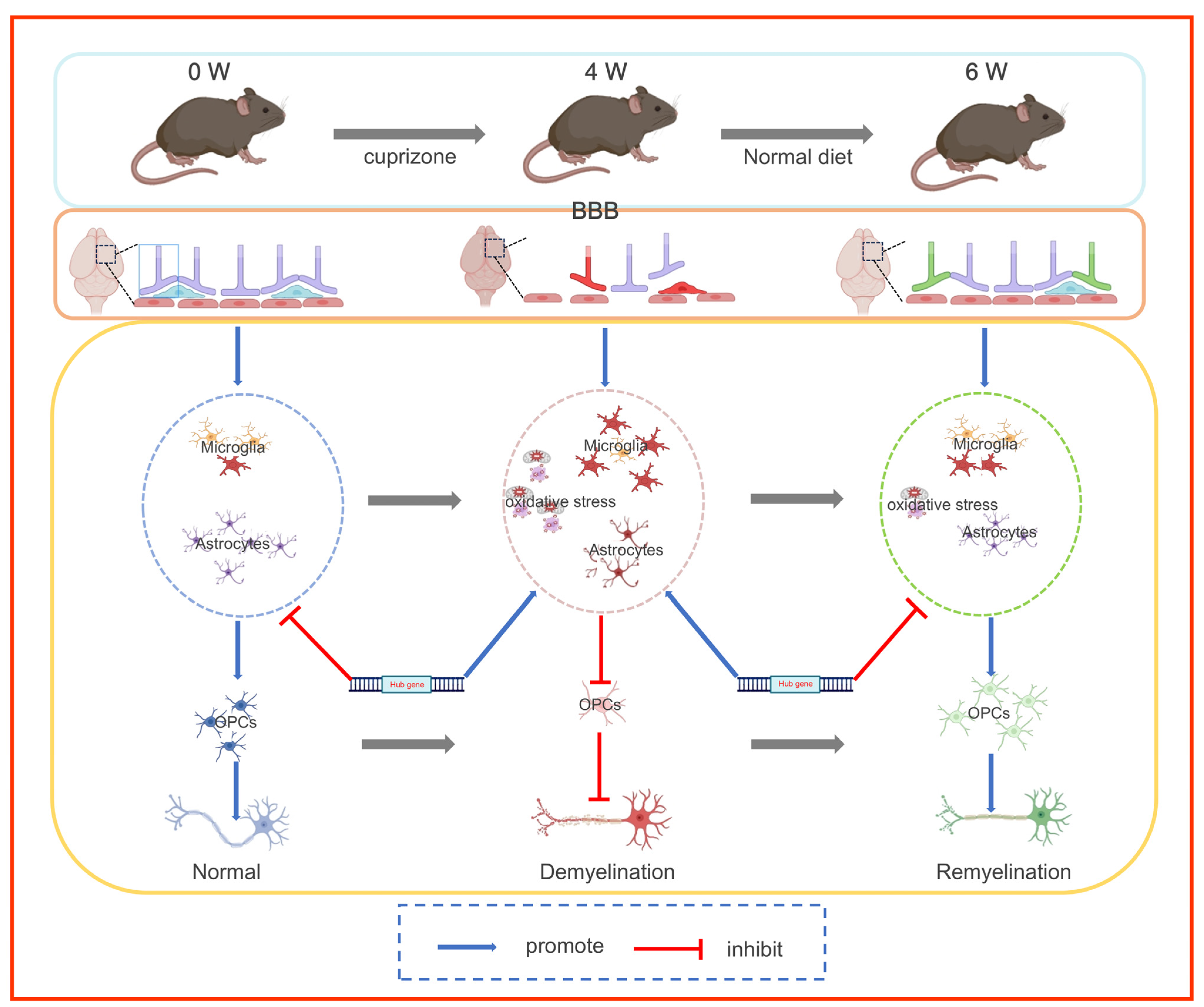
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Faria, O., Jr.; Pivonkova, H.; Varga, B.; Timmler, S.; Evans, K.A.; Káradóttir, R.T. Periods of synchronized myelin changes shape brain function and plasticity. Nat. Neurosci. 2021, 24, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.N.; Appel, B. Oligodendrocytes express synaptic proteins that modulate myelin sheath formation. Nat. Commun. 2019, 10, 4125. [Google Scholar] [CrossRef] [PubMed]
- Bergles, D.E.; Richardson, W.D. Oligodendrocyte Development and Plasticity. Cold Spring Harb. Perspect. Biol. 2015, 8, a020453. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Joseph, C.; Wang, C.; Gupta, V.; Liu, S.; Yiannikas, C.; Chua, B.E.; Chitranshi, N.; Shen, T.; Dheer, Y.; et al. Demyelination precedes axonal loss in the transneuronal spread of human neurodegenerative disease. Brain 2019, 142, 426–442. [Google Scholar] [CrossRef]
- Granberg, T.; Fan, Q.; Treaba, C.A.; Ouellette, R.; Herranz, E.; Mangeat, G.; Louapre, C.; Cohen-Adad, J.; Klawiter, E.C.; Sloane, J.A.; et al. In vivo characterization of cortical and white matter neuroaxonal pathology in early multiple sclerosis. Brain 2017, 140, 2912–2926. [Google Scholar] [CrossRef]
- Saporta, M.A.; Katona, I.; Zhang, X.; Roper, H.P.; McClelland, L.; Macdonald, F.; Brueton, L.; Blake, J.; Suter, U.; Reilly, M.M.; et al. Neuropathy in a human without the PMP22 gene. Arch. Neurol. 2011, 68, 814–821. [Google Scholar] [CrossRef]
- Croxford, J.L.; Anger, H.A.; Miller, S.D. Viral delivery of an epitope from Haemophilus influenzae induces central nervous system autoimmune disease by molecular mimicry. J. Immunol. 2005, 174, 907–917. [Google Scholar] [CrossRef]
- Karsai, G.; Kraft, F.; Haag, N.; Korenke, G.C.; Hänisch, B.; Othman, A.; Suriyanarayanan, S.; Steiner, R.; Knopp, C.; Mull, M.; et al. DEGS1-associated aberrant sphingolipid metabolism impairs nervous system function in humans. J. Clin. Investig. 2019, 129, 1229–1239. [Google Scholar] [CrossRef]
- McLaughlin, K.A.; Wucherpfennig, K.W. B cells and autoantibodies in the pathogenesis of multiple sclerosis and related inflammatory demyelinating diseases. Adv. Immunol. 2008, 98, 121–149. [Google Scholar] [CrossRef]
- Sen, M.K.; Mahns, D.A.; Coorssen, J.R.; Shortland, P.J. Behavioural phenotypes in the cuprizone model of central nervous system demyelination. Neurosci. Biobehav. Rev. 2019, 107, 23–46. [Google Scholar] [CrossRef]
- Solti, I.; Kvell, K.; Talaber, G.; Veto, S.; Acs, P.; Gallyas, F., Jr.; Illes, Z.; Fekete, K.; Zalan, P.; Szanto, A.; et al. Thymic Atrophy and Apoptosis of CD4+CD8+ Thymocytes in the Cuprizone Model of Multiple Sclerosis. PLoS ONE 2015, 10, e0129217. [Google Scholar] [CrossRef] [PubMed]
- Matute, C.; Pérez-Cerdá, F. Multiple sclerosis: Novel perspectives on newly forming lesions. Trends Neurosci. 2005, 28, 173–175. [Google Scholar] [CrossRef]
- Mekhail, M.; Almazan, G.; Tabrizian, M. Oligodendrocyte-protection and remyelination post-spinal cord injuries: A review. Prog. Neurobiol. 2012, 96, 322–339. [Google Scholar] [CrossRef]
- Sen, M.K.; Mahns, D.A.; Coorssen, J.R.; Shortland, P.J. The roles of microglia and astrocytes in phagocytosis and myelination: Insights from the cuprizone model of multiple sclerosis. Glia 2022, 70, 1215–1250. [Google Scholar] [CrossRef]
- Gudi, V.; Gingele, S.; Skripuletz, T.; Stangel, M. Glial response during cuprizone-induced de- and remyelination in the CNS: Lessons learned. Front. Cell Neurosci. 2014, 8, 73. [Google Scholar] [CrossRef]
- Zhan, J.; Mann, T.; Joost, S.; Behrangi, N.; Frank, M.; Kipp, M. The Cuprizone Model: Dos and Do Nots. Cells 2020, 9, 843. [Google Scholar] [CrossRef]
- Dimou, L.; Götz, M. Glial cells as progenitors and stem cells: New roles in the healthy and diseased brain. Physiol. Rev. 2014, 94, 709–737. [Google Scholar] [CrossRef]
- Kocur, M.; Schneider, R.; Pulm, A.K.; Bauer, J.; Kropp, S.; Gliem, M.; Ingwersen, J.; Goebels, N.; Alferink, J.; Prozorovski, T.; et al. IFNβ secreted by microglia mediates clearance of myelin debris in CNS autoimmunity. Acta Neuropathol. Commun. 2015, 3, 20. [Google Scholar] [CrossRef]
- das Neves, S.P.; Sousa, J.C.; Sousa, N.; Cerqueira, J.J.; Marques, F. Altered astrocytic function in experimental neuroinflammation and multiple sclerosis. Glia 2021, 69, 1341–1368. [Google Scholar] [CrossRef]
- Qin, C.; Chen, M.; Dong, M.H.; Yang, S.; Zhang, H.; You, Y.F.; Zhou, L.Q.; Chu, Y.H.; Tang, Y.; Pang, X.W.; et al. Soluble TREM2 triggers microglial dysfunction in neuromyelitis optica spectrum disorders. Brain 2024, 147, 163–176. [Google Scholar] [CrossRef]
- Calì, C.; Cantando, I.; Veloz Castillo, M.F.; Gonzalez, L.; Bezzi, P. Metabolic Reprogramming of Astrocytes in Pathological Conditions: Implications for Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 8922. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, L.; Peng, C.; Bian, C.; Ocak, P.E.; Zhang, J.H.; Yang, Y.; Zhou, D.; Chen, G.; Luo, Y. Targeting Oxidative Stress and Inflammatory Response for Blood-Brain Barrier Protection in Intracerebral Hemorrhage. Antioxid. Redox Signal. 2022, 37, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Jiang, X.; Zhang, W.; Shi, Y.; Leak, R.K.; Keep, R.F.; Ye, Q.; Yang, T.; Li, S.; Hu, X.; et al. Endothelial peroxiredoxin-4 is indispensable for blood-brain barrier integrity and long-term functional recovery after ischemic stroke. Proc. Natl. Acad. Sci. USA 2024, 121, e2400272121. [Google Scholar] [CrossRef] [PubMed]
- Lueptow, L.M. Novel Object Recognition Test for the Investigation of Learning and Memory in Mice. J. Vis. Exp. 2017, 126, e55718. [Google Scholar] [CrossRef]
- Antunes, M.; Biala, G. The novel object recognition memory: Neurobiology, test procedure, and its modifications. Cogn. Process. 2012, 13, 93–110. [Google Scholar] [CrossRef]
- Hou, J.; Zhou, Y.; Cai, Z.; Terekhova, M.; Swain, A.; Andhey, P.S.; Guimaraes, R.M.; Ulezko Antonova, A.; Qiu, T.; Sviben, S.; et al. Transcriptomic atlas and interaction networks of brain cells in mouse CNS demyelination and remyelination. Cell Rep. 2023, 42, 112293. [Google Scholar] [CrossRef]
- Popescu, B.F.; Lucchinetti, C.F. Pathology of demyelinating diseases. Annu. Rev. Pathol. 2012, 7, 185–217. [Google Scholar] [CrossRef]
- Restrepo-Martinez, M.; Bell, V.; Ramirez-Bermudez, J. Cognitive disorders in patients with neuroimmunological disease. Curr. Opin. Psychiatry 2025, 38, 126–133. [Google Scholar] [CrossRef]
- Benedict, R.H.B.; Amato, M.P.; DeLuca, J.; Geurts, J.J.G. Cognitive impairment in multiple sclerosis: Clinical management, MRI, and therapeutic avenues. Lancet Neurol. 2020, 19, 860–871. [Google Scholar] [CrossRef]
- Matsushima, G.K.; Morell, P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001, 11, 107–116. [Google Scholar] [CrossRef]
- Leo, H.; Kipp, M. Remyelination in Multiple Sclerosis: Findings in the Cuprizone Model. Int. J. Mol. Sci. 2022, 23, 16093. [Google Scholar] [CrossRef] [PubMed]
- Ghaffarian, N.; Mesgari, M.; Cerina, M.; Göbel, K.; Budde, T.; Speckmann, E.J.; Meuth, S.G.; Gorji, A. Thalamocortical-auditory network alterations following cuprizone-induced demyelination. J. Neuroinflamm. 2016, 13, 160. [Google Scholar] [CrossRef]
- Ortiz, F.C.; Habermacher, C.; Graciarena, M.; Houry, P.Y.; Nishiyama, A.; Nait Oumesmar, B.; Angulo, M.C. Neuronal activity in vivo enhances functional myelin repair. JCI Insight 2019, 5, e123434. [Google Scholar] [CrossRef]
- Gautier, H.O.; Evans, K.A.; Volbracht, K.; James, R.; Sitnikov, S.; Lundgaard, I.; James, F.; Lao-Peregrin, C.; Reynolds, R.; Franklin, R.J.; et al. Neuronal activity regulates remyelination via glutamate signalling to oligodendrocyte progenitors. Nat. Commun. 2015, 6, 8518. [Google Scholar] [CrossRef]
- Taylor, K.R.; Monje, M. Neuron-oligodendroglial interactions in health and malignant disease. Nat. Rev. Neurosci. 2023, 24, 733–746. [Google Scholar] [CrossRef]
- Monje, M.; Káradóttir, R.T. The bright and the dark side of myelin plasticity: Neuron-glial interactions in health and disease. Semin. Cell Dev. Biol. 2021, 116, 10–15. [Google Scholar] [CrossRef]
- Hui, S.P.; Nag, T.C.; Ghosh, S. Neural cells and their progenitors in regenerating zebrafish spinal cord. Int. J. Dev. Biol. 2020, 64, 353–366. [Google Scholar] [CrossRef]
- Imitola, J.; Snyder, E.Y.; Khoury, S.J. Genetic programs and responses of neural stem/progenitor cells during demyelination: Potential insights into repair mechanisms in multiple sclerosis. Physiol. Genom. 2003, 14, 171–197. [Google Scholar] [CrossRef]
- Lohrberg, M.; Winkler, A.; Franz, J.; van der Meer, F.; Ruhwedel, T.; Sirmpilatze, N.; Dadarwal, R.; Handwerker, R.; Esser, D.; Wiegand, K.; et al. Lack of astrocytes hinders parenchymal oligodendrocyte precursor cells from reaching a myelinating state in osmolyte-induced demyelination. Acta Neuropathol. Commun. 2020, 8, 224. [Google Scholar] [CrossRef]
- Faissner, S.; Plemel, J.R.; Gold, R.; Yong, V.W. Progressive multiple sclerosis: From pathophysiology to therapeutic strategies. Nat. Rev. Drug Discov. 2019, 18, 905–922. [Google Scholar] [CrossRef]
- Correale, J.; Gaitán, M.I.; Ysrraelit, M.C.; Fiol, M.P. Progressive multiple sclerosis: From pathogenic mechanisms to treatment. Brain 2017, 140, 527–546. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Korhonen, E.A.; Merlini, A.; Strauss, J.; Wihuri, E.; Nurmi, H.; Antila, S.; Paech, J.; Deutsch, U.; Engelhardt, B.; et al. Angiopoietin-2 blockade ameliorates autoimmune neuroinflammation by inhibiting leukocyte recruitment into the CNS. J. Clin. Investig. 2020, 130, 1977–1990. [Google Scholar] [CrossRef]
- Na, S.Y.; Janakiraman, M.; Leliavski, A.; Krishnamoorthy, G. High-salt diet suppresses autoimmune demyelination by regulating the blood-brain barrier permeability. Proc. Natl. Acad. Sci. USA 2021, 118, e2025944118. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ma, Q.; Khatibi, N.; Chen, W.; Sozen, T.; Cheng, O.; Tang, J. Ac-YVAD-CMK Decreases Blood-Brain Barrier Degradation by Inhibiting Caspase-1 Activation of Interleukin-1β in Intracerebral Hemorrhage Mouse Model. Transl. Stroke Res. 2010, 1, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, F.; Li, Z.; Mu, Y.; Yong, V.W.; Xue, M. Neuroprotective Effects of Chlorogenic Acid in a Mouse Model of Intracerebral Hemorrhage Associated with Reduced Extracellular Matrix Metalloproteinase Inducer. Biomolecules 2022, 12, 1020. [Google Scholar] [CrossRef]
- Seo, J.H.; Miyamoto, N.; Hayakawa, K.; Pham, L.D.; Maki, T.; Ayata, C.; Kim, K.W.; Lo, E.H.; Arai, K. Oligodendrocyte precursors induce early blood-brain barrier opening after white matter injury. J. Clin. Investig. 2013, 123, 782–786. [Google Scholar] [CrossRef]
- Standiford, M.M.; Grund, E.M.; Howe, C.L. Citrullinated myelin induces microglial TNFα and inhibits endogenous repair in the cuprizone model of demyelination. J. Neuroinflamm. 2021, 18, 305. [Google Scholar] [CrossRef]
- Gao, H.; Danzi, M.C.; Choi, C.S.; Taherian, M.; Dalby-Hansen, C.; Ellman, D.G.; Madsen, P.M.; Bixby, J.L.; Lemmon, V.P.; Lambertsen, K.L.; et al. Opposing Functions of Microglial and Macrophagic TNFR2 in the Pathogenesis of Experimental Autoimmune Encephalomyelitis. Cell Rep. 2017, 18, 198–212. [Google Scholar] [CrossRef]
- Rawji, K.S.; Young, A.M.H.; Ghosh, T.; Michaels, N.J.; Mirzaei, R.; Kappen, J.; Kolehmainen, K.L.; Alaeiilkhchi, N.; Lozinski, B.; Mishra, M.K.; et al. Niacin-mediated rejuvenation of macrophage/microglia enhances remyelination of the aging central nervous system. Acta Neuropathol. 2020, 139, 893–909. [Google Scholar] [CrossRef]
- Gharibani, P.; Abramson, E.; Shanmukha, S.; Smith, M.D.; Godfrey, W.H.; Lee, J.J.; Hu, J.; Baydyuk, M.; Dorion, M.F.; Deng, X.; et al. The protein kinase C modulator bryostatin-1 therapeutically targets microglia to attenuate neuroinflammation and promote remyelination. Sci. Transl. Med. 2025, 17, eadk3434. [Google Scholar] [CrossRef]
- Hu, G.; Huang, K.; Hu, Y.; Du, G.; Xue, Z.; Zhu, X.; Fan, G. Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci. Rep. 2016, 6, 31851. [Google Scholar] [CrossRef] [PubMed]
- Spurgat, M.S.; Tang, S.J. Single-Cell RNA-Sequencing: Astrocyte and Microglial Heterogeneity in Health and Disease. Cells 2022, 11, 2021. [Google Scholar] [CrossRef]
- Wu, Z.; Li, G.; Wang, S.; Zhang, N.; Li, X.; Zhang, F.; Niu, J.; Wang, N.; Zu, J.; Wang, Y. Single-cell analysis of spinal cord injury reveals functional heterogeneity of oligodendrocyte lineage cells. Gene 2023, 886, 147713. [Google Scholar] [CrossRef]
- Smith, B.M.; Giddens, M.M.; Neil, J.; Owino, S.; Nguyen, T.T.; Duong, D.; Li, F.; Hall, R.A. Mice lacking Gpr37 exhibit decreased expression of the myelin-associated glycoprotein MAG and increased susceptibility to demyelination. Neuroscience 2017, 358, 49–57. [Google Scholar] [CrossRef]
- Kim, K.Y.; Ju, W.K. cAMP/PKA pathway and mitochondrial protection in oxidative stress-induced optic nerve head astrocytes. Neural Regen. Res. 2021, 16, 108–109. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, Z.; Yin, Q.; Zhang, X. TTB Protects Astrocytes Against Oxygen-Glucose Deprivation/Reoxygenation-Induced Injury via Activation of Nrf2/HO-1 Signaling Pathway. Front. Pharmacol. 2019, 10, 792. [Google Scholar] [CrossRef]
- Qian, S.; Qian, L.; Yang, Y.; Cui, J.; Zhao, Y. Transcriptome sequencing analysis revealed the molecular mechanism of podoplanin neutralization inhibiting ischemia/reperfusion-induced microglial activation. Ann. Transl. Med. 2022, 10, 638. [Google Scholar] [CrossRef]
- Ding, Z.; Dai, C.; Zhong, L.; Liu, R.; Gao, W.; Zhang, H.; Yin, Z. Neuregulin-1 converts reactive astrocytes toward oligodendrocyte lineage cells via upregulating the PI3K-AKT-mTOR pathway to repair spinal cord injury. Biomed. Pharmacother. 2021, 134, 111168. [Google Scholar] [CrossRef]
- Sanz-Rodriguez, M.; Gruart, A.; Escudero-Ramirez, J.; de Castro, F.; Delgado-García, J.M.; Wandosell, F.; Cubelos, B. R-Ras1 and R-Ras2 Are Essential for Oligodendrocyte Differentiation and Survival for Correct Myelination in the Central Nervous System. J. Neurosci. 2018, 38, 5096–5110. [Google Scholar] [CrossRef]
- Alcover-Sanchez, B.; Garcia-Martin, G.; Wandosell, F.; Cubelos, B. R-Ras GTPases Signaling Role in Myelin Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 5911. [Google Scholar] [CrossRef]
- Healy, L.M.; Stratton, J.A.; Kuhlmann, T.; Antel, J. The role of glial cells in multiple sclerosis disease progression. Nat. Rev. Neurol. 2022, 18, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Voet, S.; Srinivasan, S.; Lamkanfi, M.; van Loo, G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 2019, 11, e10248. [Google Scholar] [CrossRef] [PubMed]
- Rawji, K.S.; Gonzalez Martinez, G.A.; Sharma, A.; Franklin, R.J.M. The Role of Astrocytes in Remyelination. Trends Neurosci. 2020, 43, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.Y.; Liu, L.; Yang, Q.W. Functions and mechanisms of microglia/macrophages in neuroinflammation and neurogenesis after stroke. Prog. Neurobiol. 2016, 142, 23–44. [Google Scholar] [CrossRef]
- Fu, S.P.; Chen, S.Y.; Pang, Q.M.; Zhang, M.; Wu, X.C.; Wan, X.; Wan, W.H.; Ao, J.; Zhang, T. Advances in the research of the role of macrophage/microglia polarization-mediated inflammatory response in spinal cord injury. Front. Immunol. 2022, 13, 1014013. [Google Scholar] [CrossRef]
- Sun, W.; Wang, Q.; Zhang, R.; Zhang, N. Ketogenic diet attenuates neuroinflammation and induces conversion of M1 microglia to M2 in an EAE model of multiple sclerosis by regulating the NF-κB/NLRP3 pathway and inhibiting HDAC3 and P2X7R activation. Food Funct. 2023, 14, 7247–7269. [Google Scholar] [CrossRef]
- Hemmati, S.; Sadeghi, M.A.; Yousefi-Manesh, H.; Eslamiyeh, M.; Vafaei, A.; Foroutani, L.; Donyadideh, G.; Dehpour, A.; Rezaei, N. Protective Effects of Leukadherin1 in a Rat Model of Targeted Experimental Autoimmune Encephalomyelitis (EAE): Possible Role of P47phox and MDA Downregulation. J. Inflamm. Res. 2020, 13, 411–420. [Google Scholar] [CrossRef]
- Fiorini, M.; Piovani, G.; Schumacher, R.F.; Magri, C.; Bertini, V.; Mazzolari, E.; Notarangelo, L.; Notarangelo, L.D.; Barlati, S. ITGB2 mutation combined with deleted ring 21 chromosome in a child with leukocyte adhesion deficiency. J. Allergy Clin. Immunol. 2009, 124, 1356–1358. [Google Scholar] [CrossRef]
- Glawe, J.D.; Patrick, D.R.; Huang, M.; Sharp, C.D.; Barlow, S.C.; Kevil, C.G. Genetic deficiency of Itgb2 or ItgaL prevents autoimmune diabetes through distinctly different mechanisms in NOD/LtJ mice. Diabetes 2009, 58, 1292–1301. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Liu, T.; Li, Z.; Wei, W.; Zhao, Q.; Wang, S. Dynamic Transcriptomic and Cellular Remodeling Underlie Cuprizone-Induced Demyelination and Endogenous Repair in the CNS. Antioxidants 2025, 14, 692. https://doi.org/10.3390/antiox14060692
Ma Y, Liu T, Li Z, Wei W, Zhao Q, Wang S. Dynamic Transcriptomic and Cellular Remodeling Underlie Cuprizone-Induced Demyelination and Endogenous Repair in the CNS. Antioxidants. 2025; 14(6):692. https://doi.org/10.3390/antiox14060692
Chicago/Turabian StyleMa, Yantuanjin, Tianyi Liu, Zhipeng Li, Wei Wei, Qiting Zhao, and Shufen Wang. 2025. "Dynamic Transcriptomic and Cellular Remodeling Underlie Cuprizone-Induced Demyelination and Endogenous Repair in the CNS" Antioxidants 14, no. 6: 692. https://doi.org/10.3390/antiox14060692
APA StyleMa, Y., Liu, T., Li, Z., Wei, W., Zhao, Q., & Wang, S. (2025). Dynamic Transcriptomic and Cellular Remodeling Underlie Cuprizone-Induced Demyelination and Endogenous Repair in the CNS. Antioxidants, 14(6), 692. https://doi.org/10.3390/antiox14060692





