Detection of Cereibacter azotoformans-YS02 as a Novel Source of Coenzyme Q10 and Its Metabolic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Reagents, and Culture Media
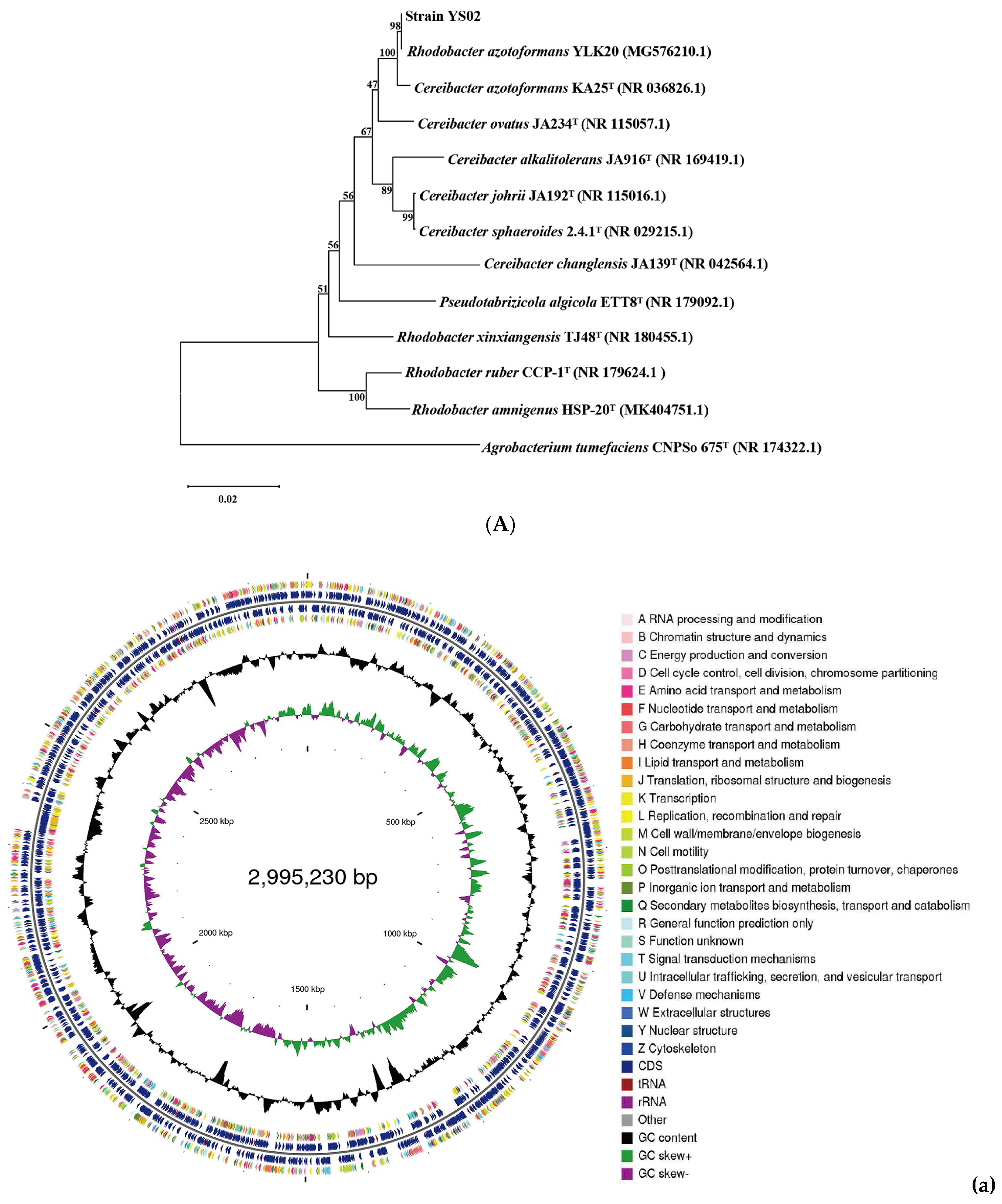
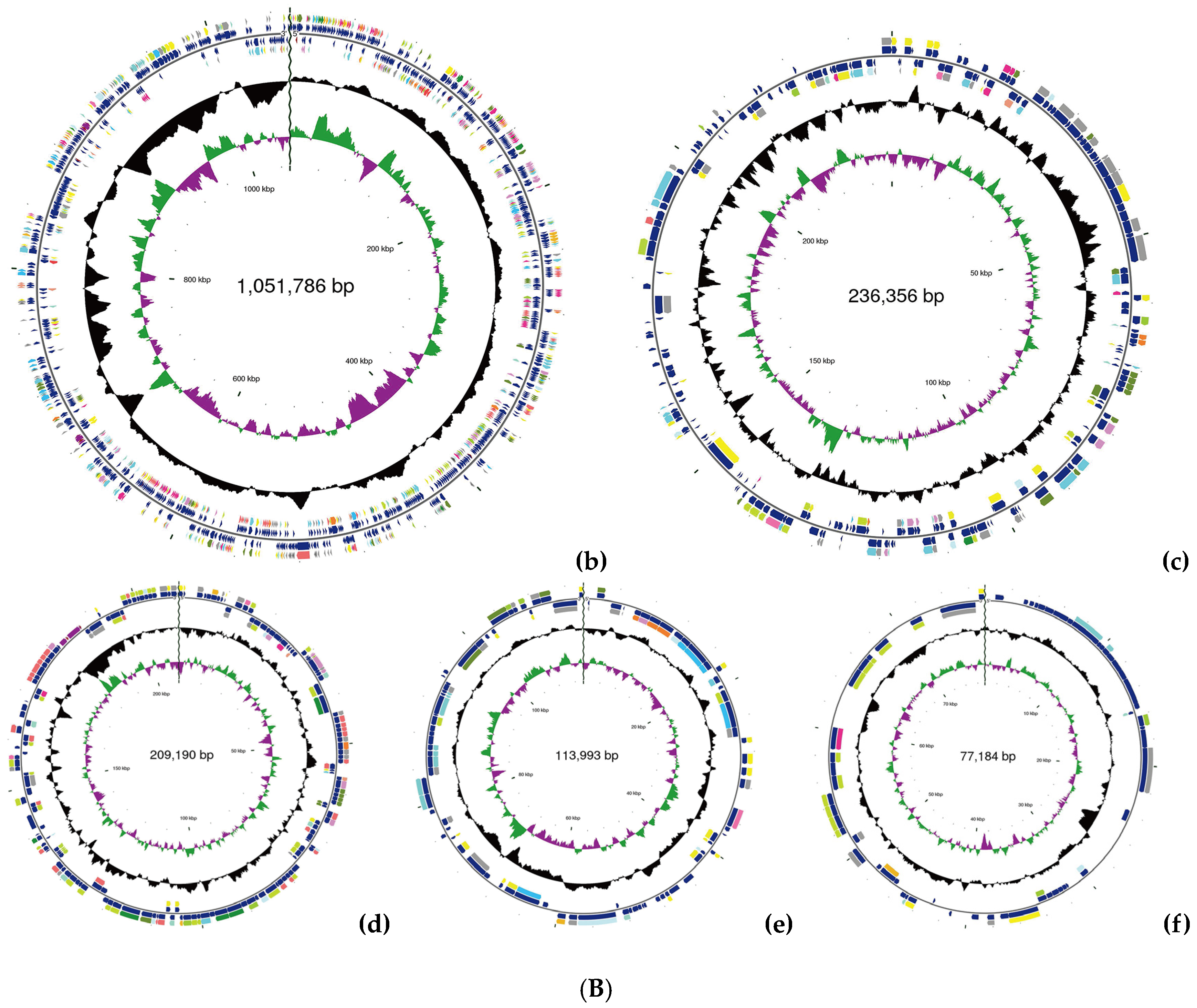
2.2. WGS Analysis of C. azotoformans-YS02
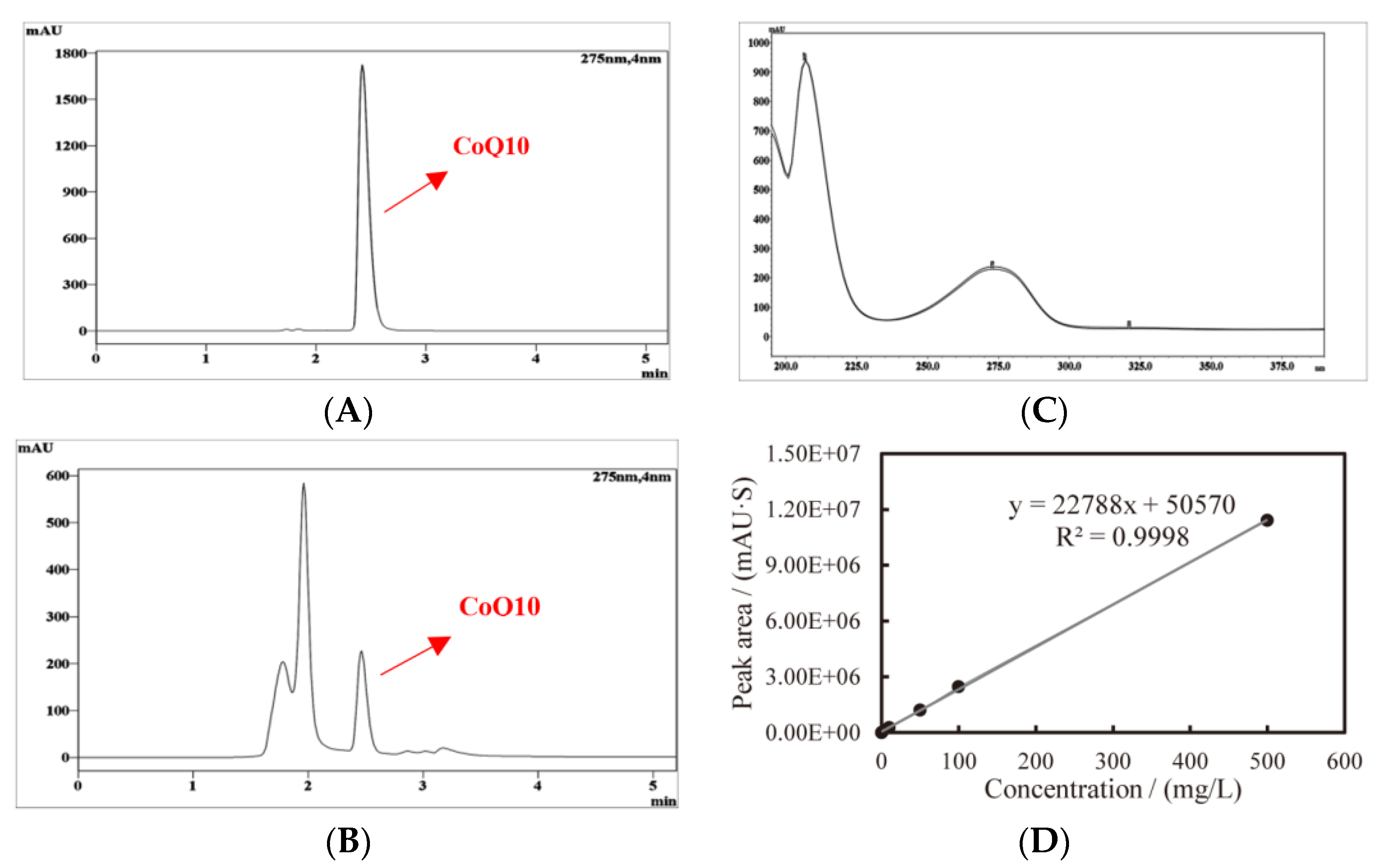
2.3. Extraction and Quantification of CoQ10
2.4. Metabolomics Analysis by UPLC-Q-Exactive Orbitrap MS
2.5. Statistical Analysis of Data
3. Results and Discussion
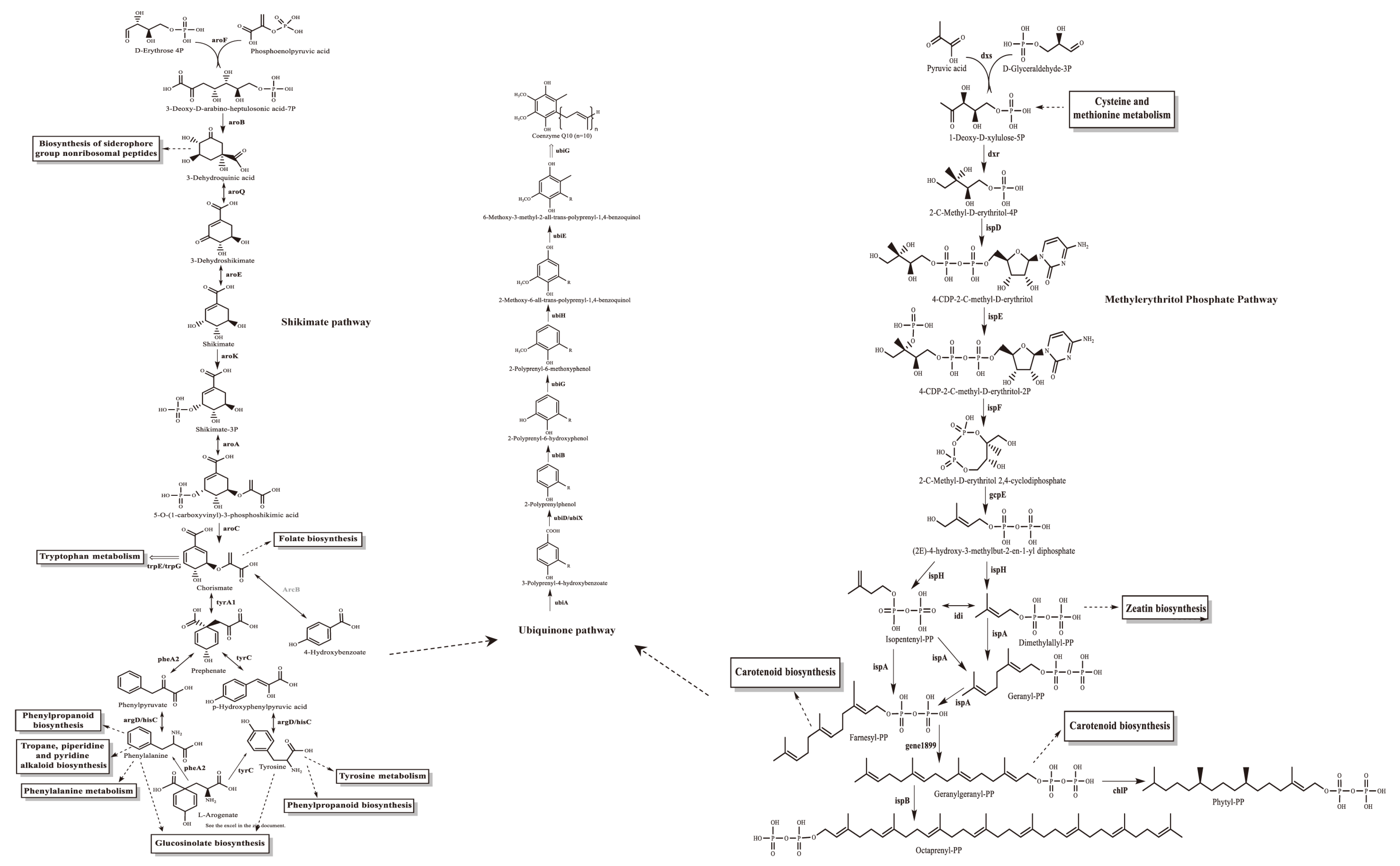
3.1. Overview of C. azotoformans-YS02 Genome and Molecular Mechanism for CoQ10 Production
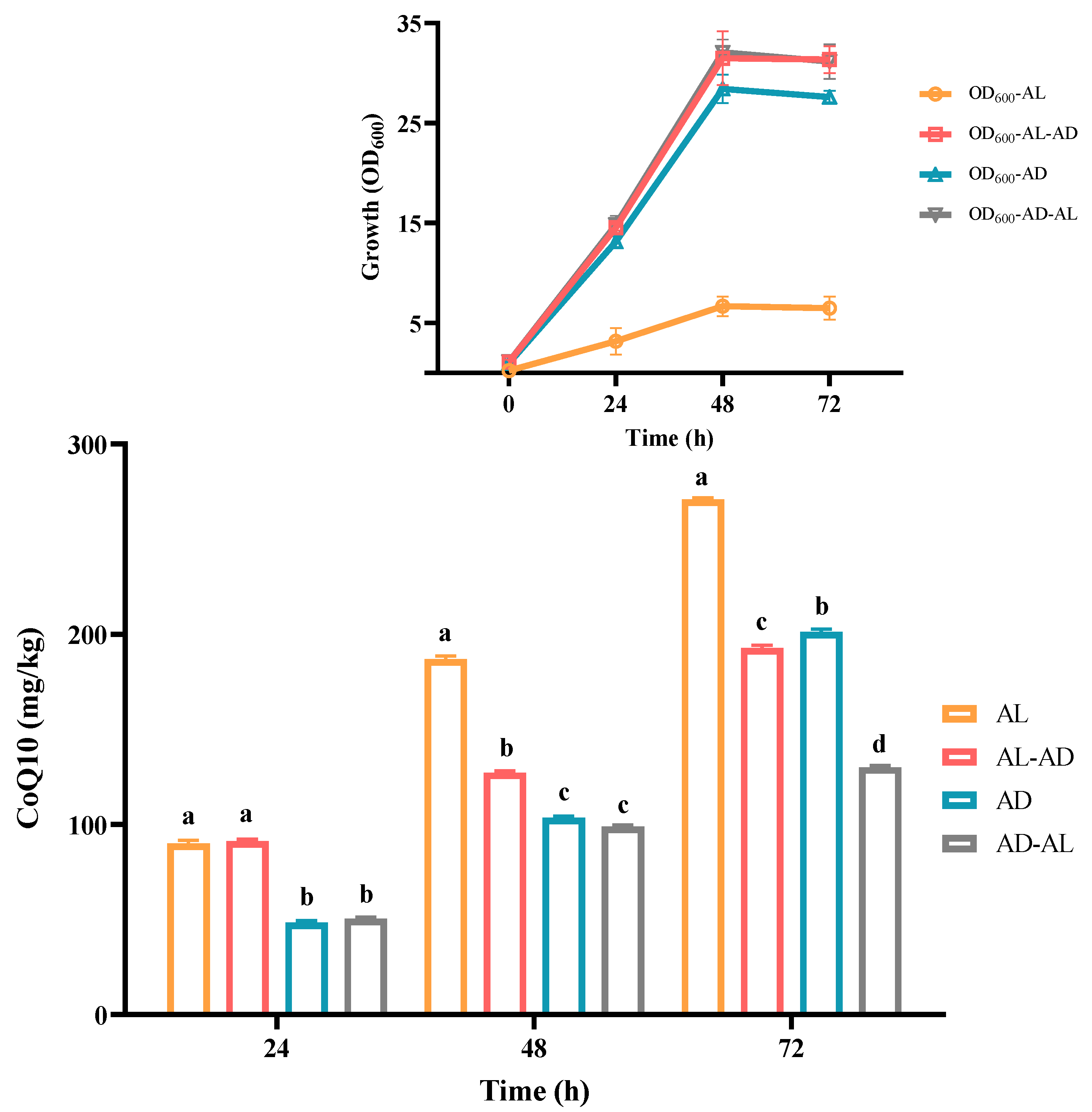
3.2. Determination of CoQ10 in C. azotoformans-YS02 Under Four Different Culture Modes
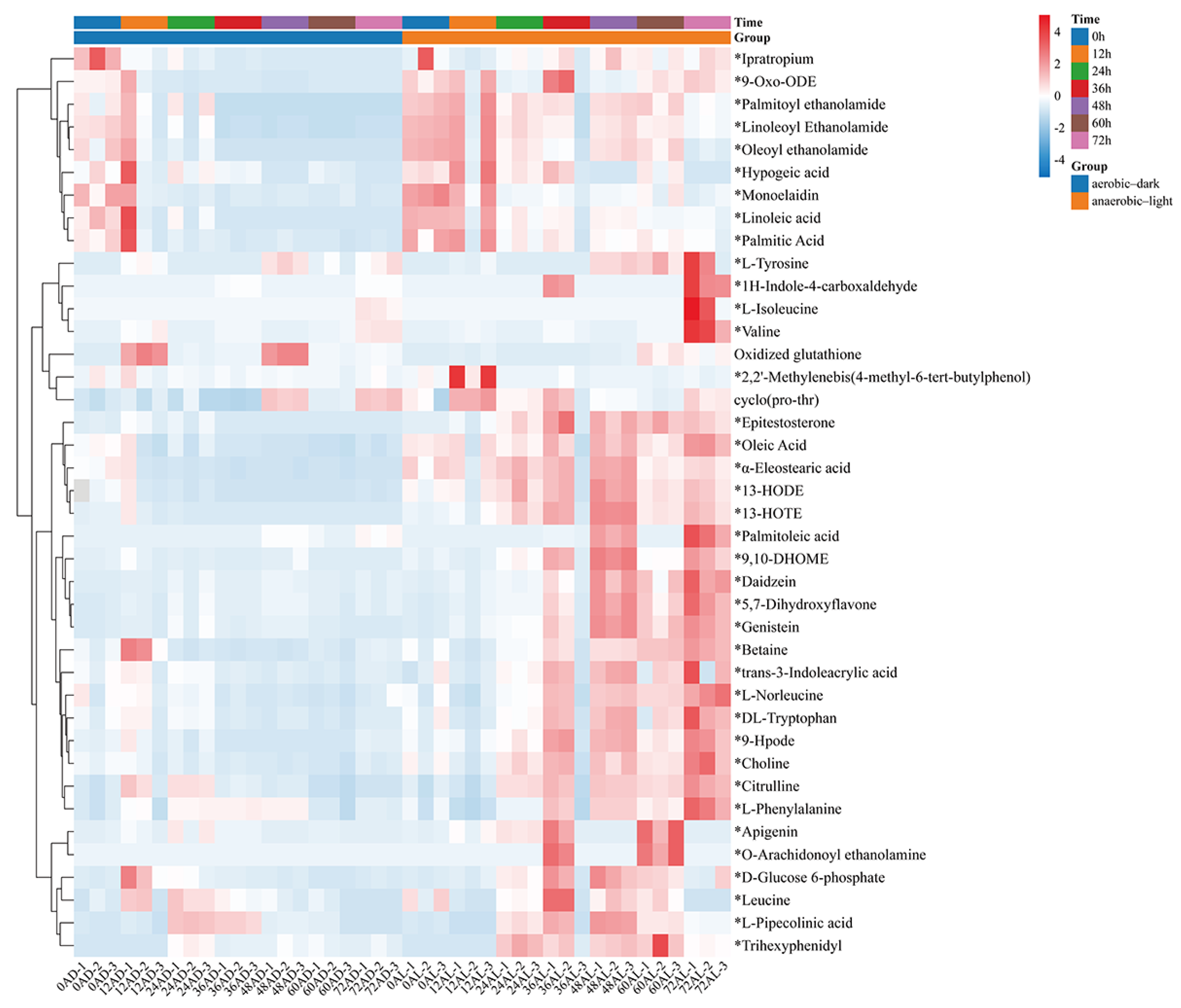
3.3. Analysis of Differential Metabolites in C. azotoformans-YS02 Under Different Culture Conditions
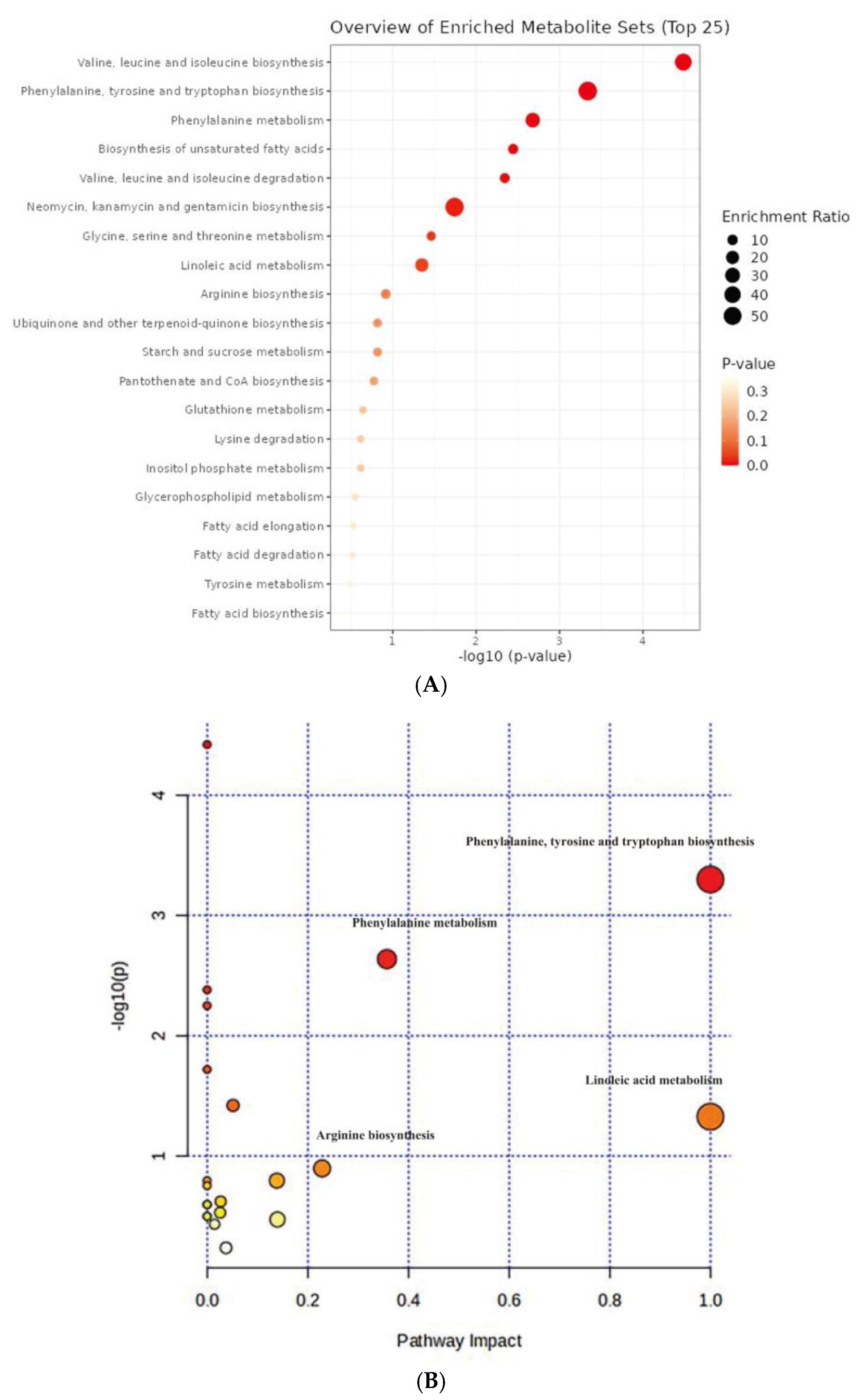
3.4. Enrichment Analysis and Pathway Analysis Based on Differential Metabolites in C. azotoformans-YS02
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hiraishi, A.; Muramatsu, K.; Urata, K. Characterization of new denitrifying Rhodobacter strains isolated from photosynthetic sludge for wastewater treatment. J. Ferment. Bioeng. 1995, 79, 39–44. [Google Scholar] [CrossRef]
- Chung Han Chua, E.; Wee, S.K.; Kansedo, J.; Lau, S.Y.; Lim, K.H.; Dol, S.S.; Lipton, A.N. Biological Hydrogen Energy Production by Novel Strains Bacillus paramycoides and Cereibacter azotoformans through Dark and Photo Fermentation. Energies 2023, 16, 9807. [Google Scholar] [CrossRef]
- Barcelo, L.A.F.; Lantican, N.B.; Ventura, R.L.G.; Ventura, J.-R.S. Genetic characterization of biohydrogen-producing purple non-sulfur bacteria Rhodobacter johrii MAY2 isolate via whole genome analysis. Gene 2024, 924, 148597. [Google Scholar] [CrossRef]
- Wang, H.; Sha, X.; Li, R.; Li, Y.; Khaleque, H.N.; Zhang, Y.; Bohu, T.; Bai, Z.; Zhuang, X. Comparative Genome Analysis Provides Molecular Evidence for Reclassification of the Photosynthetic Bacterium Rhodobacter sphaeroides EBL0706 as a Strain of Luteovulum azotoformans. Microorganisms 2021, 9, 1754. [Google Scholar] [CrossRef]
- Mehić, S.; Saltikov, C. Genome sequence and characterisation of a freshwater photoarsenotroph, Cereibacter azotoformans strain ORIO, isolated from sediments capable of cyclic light–dark arsenic oxidation and reduction. Environ. Microbiol. 2023, 25, 3738–3752. [Google Scholar] [CrossRef]
- Morrison, H.M.; Bose, A. Purple non-sulfur bacteria for biotechnological applications. J. Ind. Microbiol. Biotechnol. 2025, 52, kuae052. [Google Scholar] [CrossRef]
- Qi, Z.; Gao, A.; Li, L.; Li, Z.; Zhang, W.; Dong, S.; Liu, X. A novel strategy to improving Rhodobacter azotoformans denitrification efficiency: Insight into the role of a two-component system NtrX/Y in denitrification regulation. Bioresour. Technol. 2023, 368, 128439. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, S.; An, J.; Diao, J.; Xu, L.; Gai, C. Rhodobacter azotoformans supplementation improves defense ability of Chinese mitten crab Eriocheir sinensis against citrobacteriosis. Fish Shellfish. Immunol. 2022, 131, 991–998. [Google Scholar] [CrossRef]
- Li, Z.; Li, L.; Sun, H.; Wang, W.; Yang, Y.; Qi, Z.; Liu, X. Ammonia assimilation: A double-edged sword influencing denitrification of Rhodobacter azotoformans and for nitrogen removal of aquaculture wastewater. Bioresour. Technol. 2022, 345, 126495. [Google Scholar] [CrossRef]
- Murakami, K.; Kamimura, D.; Hasebe, R.; Uchida, M.; Abe, N.; Yamamoto, R.; Jiang, J.-J.; Hidaka, Y.; Nakanishi, Y.; Fujita, S.; et al. Rhodobacter azotoformans LPS (RAP99-LPS) Is a TLR4 Agonist That Inhibits Lung Metastasis and Enhances TLR3-Mediated Chemokine Expression. Front. Immunol. 2021, 12, 675909. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, L.; Yin, L.; Xie, S.; Xiao, M. Carotenogenesis gene cluster and phytoene desaturase catalyzing both three- and four-step desaturations from Rhodobacter azotoformans. FEMS Microbiol. Lett. 2012, 333, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Kanie, Y.; Yamaguchi, Y.; Hayashi, A.; Uzawa, J.; Hatakeyama, M.; Hidaka, Y.; Toda, N.; Nakamura, S.; Kanie, O. Structural analysis of a novel lipooligosaccharide (LOS) from Rhodobacter azotoformans. Carbohydr. Res. 2019, 473, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Raizner, A.E.; Quiñones, M.A. Coenzyme Q10 for Patients with Cardiovascular Disease. J. Am. Coll. Cardiol. 2021, 77, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xia, M.; Zheng, C.; Yang, Y.; Bao, J.; Dai, W.; Mei, X. The Cocrystal of Ubiquinol: Improved Stability and Bioavailability. Pharmaceutics 2023, 15, 2499. [Google Scholar] [CrossRef]
- Vetvicka, V.; Vetvickova, J. Combination Therapy with Glucan and Coenzyme Q10 in Murine Experimental Autoimmune Disease and Cancer. Anticancer Res. 2018, 38, 3291–3297. [Google Scholar] [CrossRef]
- Parmar, S.S.; Jaiwal, A.; Dhankher, O.P.; Jaiwal, P.K. Coenzyme Q10 production in plants: Current status and future prospects. Crit. Rev. Biotechnol. 2013, 35, 152–164. [Google Scholar] [CrossRef]
- Gasmi, A.; Bjørklund, G.; Mujawdiya, P.K.; Semenova, Y.; Piscopo, S.; Peana, M. Coenzyme Q10 in aging and disease. Crit. Rev. Food Sci. Nutr. 2022, 64, 3907–3919. [Google Scholar] [CrossRef]
- Testai, L.; Martelli, A.; Flori, L.; Cicero, A.; Colletti, A. Coenzyme Q10: Clinical Applications beyond Cardiovascular Diseases. Nutrients 2021, 13, 1697. [Google Scholar] [CrossRef]
- Matamoros, J.A.; Rubio-Casado, S.; Fernández-Albarral, J.A.; Martínez-López, M.A.; Salobrar-García, E.; Marco, E.M.; Paleo-García, V.; de Hoz, R.; López-Cuenca, I.; Elvira-Hurtado, L.; et al. Neuroprotective Effect of the Combination of Citicoline and CoQ10 in a Mouse Model of Ocular Hypertension. Antioxidants 2024, 14, 4. [Google Scholar] [CrossRef]
- Arenas-Jal, M.; Suñé-Negre, J.M.; García-Montoya, E. Coenzyme Q10 supplementation: Efficacy, safety, and formulation challenges. Compr. Rev. Food Sci. Food Saf. 2020, 19, 574–594. [Google Scholar] [CrossRef]
- Lee, S.Q.E.; Tan, T.S.; Kawamukai, M.; Chen, E.S. Cellular factories for coenzyme Q10 production. Microb. Cell Factories 2017, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Bank, G.; Kagan, D.; Madhavi, D. Coenzyme Q10: Clinical Update and Bioavailability. J. Evid. Based Complement. Altern. Med. 2011, 16, 129–137. [Google Scholar] [CrossRef]
- Xiao, Y.; Zheng, Y.; Zhou, Y.; Yu, C.; Ye, T.-E. Metabolic flux analysis of coenzyme Q10 synthesized by Rhodobacter sphaeroides under the influence of different pH regulators. Microb. Cell Fact. 2023, 22, 206. [Google Scholar] [CrossRef]
- Yen, H.-W.; Chiu, C.-H. The influences of aerobic-dark and anaerobic-light cultivation on CoQ10 production by Rhodobacter sphaeroides in the submerged fermenter. Enzym. Microb. Technol. 2007, 41, 600–604. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, L.; Wang, K.-F.; Xu, L.; Zhou, L.; Wang, W.; Li, C.; Xu, Z.; Shi, T.; Chen, H.; et al. Phosphate limitation increases coenzyme Q10 production in industrial Rhodobacter sphaeroides HY01. Synth. Syst. Biotechnol. 2019, 4, 212–219. [Google Scholar] [CrossRef]
- Hiraishi, A.; Muramatsu, K.; Ueda, Y. Molecular genetic analyses of Rhodobacter azotoformans sp. nov. and related species of phototrophic bacteria. Syst. Appl. Microbiol. 1996, 19, 168–177. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, M.; Chen, P.; Harnly, J.M.; Sun, J. Mass Spectrometry-Based Nontargeted and Targeted Analytical Approaches in Fingerprinting and Metabolomics of Food and Agricultural Research. J. Agric. Food Chem. 2022, 70, 11138–11153. [Google Scholar] [CrossRef]
- Chen, W.-C.; Liu, T.-H.; Wang, L.-F.; Chien, C.-C.; Chen, S.-Y.; Wei, Y.-H. Enhanced production and characterization of coenzyme Q10 from Rhodobacter sphaeroides using a potential fermentation strategy. J. Taiwan Inst. Chem. Eng. 2022, 137, 104201. [Google Scholar] [CrossRef]
- He, S.; Lu, H.; Zhang, G.; Ren, Z. Production of coenzyme Q10 by purple non-sulfur bacteria: Current development and future prospect. J. Clean. Prod. 2021, 307, 127326. [Google Scholar] [CrossRef]
- Woronowicz, K.; Duncan, K.J.; Nagelli, E.A.; Burpo, F.J. Chapter 13—Energy harvesting, charge, and mass transport considerations for reaction center-nanomaterial composites. In Photosynthesis; Hou, H.J.M., Allakhverdiev, S.I., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 293–332. [Google Scholar]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Pierrel, F.; Burgardt, A.; Lee, J.-H.; Pelosi, L.; Wendisch, V.F. Recent advances in the metabolic pathways and microbial production of coenzyme Q. World J. Microbiol. Biotechnol. 2022, 38, 58. [Google Scholar] [CrossRef] [PubMed]
- Igor, P.; Katja, Z.; Janko, Z. Coenzyme Q10 contents in foods and fortification strategies. Crit. Rev. Food Sci. Nutr. 2010, 50, 269–280. [Google Scholar]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-H.; Ryu, Y.-W.; Park, Y.-C.; Seo, J.-H. Synergistic effects of chromosomal ispB deletion and dxs overexpression on coenzyme Q10 production in recombinant Escherichia coli expressing Agrobacterium tumefaciens dps gene. J. Biotechnol. 2009, 144, 64–69. [Google Scholar] [CrossRef]
- Shukla, S.; Dubey, K.K. CoQ10 a super-vitamin: Review on application and biosynthesis. 3 Biotech 2018, 8, 249. [Google Scholar] [CrossRef]
- White, M.D.; Payne, K.A.P.; Fisher, K.; Marshall, S.A.; Parker, D.; Rattray, N.J.W.; Trivedi, D.K.; Goodacre, R.; Rigby, S.E.J.; Scrutton, N.S.; et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 2015, 522, 502–506. [Google Scholar] [CrossRef]
- Burgardt, A.; Moustafa, A.; Persicke, M.; Sproß, J.; Patschkowski, T.; Risse, J.M.; Peters-Wendisch, P.; Lee, J.-H.; Wendisch, V.F. Coenzyme Q10 Biosynthesis Established in the Non-Ubiquinone Containing Corynebacterium glutamicum by Metabolic Engineering. Front. Bioeng. Biotechnol. 2021, 9, 650961. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, J.Y.; Wu, J.; Wang, J.; Poplawsky, A.; Lin, S.; Zhu, B.; Chang, C.; Zhou, T.; Zhang, L.H.; et al. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol. Microbiol. 2013, 87, 80–93. [Google Scholar] [CrossRef]
- Schöner, T.A.; Fuchs, S.W.; Reinhold-Hurek, B.; Bode, H.B. Identification and biosynthesis of a novel xanthomonadin-dialkylresorcinol-hybrid from Azoarcus sp. BH72. PLoS ONE 2017, 9, e90922. [Google Scholar] [CrossRef]
- Simeonidis, E.; Price, N.D. Genome-scale modeling for metabolic engineering. J. Ind. Microbiol. Biotechnol. 2015, 42, 327–338. [Google Scholar] [CrossRef]

| Parameters | Values |
|---|---|
| Clean bases | 1,217,884,848 bp |
| Clean Q30 | 94.98% |
| largest reads length | 24,752 bp |
| Reads N50 | 9417 bp |
| Chromosome | 1 |
| Plasmid | 5 |
| Genome Size | 4,683,739 bp |
| G + C | 68.02% |
| Coding gene | 4432 |
| Gene total length | 4,071,321 bp |
| Gene average length | 918.62 bp |
| Gene/Genome | 86.92% |
| tRNA | 55 |
| rRNA | 12 |
| sRNA | 33 |
| Repeat DNAs | 40 |
| SINEs a | 16 |
| LINEs b | 8 |
| Genes of KEGG | 3252 |
| Genes of COG | 3571 |
| Gene ID | Gene Name | Location | Length (bp) | NR Description | Swiss-Prot Description | Pfam ID | COG ID | GO ID | KO ID | KO Description |
|---|---|---|---|---|---|---|---|---|---|---|
| gene1887 | dxs | Chromosome (1,871,388–1,869,442) | 1947 | MULTISPECIES: 1-deoxy-D-xylulose-5-phosphate synthase | 1-deoxy-D-xylulose-5-phosphate synthase 1 OS = Cereibacter sphaeroides (strain ATCC 17023/DSM 158/JCM 6121/CCUG 31486/LMG 2827/NBRC 12203/NCIMB 8253/ATH 2.4.1.) OX = 272,943 GN = dxs1 PE = 3 SV = 1 | PF13292; PF02779; PF02780 | COG1154 | GO: 0016114; GO: 0009228;GO: 0052865; GO: 0000287; GO: 0030976;GO: 0008661 | K01662 | 1-deoxy-D-xylulose-5-phosphate synthase [EC: 2.2.1.7] |
| gene1977 | dxr | Chromosome (1,960,897–1,959,713) | 1185 | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 1-deoxy-D-xylulose 5-phosphate reductoisomerase OS = Cereibacter sphaeroides (strain ATCC 17025/ATH 2.4.3) OX = 349,102 GN = dxr PE = 3 SV = 1 | PF08436; PF02670; PF13288 | COG0743 | - | K00099 | 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC: 1.1.1.267] |
| gene2789 | dxs | Chromosome (2,788,800–2,786,887) | 1914 | MULTISPECIES: 1-deoxy-D-xylulose-5-phosphate synthase | 1-deoxy-D-xylulose-5-phosphate synthase 2 OS = Cereibacter sphaeroides (strain ATCC 17023/DSM 158/JCM 6121/CCUG 31486/LMG 2827/NBRC 12203/NCIMB 8253/ATH 2.4.1.) OX = 272,943 GN = dxs2 PE = 3 SV = 1 | PF13292; PF02779; PF02780 | COG1154 | GO: 0016114; GO: 0009228; GO: 0052865; GO: 0000287; GO: 0030976; GO: 0008661 | K01662 | 1-deoxy-D-xylulose-5-phosphate synthase [EC: 2.2.1.7] |
| gene0908 | idi | Chromosome (915,412–915,945) | 534 | isopentenyl-diphosphate Delta-isomerase | Isopentenyl-diphosphate Delta-isomerase OS = Cereibacter sphaeroides (strain ATCC 17025/ATH 2.4.3) OX = 349,102 GN = idi PE = 3 SV = 2 | PF00293 | COG1443 | - | K01823 | isopentenyl-diphosphate Delta-isomerase [EC:5.3.3.2] |
| gene2309 | ispB | Chromosome (2,300,848–2,301,849) | 1002 | MULTISPECIES: polyprenyl synthetase family protein | Octaprenyl diphosphate synthase OS = Shigella flexneri OX = 623 GN = ispB PE = 3 SV = 1 | PF00348 | COG0142 | GO: 0008299; GO: 0016740 | K02523 | octaprenyl-diphosphate synthase [EC: 2.5.1.90] |
| gene0203 | ubiA | Chromosome (198,354–199,346) | 993 | 4-hydroxybenzoate octaprenyltransferase | 4-hydroxybenzoate polyprenyltransferase, mitochondrial OS = Dictyostelium discoideum OX = 44,689 GN = coq2 PE = 3 SV = 1 | PF01040 | COG0382 | - | K03179 | 4-hydroxybenzoate polyprenyltransferase [EC: 2.5.1.39] |
| gene2225 | ubiD | Chromosome (2,217,921–2,219,414) | 1494 | MULTISPECIES: UbiD family decarboxylase | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase OS = Pseudomonas syringae pv. syringae (strain B728a) OX = 205,918 GN = ubiD PE = 3 SV = 2 | PF01977; PF20696; PF20695 | COG0043 | - | K03182 | 4-hydroxy-3-polyprenylbenzoate decarboxylase [EC: 4.1.1.98] |
| gene2993 | ubiE | Chromosome (2,992,547–2,991,795) | 753 | MULTISPECIES: bifunctional demethylmenaquinone methyltransferase/2-methoxy-6-polyprenyl-1,4-benzoquinol methylase UbiE | Ubiquinone/menaquinone biosynthesis C-methyltransferase UbiE OS = Cereibacter sphaeroides (strain ATCC 17025/ATH 2.4.3) OX = 349,102 GN = ubiE PE = 3 SV = 1 | PF01209; PF13649; PF08241; PF13847; PF08242; PF13489; PF05175 | COG2226 | GO: 0032259; GO: 0006744; GO: 0009060; GO: 0009234; GO: 0043770; GO: 0102955; GO: 0043333; GO: 0102027; GO: 0102094 | K03183 | demethylmenaquinone methyltransferase/2-methoxy-6-polyprenyl-1,4-benzoquinol methylase [EC: 2.1.1.163 2.1.1.201] |
| gene2814 | ubiG | Chromosome (2,811,344–2,810,601) | 744 | MULTISPECIES: bifunctional 2-polyprenyl-6-hydroxyphenol methylase/3-demethylubiquinol 3-O-methyltransferase UbiG | Ubiquinone biosynthesis O-methyltransferase OS = Cereibacter sphaeroides (strain ATCC 17025/ATH 2.4.3) OX = 349,102 GN = ubiG PE = 3 SV = 1 | PF13489; PF08241; PF13649; PF13847; PF08242; PF02353; PF08003; PF06325; PF01209 | COG2227 | GO: 0032259; GO: 0006744; GO: 0005737; GO: 0102208; GO: 0008425; GO: 0008689 | K00568 | 2-polyprenyl-6-hydroxyphenyl methylase/3-demethylubiquinone-9 3-methyltransferase [EC: 2.1.1.222 2.1.1.64] |
| gene0627 | ubiH | Chromosome (626,189–624,990) | 1200 | MULTISPECIES: UbiH/UbiF family hydroxylase | Ubiquinone hydroxylase UbiL OS = Rhodospirillum rubrum (strain ATCC 11170/ATH 1.1.1/DSM 467/LMG 4362/NCIMB 8255/S1) OX = 269,796 GN = ubiL PE = 1 SV = 1 | PF01494; PF00890 | COG0654 | GO: 0006744; GO: 0071949; GO: 0016705 | K03185 | 2-octaprenyl-6-methoxyphenol hydroxylase [EC: 1.14.13.-] |
| gene2678 | ubiH | Chromosome (2,676,978–2,678,204) | 1227 | UbiH/UbiF/VisC/COQ6 family ubiquinone biosynthesis hydroxylase | Ubiquinone hydroxylase UbiL OS = Rhodospirillum rubrum (strain ATCC 11170/ATH 1.1.1/DSM 467/LMG 4362/NCIMB 8255/S1) OX = 269,796 GN = ubiL PE = 1 SV = 1 | PF01494 | COG0654 | - | K03185 | 2-octaprenyl-6-methoxyphenol hydroxylase [EC: 1.14.13.-] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Xu, Q.; Raka, R.N.; Yin, C.; Liu, X.; Yan, H. Detection of Cereibacter azotoformans-YS02 as a Novel Source of Coenzyme Q10 and Its Metabolic Analysis. Antioxidants 2025, 14, 429. https://doi.org/10.3390/antiox14040429
Song M, Xu Q, Raka RN, Yin C, Liu X, Yan H. Detection of Cereibacter azotoformans-YS02 as a Novel Source of Coenzyme Q10 and Its Metabolic Analysis. Antioxidants. 2025; 14(4):429. https://doi.org/10.3390/antiox14040429
Chicago/Turabian StyleSong, Meijie, Qianqian Xu, Rifat Nowshin Raka, Chunhua Yin, Xiaolu Liu, and Hai Yan. 2025. "Detection of Cereibacter azotoformans-YS02 as a Novel Source of Coenzyme Q10 and Its Metabolic Analysis" Antioxidants 14, no. 4: 429. https://doi.org/10.3390/antiox14040429
APA StyleSong, M., Xu, Q., Raka, R. N., Yin, C., Liu, X., & Yan, H. (2025). Detection of Cereibacter azotoformans-YS02 as a Novel Source of Coenzyme Q10 and Its Metabolic Analysis. Antioxidants, 14(4), 429. https://doi.org/10.3390/antiox14040429








