Roles of ROS and NO in Plant Responses to Individual and Combined Salt Stress and Waterlogging
Abstract
1. Introduction
2. Mechanisms Underlying Responses of Plants to Salt Stress Associated with ROS-Regulatory Systems and NO Signaling in Plants
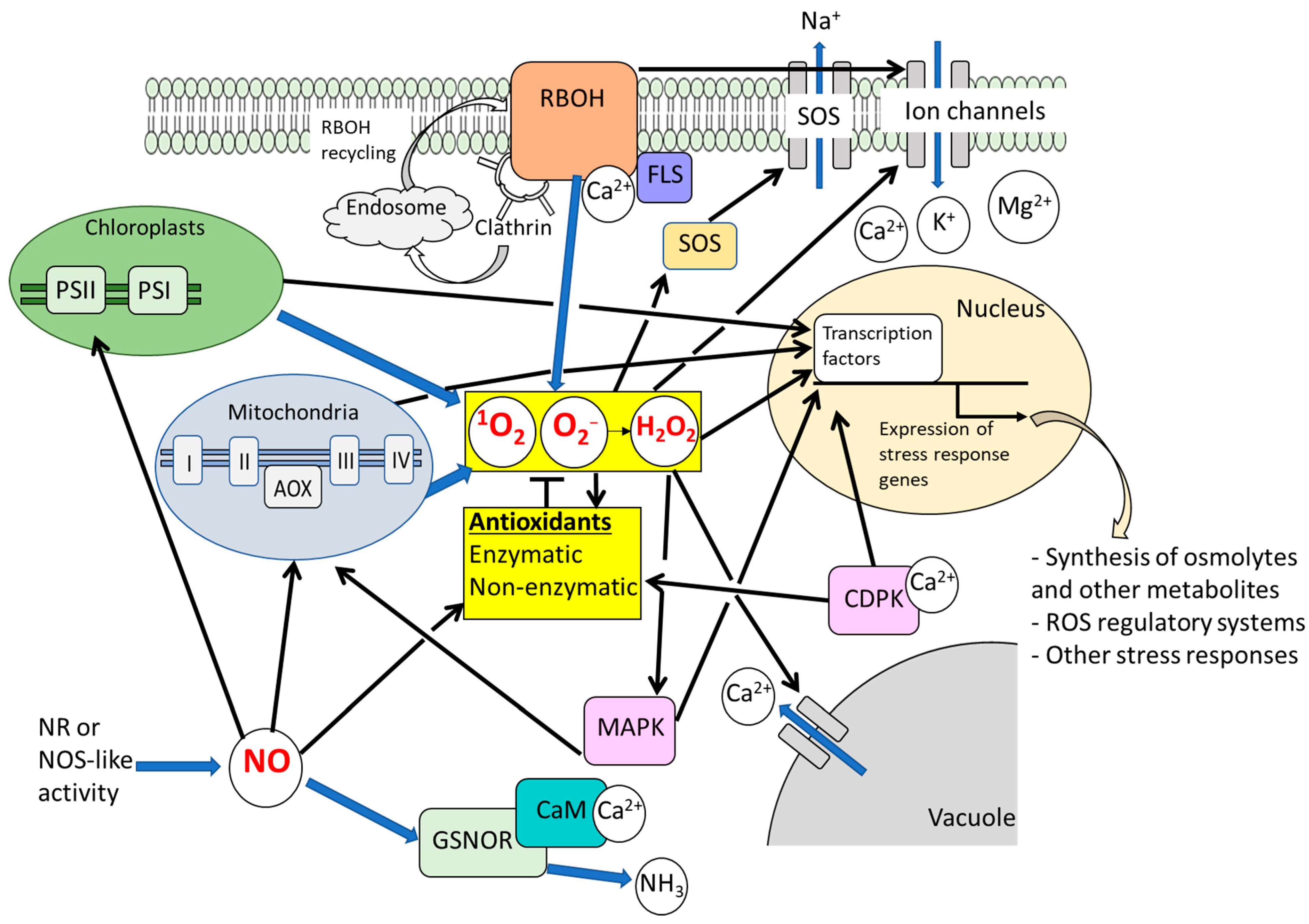
2.1. Significance of ROS-Regulatory Systems and ROS Signaling Under Salt Stress
2.2. Significance of NO Signaling Under Salt Stress
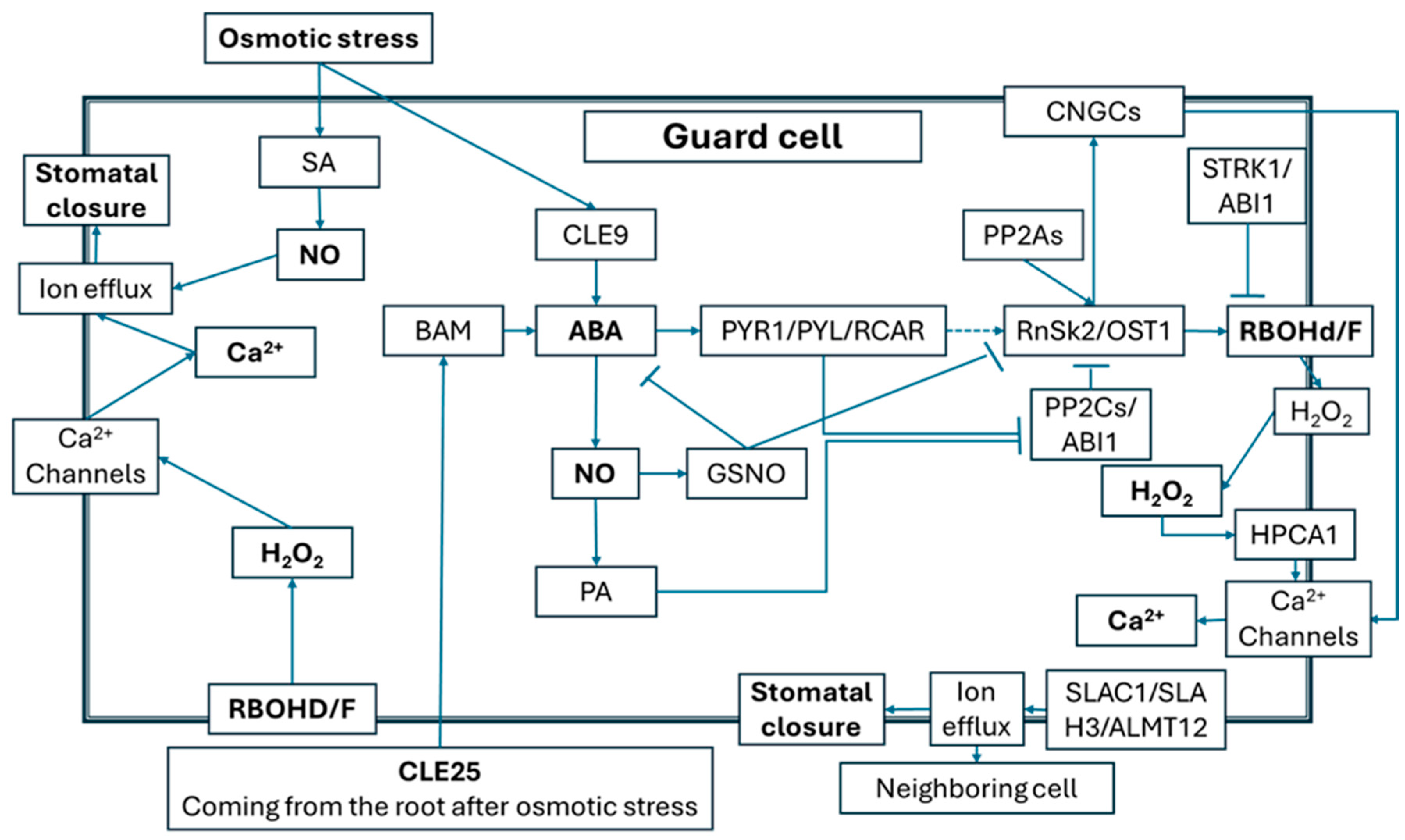
2.3. Roles of ROS and NO Signals in Alleviation of Osmotic Effects Caused by Salt Stress
2.4. ROS/NO-Related Signaling Events Mediating Plant Responses to Salt Stress-Induced Ionic Toxicity
3. Mechanisms Underlying Responses of Plants to Waterlogging Associated with ROS-Regulatory Systems and NO Signaling
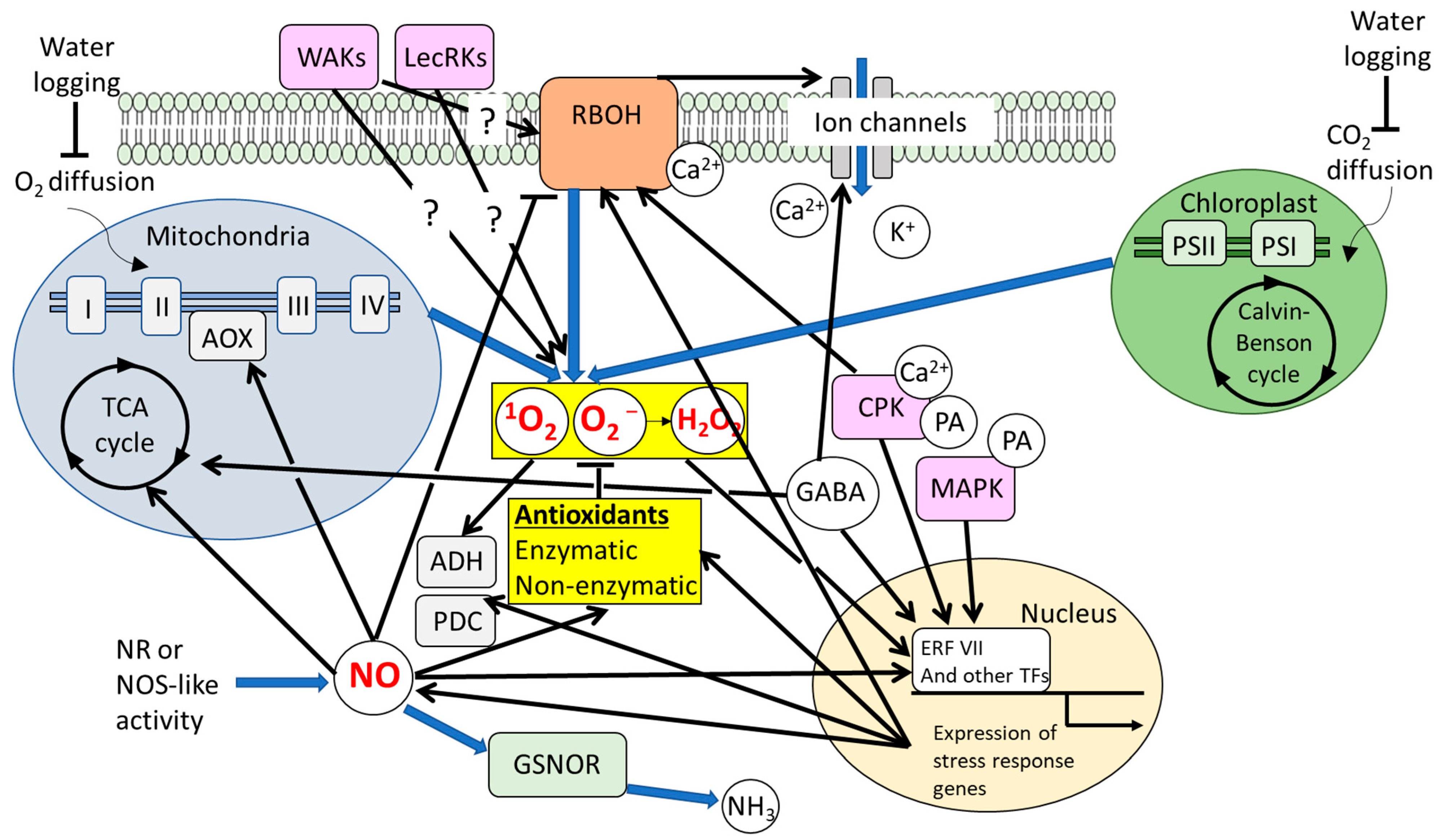
3.1. Significance of ROS-Regulatory Systems and ROS Signaling Under Waterlogging
3.2. Significance of NO Signaling Under Waterlogging
3.3. ROS and NO-Dependent Signals Involved in Formation of Aerenchyma, Adventitious Roots, and ROL Barrier
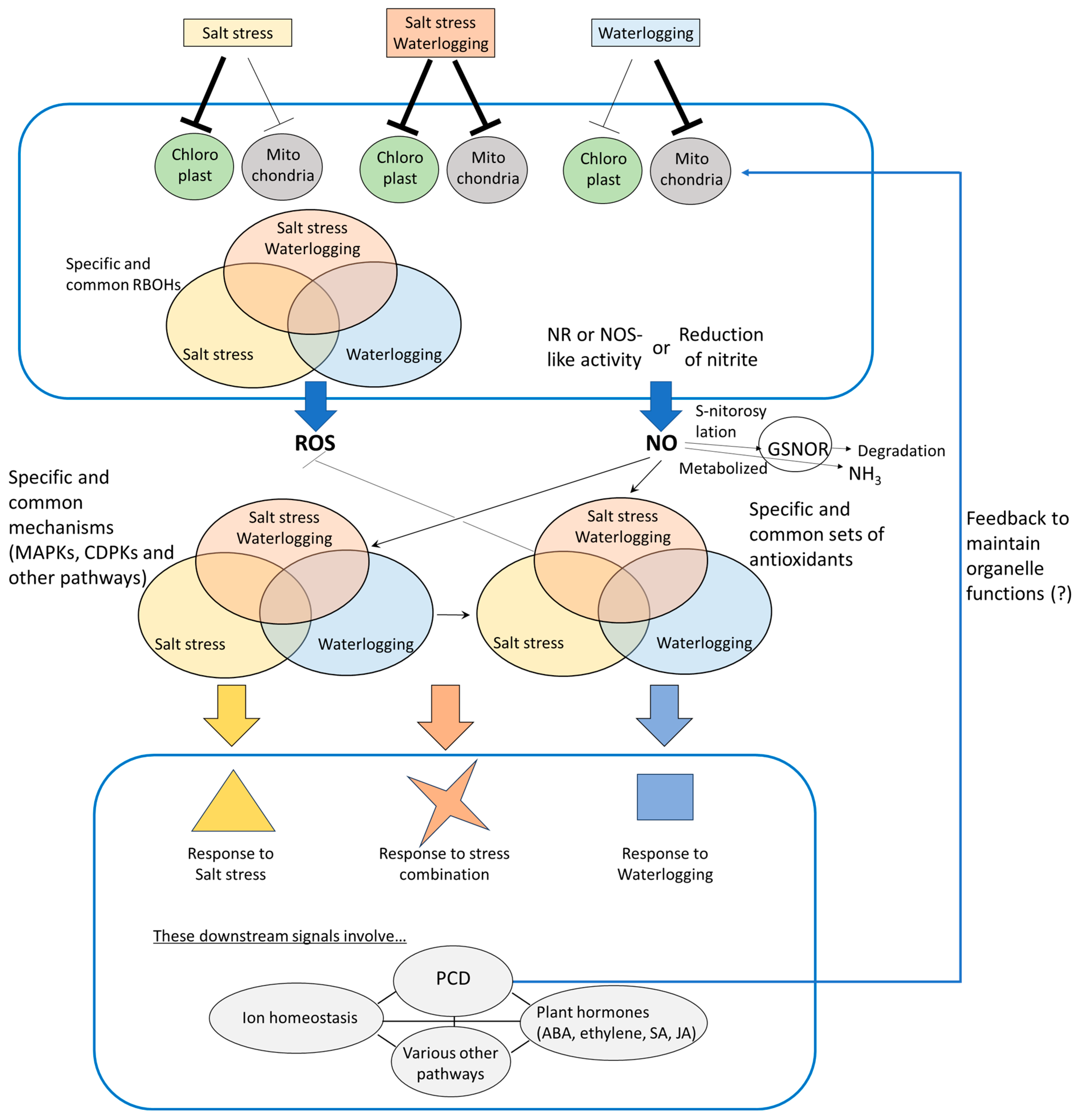
4. Signaling Pathways Involved in ROS- and NO-Mediated Plant Responses to Combined Salinity and Waterlogging Stresses
4.1. Sources of ROS and NO Under Combined Salt and Waterlogging Stress
4.2. ROS- and NO-Related Signals Underlying Responses of Plants to Salt Stress and Waterlogging Applied Individually or in Combination
| Plant Species | Stress Type & Duration | Oxidative Responses | Antioxidative/Other Responses | Reference |
|---|---|---|---|---|
| Cicer arietinum L. | NaCl 100 mM; 45 d | MDA, H2O2 ↑ | SOD, CAT, APX, GR ↑ | [6] |
| Lycopersicum esculentum L. cv. Micro-tom | NaCl 50 mM + waterlogging; 14 d | MDA, H2O2 ↑ | CAT, GPX, GST ↓ APX, MDHAR, DHAR, GR ↓ | [58] |
| Zea mays | NaCl 10 dSm−1 + waterlogging; 7 d | H2O2 ↑ | SOD, CAT, APX ↑ | [59] |
| Suaeda glauca | NaCl 400 mM; 10 d | MDA, O2•−, H2O2 ↑ | SOD, APX ↓ | [60] |
| Elaeagnus angustifolia | NaCl 0.6% + waterlogging; 14 d | MDA, H2O2 ↑ | SOD, APX, POD, GR ↑ | [62] |
| Cucumis sativus | Waterlogging; 5 d | MDA, H2O2, NO ↑ | NR activity ↑ RBOH9, NRT1.8, REP2.3, HEM3 ↑ | [183] |
| Arabidopsis thaliana L. | Anoxia; 4 h | H2O2, NO ↑ | ASC ↓ DHA↑ GR, POD, CAT, APX ↓ | [202] |
| Momordica charantia | NaCl 25 mM; 7 d | MDA, O2•−, H2O2, NO ↑ | CAT, APX, GR, GST ↑ | [209] |
| Cajanus cajan L. Millsp. | NaCl 30 mM + waterlogging; 12 d | Membrane injury ↑ Lipid peroxidation ↑ | Proline ↑ | [214] |
| Mentha aquatica L. | NaCl 150 mM + waterlogging; 30 d | MDA, H2O2 ↑ | SOD, CAT, APX ↑ | [215] |
| Triticum aestivum | NaCl 195 mM + waterlogging; 5 d | MDA, O2•−, H2O2 ↑ | SOD, CAT, APX ↓ | [216] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | ascorbic acid |
| ABA | abscisic acid |
| ADH | alcohol dehydrogenase |
| AOX | ascorbate oxidase |
| APX | ascorbate peroxidase |
| ASA | ascorbate |
| BADH | betaine aldehyde dehydrogenase |
| BAM | barely any meristem |
| CAT | catalase |
| CDLC | calcium-dependent protein kinases |
| CHC | clathrin heavy chain |
| CLC | clathrin light chain |
| DHAR | dehydroascorbate reductase |
| ERF | ethylene-responsive factor |
| FLS | flagellin sensitive |
| GABA | γ-aminobutyric acid |
| GB | gibberellin |
| GBF | g-box binding factor |
| GR | glutathione reductase |
| GSH | glutathione |
| GSNOR | S-nitrosoglutathione reductase |
| GST | glutathione s-transferase |
| HB | hemoglobin |
| MDA | malondialdehyde |
| MDHAR | monodehydroascorbate reductase |
| MJ | methyl jasmonate |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NF | nuclear factory |
| NO | nitric oxide |
| NR | nitrate reductase |
| OST | open stomata |
| PDC | pyruvate decarboxylase |
| PIF | phytochrome interacting factor |
| POD | peroxidase |
| RBOH | respiratory burst oxidase homolog |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
| SOS | salt overlay sensitive |
| TPC | two pore channel |
References
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum. 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.-Y.; Yun, A.D.-J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Ziska, L.H.; Bunce, J.A.; Shimono, H.; Gealy, D.R.; Baker, J.T.; Newton, P.C.D.; Reynolds, M.P.; Jagadish, K.S.V.; Zhu, C.; Howden, M.; et al. Food security and climate change: On the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc. R. Soc. B Boil. Sci. 2012, 279, 4097–4105. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Ahmad, P.; Abdel Latef, A.A.; Hashem, A.; Abd Allah, E.F.; Gucel, S.; Tran, L.-S.P. Nitric Oxide Mitigates Salt Stress by Regulating Levels of Osmolytes and Antioxidant Enzymes in Chickpea. Front. Plant Sci. 2016, 7, 347. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2017, 217, 523–539. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Pinheiro, J.F.C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.X.; Zhu, X.H.; Chen, J.L.; Yang, S.J.; Ding, H.D.; Zha, D.S. Nitric oxide alleviates adverse salt-induced effects by improving the photosynthetic performance and increasing the anti-oxidant capacity of eggplant (Solanum melongena L.). J. Hort. Sci. Biotech. 2015, 88, 352–360. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Alshammari, W.B.; Alshammery, K.; Lotfi, S.; Altamimi, H.; Alshammari, A.; Al-Harbi, N.A.; Jakovljević, D.; Alharbi, M.H.; Moustapha, M.E.; El-Moneim, D.A.; et al. Improvement of morphophysiological and anatomical attributes of plants under abiotic stress conditions using plant growth-promoting bacteria and safety treatments. PeerJ 2024, 12, e17286. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Dietz, K.J. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 2016, 7, 548. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Satheesh, N.; Kherawat, B.S.; Kumar, A.; Kim, H.-U.; Chung, S.-M.; Kumar, M. Regulation of reactive oxygen species during salt stress in plants and their crosstalk with other signaling molecules-current perspectives and future directions. Plants 2023, 12, 864. [Google Scholar] [CrossRef]
- Mariyam, S.; Bhardwaj, R.; Khan, N.A.; Sahi, S.V.; Seth, C.S. Review on nitric oxide at the forefront of rapid systemic signaling in mitigation of salinity stress in plants: Crosstalk with calcium and hydrogen peroxide. Plant Sci. 2023, 336, 111835. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A.; Alam, I.; Khoso, M.A.; Ali, Q.; Liu, F. Waterlogging stress in plants: Unraveling the mechanisms and impacts on growth, development, and productivity. Environ. Exp. Bot. 2024, 224, 105824. [Google Scholar] [CrossRef]
- Colmer, T.D.; Greenway, H. Ion transport in seminal and adventitious roots of cereals during O2 deficiency. J. Exp. Bot. 2011, 62, 39–57. [Google Scholar] [CrossRef]
- Leon, J.; Castillo, M.C.; Gaybas, B. The hypoxia-reoxygenation stress in plants. J. Exp. Bot. 2021, 72, 5841–5846. [Google Scholar] [CrossRef]
- Pan, J.; Sharif, R.; Xu, X.; Chen, X. Mechanisms of waterlogging tolerance in plants: Research progress and prospects. Front. Plant Sci. 2021, 11, 627331. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and management. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Ali, S.; Park, S.; Bae, H. Exploring the potential of multiomics and other integrative approaches for improving waterlogging tolerance in plants. Plants 2023, 12, 1544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, X.; Geng, X.; Zhang, X. A review of soil waterlogging impacts, mechanisms, and adaptive strategies. Front. Plant Sci. 2025, 16, 1545912. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Kunert, K. The ascorbate–glutathione cycle coming of age. J. Exp. Bot. 2024, 75, 2682–2699. [Google Scholar] [CrossRef]
- Rao, M.J.; Duan, M.; Zhou, C.; Jiao, J.; Cheng, P.; Yang, L.; Wei, W.; Shen, Q.; Ji, P.; Yang, Y.; et al. Antioxidant defense system in plants: Reactive oxygen species production, signaling, and scavenging during abiotic stress-induced oxidative damage. Horticulturae 2025, 11, 477. [Google Scholar] [CrossRef]
- Xing, L.; Zhu, M.; Luan, M.; Zhang, M.; Jin, L.; Liu, Y.; Zou, J.; Wang, L.; Xu, M. miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 2022, 188, 608–623. [Google Scholar] [CrossRef]
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D.; et al. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Liu, C.; Lin, J.-Z.; Wang, Y.; Tian, Y.; Zheng, H.-P.; Zhou, Z.-K.; Zhou, Y.-B.; Tang, X.-D.; Zhao, X.-H.; Wu, T.; et al. The protein phosphatase PC1 dephosphorylates and deactivates CatC to negatively regulate H2O2 homeostasis and salt tolerance in rice. Plant Cell 2023, 35, 3604–3625. [Google Scholar] [CrossRef]
- Jiang, L.; Xiao, M.; Huang, R.; Wang, J. The regulation of ROS and phytohormones in balancing crop yield and salt tolerance. Antioxidants 2025, 14, 63. [Google Scholar] [CrossRef]
- Wang, L.-N.; Wang, W.-C.; Liao, K.; Xu, L.-J.; Xie, D.-X.; Xie, R.-H.; Xiao, S. Survival mechanisms of plants under hypoxic stress: Physiological acclimation and molecular regulation. J. Integr. Plant. Biol. 2025, 67, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Lin, Z.; Liu, D.; Li, C.; Zhou, Z.; Mei, F.; Li, J.; Deng, X.-Y. Effect of waterlogging-induced autophagy on programmed cell death in Arabidopsis roots. Front. Plant Sci. 2019, 10, 468. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Cui, C.; Liu, Y.; Wu, K.; Du, Z.; Jiang, X.; Zhao, F.; Zhang, R.; Wang, J.; Mei, H.; et al. Physiological and transcriptional responses of sesame (Sesamum indicum L.) to waterlogging stress. Int. J. Mol. Sci. 2025, 26, 2603. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2011, 35, 259–270. [Google Scholar] [CrossRef]
- Sewelam, N.; Kazan, K.; Schenk, P. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 00187. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Dietz, K.-J.; Vogelsang, L. A general concept of quantitative abiotic stress sensing. Trend. Plant Sci. 2024, 29, 319–328. [Google Scholar] [CrossRef]
- Hendrix, S.; Vanbuel, I.; Colemont, J.; Bos Calderó, L.; Hamzaoui, M.A.; Kunnen, K.; Huybrechts, M.; Cuypers, A. Jack of all trades: Reactive oxygen species in plant responses to stress combinations and priming-induced stress tolerance. J. Exp. Bot. 2025, 76, 3686–3705. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Borhannuddin Bhuyan, M.H.M.; Parvin, K.; Bhuiyanm, T.F.; Anee, T.I.; Nahar, K.; Hossen, M.S.; Zulfiqar, F.; Alam, M.M.; Fujita, M. Regulation of ROS metabolism in plants under environmental stress: A review of recent experimental evidences. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2011, 63, 305–317. [Google Scholar] [CrossRef]
- Jiang, C.; Belfield, E.J.; Mithani, A.; Visscher, A.M.; Jiannis Ragoussis, J.; Mott, R.; Smith, A.C.; Harberd, N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012, 31, 4359–4370. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, C.; Wu, R.; Hu, Y.; Zhou, Y.; Wang, J.; Yu, X.; Zhang, Y.; Bawa, G.; Sun, X. FLS2-RBOHD-PIF4 module regulates plant response to drought and salt stress. Int. J. Mol. Sci. 2022, 23, 1080. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Z.; Qin, A.; Zhou, Y.; Zhao, Z.; Yang, J.; Hu, M.; Liu, H.; Liu, Y.; Sun, S.; et al. FLS2-RBOHD module regulates changes in the metabolome of Arabidopsis in response to abiotic stress. Plant Environ. Interact. 2023, 4, 36–54. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Gu, Q.; Zhao, G.; Zhang, Y.; Cui, W.; Xu, X.; Wang, R.; Shen, W. The AtrbohF-dependent regulation of ROS signaling is required for melatonin-induced salinity tolerance in Arabidopsis. Free Radic. Biol. Med. 2017, 108, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, L.; Ma, L.; Hao, F.-H. Both AtrbohD and AtrbohF are essential for mediating responses to oxygen deficiency in Arabidopsis. Plant Cell Rep. 2017, 36, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Jin, L.; Peng, R. Crosstalk between Ca2+ and other regulators assists plants in responding to abiotic stress. Plants 2022, 19, 1351. [Google Scholar] [CrossRef]
- Ikram, M.; Batool, M.; Ullah, M.; Khalid, B.; El-Badri, A.M.; Mohamed, I.A.A.; Zhang, L.; Kuai, J.; Xu, Z.; Zhao, J.; et al. Molecular alchemy: Converting stress into resilience via secondary metabolites and calcium signaling in rice. Rice 2025, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.-X.; Li, X.; Li, C.; Zhao, L. The role of nitric oxide in plant responses to salt stress. Int. J. Mol. Sci. 2022, 23, 6167. [Google Scholar] [CrossRef]
- Gupta, K.J.; Mur, L.A.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol. 2019, 225, 1143–1151. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Zhang, J.; Zhang, W.; Zheng, L.; Borjigin, T.; Wang, Y. Nitric oxide alleviates salt-induced stress damage by regulating the ascorbate-glutathione cycle and Na+/K+ homeostasis in Nitraria tangutorum Bobr. Plant Physiol. Biochem. 2022, 173, 46–58. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, W.; He, J.; Zhang, L.; Wei, Y.; Yang, M. Nitric oxide alleviates salt stress in seed germination and early seedling growth of pakchoi (Brassica chinensis L.) by enhancing physiological and biochemical parameters. Ecotoxicol. Environ. Saf. 2020, 187, 109785. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; Veen, H.; Yeung, E.; Voesenek, L.A.C.J. Signal dynamics and interactions during flooding stress. Plant Physiol. 2018, 176, 1106–1117. [Google Scholar] [CrossRef]
- Chen, T.; Yuan, F.; Song, J.; Wang, B. Nitric oxide participates in waterlogging tolerance through enhanced adventitious root formation in the euhalophyte Suaeda salsa. Funct. Plant Biol. 2016, 43, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Wany, A.; Kumari, A.; Gupta, K.J. Nitric oxide is essential for the development of aerenchyma in wheat roots under hypoxic stress. Plant Cell Environ. 2017, 40, 3002–3017. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Shabala, L.; Zhou, M.; Zhang, G.; Shabala, S. Barley responses to combined waterlogging and salinity stress: Separating effects of oxygen deprivation and elemental toxicity. Front. Plant Sci. 2013, 4, 313. [Google Scholar] [CrossRef] [PubMed]
- Anee, T.I.; Rachman, R.R.; Ziqi, Z.; Suzuki, N. A combination of salt stress and waterlogging provides protection to tomato plants against the negative effects of waterlogging individually applied. Physiol. Plant. 2025, 177, e70116. [Google Scholar] [CrossRef]
- Mahmood, U.; Hussain, S.; Hussain, S.; Ali, B.; Ashraf, U.; Zamir, S.; Al-Robai, S.A.; Alzahrani, F.O.; Hano, C.; El-Esawi, M.A. Morpho-physio-biochemical and molecular responses of maize hybrids to salinity and waterlogging during stress and recovery phase. Plants 2021, 10, 1345. [Google Scholar] [CrossRef]
- Duan, H.; Ma, Y.; Liu, R.; Li, Q.; Yang, Y.; Song, J. Effect of combined waterlogging and salinity stresses on euhalophyte Suaeda glauca. Plant Physiol. Biochem. 2018, 127, 231–237. [Google Scholar] [CrossRef]
- Putra, S.P.; Santosa; Salsinha, Y.C.F. Waterlogging and salinity stress affecting growth and morphological character changes of Limnocharis flava. Biodiver. J. Biol. Diver. 2023, 24, 333–340. [Google Scholar]
- Liu, X.; Chen, C.; Liu, Y.; Liu, Y.; Zhao, Y.; Chen, M. The presence of moderate salt can increase tolerance of Elaeagnus angustifolia seedlings to waterlogging stress. Plant Signal. Behav. 2020, 15, 1743518. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 00276. [Google Scholar] [CrossRef]
- Feki, K.; Tounsi, S.; Kamoun, H.; Al-Hashimi, A.; Brini, F. Decoding the role of durum wheat ascorbate peroxidase TdAPX7B-2 in abiotic stress response. Funct. Integr. Genom. 2024, 24, 223. [Google Scholar] [CrossRef] [PubMed]
- Ghorbel, M.; Feki, K.; Tounsi, S.; Bouali, N.; Besbes, M.; Brini, F. The putative auto-inhibitory domain of durum wheat catalase (TdCAT1) positively regulates bacteria cells in response to different stress conditions. Antioxidants 2022, 11, 1820. [Google Scholar] [CrossRef]
- Tounsi, S.; Kamoun, Y.; Feki, K.; Jemli, S.; Saïdi, M.N.; Ziadi, H.; Alcon, C.; Brini, F. Localization and expression analysis of a novel catalase from Triticum monococcum TmCAT1 involved in response to different environmental stresses. Plant Physiol. Biochem. 2019, 139, 366–378. [Google Scholar] [CrossRef]
- Soostani, S.B.; Ranjbar, M.; Memarian, A.; Mohammadi, M.; Yaghini, Z. Regulation of APX, SOD, and PAL genes by chitosan under salt stress in rapeseed (Brassica napus L.). BMC Plant Biol. 2025, 25, 824. [Google Scholar] [CrossRef]
- Turfan, N.; Khubalıyev, I.; Tekşen, K.; Altuner, E.M. Investigating exogenous tyrosine supplements on the responses of the kale plant to salinity stress. Food Sci. Nutr. 2025, 13, e70660. [Google Scholar] [CrossRef]
- Ashraf, H.; Ramzan, M.; Ahmad, M.Z.; Naz, G.; Usman, S.; Shah, A.A.; Shaffique, S.; Alataway, A.; Elansary, H.O. Sargassum-synthesized ZnO nanoparticles induce salt tolerance in maize plants through enhanced physiological and biochemical mechanisms. Sci. Rep. 2025, 15, 30633. [Google Scholar] [CrossRef]
- Nie, R.; Wu, C.; Ji, X.; Li, A.; Zheng, X.; Tang, J.; Sun, L.; Su, Y.; Zhang, J. Methyl jasmonate orchestrates multi-pathway antioxidant defense to enhance salt stress tolerance in walnut (Juglans regia L.). Antioxidants 2025, 14, 974. [Google Scholar] [CrossRef] [PubMed]
- Tomar, R.S.; Kataria, S.; Jajoo, A. Behind the scene: Critical role of reactive oxygen species and reactive nitrogen species in salt stress tolerance. J. Agron. Crop Sci. 2021, 207, 577–588. [Google Scholar] [CrossRef]
- Wahid, A.; Perveen, M.; Gelani, S.; Basra, S.M.A. Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J. Plant Physiol. 2007, 164, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Araújo, G.S.; de Oliveira Paula-Marinho, S.; de Paiva Pinheiro, S.K.; de Castro Miguel, E.; de Sousa Lopes, L.; Marques, C.E.; de Carvalho, H.H.; Gomes-Filho, E. H2O2 priming promotes salt tolerance in maize by protecting chloroplasts ultrastructure and primary metabolites modulation. Plant Sci. 2021, 303, 110774. [Google Scholar] [CrossRef]
- Veen, E.V.; Küpers, J.J.; Gommers, C.M.M. Plastids in a pinch: Coordinating stress and developmental responses through retrograde signalling. Plant Cell Environ. 2025, 48, 6897–6911. [Google Scholar] [CrossRef]
- Ramachandran, P.; Pandey, N.K.; Yadav, R.M.; Suresh, P.; Kumar, A.; Subramanyam, R. Photosynthetic efficiency and transcriptome analysis of Dunaliella salina under hypersaline: A retrograde signaling mechanism in the chloroplast. Front. Plant Sci. 2023, 14, 1192258. [Google Scholar] [CrossRef]
- Latef, A.; Kordrostami, M.; Zakir, A.; Zaki, H.; Saleh, O. Eustress with H2O2 Facilitates plant growth by improving tolerance to salt stress in two wheat cultivars. Plants 2019, 8, 303. [Google Scholar] [CrossRef]
- Sun, M.Y.; Liu, X.; Zheng, H.; Li, L.; Lv, Q.M.; Wang, G.P. How plants respond to salt stress: Lessons from RBOHs. Plant Sci. 2025, 360, 112710. [Google Scholar] [CrossRef]
- Lee, J.; Nguyen, H.H.; Park, Y.; Lin, J.; Hwang, I. Spatial regulation of RBOHD via AtECA4-mediated recycling and clathrin-mediated endocytosis contributes to ROS accumulation during salt stress response but not flg22-induced immune response. Plant J. 2022, 109, 816–830. [Google Scholar] [CrossRef]
- Wang, G.F.; Li, W.-Q.; Li, W.Y.; Wu, G.L.; Zhou, C.Y.; Chen, K.M. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 2013, 14, 9440–9458. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Mittler, R. Rapid systemic signaling during abiotic and biotic stresses: Is the ROS wave master of all trades? Plant J. 2020, 102, 887–896. [Google Scholar] [CrossRef]
- Evans, M.J.; Choi, W.G.; Gilroy, S.; Morris, R.J. A ROS-Assisted Calcium Wave Dependent on the AtRBOHD NADPH Oxidase and TPC1 Cation Channel Propagates the Systemic Response to Salt Stress. Plant Physiol. 2016, 171, 1771–1784. [Google Scholar] [CrossRef]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant signaling networks involving Ca(2+) and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 2015, 6, 427. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Shomali, A.; Azad, N.; Hassani, B.; Lastochkina, O.; Li, T. Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal. Behav. 2019, 14, 1665455. [Google Scholar] [CrossRef]
- Hao, J.; Lou, P.; Han, Y.; Zheng, L.; Lu, J.; Chen, Z.; Xu, M. Ultraviolet-B irradiation increases antioxidant capacity of pakchoi (Brassica rapa L.) by inducing flavonoid biosynthesis. Plants 2022, 11, 766. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Salamó, I.; Papdi, C.; Rigó, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horváth, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Yang, L.; Nan, W.; Ruan, M.; Bi, Y. Involvement of active MKK9-MAPK3/MAPK6 in increasing respiration in salt-treated Arabidopsis callus. Protoplasma 2020, 257, 965–977. [Google Scholar] [CrossRef]
- Li, C.-H.; Wang, G.; Zhao, J.-L.; Zhang, L.-Q.; Ai, L.-F.; Han, Y.-F.; Sun, D.-Y.; Zhang, S.-W.; Sun, Y. The receptor-like kinase sit1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Jiang, D.; Liu, F.; Dai, T.; Liu, W.; Jing, Q.; Cao, W. Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environ. Exp. Bot. 2009, 67, 222–227. [Google Scholar] [CrossRef]
- Alnusairi, G.S.H.; Mazrou, Y.S.A.; Qari, S.H.; Elkelish, A.; Soliman, M.H.; Eweis, M.; AlKahtani, M.D.F.; El-Samad, G.A.; Khan, I.A.; Nahhas, N.E. Exogenous nitric oxide reinforces photosynthetic efficiency, osmolyte, mineral uptake, antioxidant, expression of stress-responsive genes and ameliorates the effects of salinity stress in wheat. Plants 2021, 10, 1693. [Google Scholar] [CrossRef]
- Gokce, A.; Cetinel, A.H.S.; Turkan, I. Involvement of GLR-mediated nitric oxide effects on ROS metabolism in Arabidopsis plants under salt stress. J. Plant Res. 2024, 137, 485–503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Jia, L.; Chu, H.; Wu, D.; Peng, X.; Liu, X.; Zhang, J.; Zhao, J.; Chen, K.; Zhao, L. Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting S-Nitrosoglutathione reductase via direct binding. PLoS Genet. 2016, 12, e1006255. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.C.S.; Silva, D.M.R.; Bispo, G.L.; Oliveira, F.A.; Ono, E.O.; Rodrigues, J.D. Nitric Oxide Modulates Salt Stress Tolerance in Lettuce. Stresses 2023, 3, 701–716. [Google Scholar] [CrossRef]
- Kong, X.; Wang, T.; Li, W.; Tang, W.; Zhang, D.; Dong, H. Exogenous nitric oxide delays salt-induced leaf senescence in cotton (Gossypium hirsutum L.). Acta Physiol. Plant. 2016, 38, 61. [Google Scholar] [CrossRef]
- Fatma, M.; Masood, A.; Per, T.S.; Khan, N.A. Nitric oxide alleviates salt stress inhibited photosynthetic performance by interacting with sulfur assimilation in mustard. Front. Plant Sci. 2016, 7, 521. [Google Scholar] [CrossRef]
- Shen, Z.J.; Chen, J.; Ghoto, K.; Hu, W.J.; Gao, G.F.; Luo, M.R.; Li, Z.; Simon, M.; Zhu, X.Y.; Zheng, H.L. Proteomic analysis on mangrove plant Avicennia marina leaves reveals nitric oxide enhances the salt tolerance by up-regulating photosynthetic and energy metabolic protein expression. Tree Physiol. 2018, 38, 1605–1622. [Google Scholar] [CrossRef]
- Leitner, M.; Vandelle, E.; Gaupels, F.; Bellin, D.; Delledonne, M. NO signals in the haze. Nitric oxide signalling in plant defence. Curr. Opin. Plant Biol. 2009, 12, 451–458. [Google Scholar] [CrossRef]
- Baghour, M.; Gálvez, F.J.; Sánchez, M.E.; Aranda, M.N.; Venema, K.; Rodríguez-Rosales, M.P. Overexpression of LeNHX2 and SlSOS2 increases salt tolerance and fruit production in double transgenic tomato plants. Plant Physiol. Biochem. 2019, 135, 77–86. [Google Scholar] [CrossRef]
- Sun, L.R.; Yue, C.M.; Hao, F.S. Update on roles of nitric oxide in regulating stomatal closure. Plant Signal. Behav. 2019, 14, e1649569. [Google Scholar] [CrossRef] [PubMed]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Molassiotis, A.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Molassiotis, A.; Job, D.; Ziogas, V.; Tanou, G. Citrus Plants: A model system for unlocking the secrets of NO and ROS-inspired priming against salinity and drought. Front. Plant Sci. 2016, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Inafuku, M.; Nahar, K.; Fujita, M.; Oku, H. Nitric oxide regulates plant growth, physiology, antioxidant defense, and ion homeostasis to confer salt tolerance in the mangrove species, Kandelia obovata. Antioxidants 2021, 10, 611. [Google Scholar] [CrossRef]
- Khan, M.; Ali, S.; Azzawi, A.; Yun, B. Nitric oxide acts as a key signaling molecule in plant development under stressful conditions. Int. J. Mol. Sci. 2023, 24, 4782. [Google Scholar] [CrossRef]
- Szepesi, Á.; Bakacsy, L.; Fehér, A.; Kovács, H.; Pálfi, P.; Poór, P.; Szőllősi, R.; Gondor, O.K.; Janda, T.; Szalai, G.; et al. L-aminoguanidine induces imbalance of ROS/RNS homeostasis and polyamine catabolism of tomato roots after short-term salt exposure. Antioxidants 2023, 12, 1614. [Google Scholar] [CrossRef]
- Tanou, G.; Filippou, P.; Belghazi, M.; Job, D.; Diamantidis, G.; Fotopoulos, V.; Molassiotis, A. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 2012, 72, 585–599. [Google Scholar] [CrossRef]
- Ismail, A.; Takeda, S.; Nick, P. Life and death under salt stress: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, R.; Shabala, S. Stomata in a saline world. Curr. Opin. Plant Biol. 2018, 46, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Blatt, M.R. Ca(2+) signalling and control of guard-cell volume in stomatal movements. Curr. Opin. Plant Biol. 2000, 2, 196–204. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Allen, G.J.; Hugouvieux, V.; Kwak, J.M.; Waner, D. Guard Cell Signal Transduction. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 627–658. [Google Scholar] [CrossRef]
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.C.; Harberd, N.P. An Arabidopsis Soil-Salinity–Tolerance Mutation Confers Ethylene-Mediated Enhancement of Sodium/Potassium Homeostasis. Plant Cell 2013, 25, 3535–3552. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Ann. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Waadt, R.; Manalansan, B.; Rauniyar, N.; Munemasa, S.; Booker, M.A.; Brandt, B.; Waadt, C.; Nusinow, D.A.; Kay, S.A.; Kunz, H.; et al. Identification of Open Stomata1-Interacting Proteins Reveals Interactions with Sucrose Non-fermenting1-Related Protein Kinases2 and with Type 2A Protein Phosphatases That Function in Abscisic Acid Responses. Plant Physiol. 2015, 169, 760–779. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.-H.; Zhang, B.; Hills, A.; Blatt, M.R. PYR/PYL/RCAR Abscisic Acid Receptors Regulate K+ and Cl− Channels through Reactive Oxygen Species-Mediated Activation of Ca2+ Channels at the Plasma Membrane of Intact Arabidopsis Guard Cells. Plant Physiol. 2013, 163, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-P.; Köster, P.; Drerup, M.M.; Scholz, M.; Li, S.; Edel, K.H.; Hashimoto, K.; Kuchitsu, K.; Hippler, M.; Kudla, J. Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 2019, 221, 1935–1949. [Google Scholar] [CrossRef]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 2020, 578, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Ren, H.-M.; Tan, Y.-Q.; Qi, G.-N.; Yao, F.-Y.; Wu, G.-L.; Yang, L.-W.; Hussain, J.; Sun, S.-J.; Wang, Y.-F. S-Type Anion Channels SLAC1 and SLAH3 Function as Essential Negative Regulators of Inward K+ Channels and Stomatal Opening in Arabidopsis. Plant Cell 2016, 28, 949–965. [Google Scholar] [CrossRef]
- Imes, D.; Mumm, P.; Böhm, J.; Al-Rasheid, K.A.S.; Marten, I.; Geiger, D.; Hedrich, R. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013, 74, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Jaślan, J.; Marten, I.; Jakobson, L.; Arjus, T.; Deeken, R.; Sarmiento, C.; Angeli, A.D.; Brosché, M.; Kollist, H.; Hedrich, R. ALMT-independent guard cell R-type anion currents. New Phytol. 2023, 239, 2225–2234. [Google Scholar] [CrossRef]
- Drerup, M.M.; Schlücking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The Calcineurin B-Like Calcium Sensors CBL1 and CBL9 Together with Their Interacting Protein Kinase CIPK26 Regulate the Arabidopsis NADPH Oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, X.; Zhang, Y.; Wang, J.; Yang, J.; Ishida, T.; Jiang, W.; Han, X.; Kang, J.; Wang, X.; et al. CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ. 2019, 42, 1033–1044. [Google Scholar] [CrossRef]
- Tan, Y.-Q.; Yang, Y.; Shen, X.; Zhu, M.; Shen, J.; Zhang, W.; Hu, H.; Wang, Y.-F. Multiple cyclic nucleotide-gated channels function as ABA-activated Ca2+ channels required for ABA-induced stomatal closure in Arabidopsis. Plant Cell 2019, 35, 239–259. [Google Scholar] [CrossRef]
- Yang, Y.; Tan, Y.-Q.; Wang, X.; Li, J.-J.; Du, B.-Y.; Zhu, M.; Wang, P.; Wang, Y.-F. OPEN STOMATA 1 phosphorylates CYCLIC NUCLEOTIDE-GATED CHANNELs to trigger Ca2+ signaling for abscisic acid-induced stomatal closure in Arabidopsis. Plant Cell 2024, 36, 2328–2358. [Google Scholar] [CrossRef]
- Garcia-Mata, C.; Gay, R.; Sokolovski, S.; Hills, A.; Lamattina, L.; Blatt, M.R. Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 11116–11121. [Google Scholar] [CrossRef]
- Laxalt, A.M.; García-Mata, C.; Lamattina, L. The dual role of nitric oxide in guard cells: Promoting and attenuating the aba and phospholipid-derived signals leading to the stomatal closure. Front. Plant Sci. 2016, 7, 476. [Google Scholar] [CrossRef]
- Wang, P.; Du, Y.; Hou, Y.-J.; Zhao, Y.; Hsu, C.-C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.-P.; Zhu, J.-K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2014, 112, 613–618. [Google Scholar] [CrossRef]
- Hao, F.; Zhao, S.; Dong, H.; Zhang, H.; Sun, L.; Miao, C. Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 298–307. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Anee, T.I.; Bhuiyan, T.F.; Nahar, K.; Fujita, M. Emerging role of osmolytes in enhancing abiotic stress tolerance in rice. In Advances in Rice Research for Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Nahar, K., Biswas, J.K., Eds.; Woodhead Publications: Amsterdam, The Netherlands, 2019; pp. 677–708. [Google Scholar]
- Singh, P.; Choudhary, K.K.; Chaudhary, N.; Gupta, S.; Sahu, M.; Tejaswini, B.; Sarkar, S. Salt stress resilience in plants mediated through osmolyte accumulation and its crosstalk mechanism with phytohormones. Front. Plant Sci. 2022, 13, 1006617. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- El-Badri, A.M.; Batool, M.; Mohamed, I.A.A.; Wang, Z.; Khatab, A.; Sherif, A.; Ahmad, H.; Khan, M.N.; Hassan, H.M.; Elrewainy, I.M.; et al. Antioxidative and metabolic contribution to salinity stress responses in two rapeseed cultivars during the early seedling stage. Antioxidants 2021, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.I.; Naikoo, M.I.; Rehman, F.; Naushin, F.; Khan, F.A. Proline accumulation in plants: Roles in stress tolerance and plant development. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Khan, N.A., Eds.; Springer: New Delhi, India, 2016; pp. 155–166. [Google Scholar]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- You, J.; Hu, H.; Xiong, L. An ornithine δ-aminotransferase gene OsOAT confers drought and oxidative stress tolerance in rice. Plant Sci. 2012, 197, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhen, W.-B.; Ma, Q.-H. Proline metabolism in response to salt stress in common reed [Phragmites australis (Cav.) Trin. Ex Steud]. Bot. Mar. 2009, 52, 341–347. [Google Scholar] [CrossRef]
- Spormann, S.; Nadais, P.; Sousa, F.; Pinto, M.; Martins, M.; Sousa, B.; Fidalgo, F.; Soares, C. Accumulation of Proline in Plants under Contaminated Soils-Are We on the Same Page? Antioxidants 2023, 12, 666. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.S.; Meena, M.; Prasad, V.; Zehra, A.; Gupta, V.K. Mannitol metabolism during pathogenic fungal–host interactions under stressed conditions. Front. Microbiol. 2015, 6, 1019. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Kundu, A. Sugars and sugar polyols in overcoming environmental stresses. In Protective Chemical Agents in the Amelioration of Plant Abiotic Stress; Roychoudhury, A., Tripathi, D.K., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 2020; pp. 71–101. [Google Scholar]
- Patel, K.G.; Mandaliya, V.B.; Mishra, G.P.; Dobaria, J.R.; Thankappan, R. Transgenic peanut overexpressing mtlD gene confers enhanced salinity stress tolerance via mannitol accumulation and differential antioxidative responses. Acta Physiol. Plant. 2016, 38, 181. [Google Scholar] [CrossRef]
- Zhai, H.; Wang, F.; Si, Z.; Huo, J.; Xing, L.; An, Y.; He, Y.; Liu, Q. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 2016, 14, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Chen, Q.; Fu, Y.; Xie, H.; Li, T.; Li, D.; Yang, Y.; Xie, Z.; Qi, K.; Zhang, S.; et al. PbGBF3 enhances salt response in pear by upregulating PbAPL2 and PbSDH1 and reducing ABA-mediated salt sensitivity. Plant J. 2024, 119, 2837–2853. [Google Scholar] [CrossRef]
- Lv, D.W.; Zhu, G.R.; Zhu, D.; Bian, Y.W.; Liang, X.N.; Cheng, Z.W.; Deng, X.; Yan, Y.-M. Proteomic and phosphoproteomic analysis reveals the response and defense mechanism in leaves of diploid wheat T. monococcum under salt stress and recovery. J. Proteom. 2016, 143, 93–105. [Google Scholar] [CrossRef]
- Wang, F.W.; Wang, M.L.; Guo, C.; Wang, N.; Li, X.W.; Chen, H.; Dong, Y.Y.; Chen, X.F.; Wang, Z.M.; Li, H.Y. Cloning and characterization of a novel betaine aldehyde dehydrogenase gene from Suaeda corniculata. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; He, G.; Li, J.; Perez-Hormaeche, J.; Becker, T.; Luo, M.; Wallrad, L.; Gao, J.; Li, J.; Pardo, J.M.; et al. A salt stress-activated GSO1-SOS2-SOS1 module protects the Arabidopsis root stem cell niche by enhancing sodium ion extrusion. EMBO J. 2023, 42, e113004. [Google Scholar] [CrossRef]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of Nine Sucrose Nonfermenting 1-related Protein Kinases 2 Activated by Hyperosmotic and Saline Stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef]
- Verslues, P.E.; Batelli, G.; Grillo, S.; Agius, F.; Kim, Y.-S.; Zhu, J.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.-K. Interaction of SOS2 with Nucleoside Diphosphate Kinase 2 and Catalases Reveals a Point of Connection between Salt Stress and H2O2 Signaling in Arabidopsis thaliana. Mol. Cell. Biol. 2007, 27, 7771–7780. [Google Scholar] [CrossRef]
- Chung, J.-S.; Zhu, J.-K.; Bressan, R.A.; Hasegawa, P.M.; Shi, H. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 2008, 53, 554–565. [Google Scholar] [CrossRef]
- Lang, T.; Deng, S.; Zhao, N.; Deng, C.; Zhang, Y.; Zhang, Y.; Zhang, H.; Sa, G.; Yao, J.; Wu, C.; et al. Salt-sensitive signaling networks in the mediation of K+/Na+ homeostasis gene expression in Glycyrrhiza uralensis roots. Front. Plant Sci. 2017, 8, 1403. [Google Scholar] [CrossRef]
- Katano, K.; Kataoka, R.; Fujii, M.; Suzuki, N. Differences between seedlings and flowers in anti-ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2. Plant Physiol. Biochem. 2018, 123, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Myers, R.J., Jr.; Grant, D.A.G.; Mittler, R. Plasmodesmata-localized proteins and ROS orchestrate light-induced rapid systemic signaling in Arabidopsis. Sci. Signal. 2021, 14, eabf0322. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2017, 69, 3465–3476. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, Q.; Dai, X.; He, M. Effects of potassium chloride and nitric oxide on growth and physiological characteristics of winter wheat under salt stress. Biol. Plant 2020, 64, 258–265. [Google Scholar] [CrossRef]
- Habib, N.; Ashraf, M. Effect of exogenously applied nitric oxide on water relations and ionic composition of rice (Oryza sativa L.) plants under salt stress. Pak. J. Bot. 2014, 46, 111–116. [Google Scholar]
- Vaishnav, A.; Jain, S.; Kasotia, A.; Kumari, S.; Gaur, R.K.; Choudhary, D.K. Effect of nitric oxide signaling in bacterial-treated soybean plant under salt stress. Arch. Microbiol. 2013, 195, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-G.; Tian, Q.-Y.; Zhang, W.-H. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007, 144, 206–217. [Google Scholar] [CrossRef]
- Teoh, E.Y.; Teo, C.H.; Baharum, N.A.; Pua, T.L.; Tan, B.C. Waterlogging stress induces antioxidant defense responses, aerenchyma formation, and alters metabolism in banana plants. Plants 2023, 12, 2052. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, M.; Liu, W.; Lian, M.; Yang, S.; Peng, F.; Xiao, Y. Waterlogging resistance and evaluation of physiological mechanisms of three peach (Prunus persica) rootstocks. Protoplasma 2023, 260, 1375–1388. [Google Scholar] [CrossRef]
- Fathi, A.; Shiade, S.R.G.; Saleem, A.; Shohani, F.; Fazeli, A.; Riaz, A.; Zulfiqar, U.; Shabaan, M.; Ahmed, I.; Rahimi, M. Reactive Oxygen Species (ROS) and Antioxidant Systems in Enhancing Plant Resilience Against Abiotic Stress. Int. J. Agron. 2025, 2025, 8834883. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, D. Lactylation constrains OXPHOS under hypoxia. Cell Res. 2024, 34, 91–92. [Google Scholar] [CrossRef]
- Niu, L.; Jiang, F.; Yin, J.; Wang, Y.; Li, Y.; Yu, X.; Song, X.; Ottosen, C.-O.; Rosenqvist, E.; Mittler, R.; et al. ROS-mediated waterlogging memory, induced by priming, mitigates photosynthesis inhibition in tomato under waterlogging stress. Front. Plant Sci. 2023, 14, 1238108. [Google Scholar] [CrossRef]
- Papdi, C.; Abrahám, E.; Joseph, M.P.; Popescu, C.; Koncz, C.; Szabados, L. Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 2008, 147, 528–542. [Google Scholar] [CrossRef]
- Hinz, M.; Wilson, I.W.; Yang, J.; Buerstenbinder, K.; Llewellyn, D.; Dennis, E.S.; Sauter, M.; Dolferus, R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010, 153, 757–772. [Google Scholar] [CrossRef]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Zhao, C.; Wang, J.; Zheng, X.; Yu, F.; Qiu, F. Genetic variations in ZmEREB179 are associated with waterlogging tolerance in maize. J. Genet. Genom. 2025, 52, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.; Juntawong, P.; Peña-Castro, J.M. Submergence and waterlogging stress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; He, S.; Hao, F. AtrbohD functions downstream of ROP2 and positively regulates waterlogging response in Arabidopsis. Plant Signal. Behav. 2018, 13, e1513300. [Google Scholar] [CrossRef]
- Sasidharan, R.; Voesenek, L.A.C.J. Ethylene-Mediated acclimations to flooding stress. Plant Physiol. 2025, 169, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, J.M.; Cardoso, A.A. When roots talk to shoots about flooding. Plant Physiol. 2023, 193, 1729–1731. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.Y.; Bian, Z.Y.; Zhou, L.N.; Cheng, W.; Hai, N.; Yang, C.Q.; Yang, T.; Wang, X.Y.; Wang, C.Y. Hydrogen sulfide enhances nitric oxide-induced tolerance of hypoxia in maize (Zea mays L.). Plant Cell Rep. 2016, 35, 2325–2340. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Huang, X.; Cui, J.; Shabala, L.; Zhou, M.; Yu, M.; Shabala, S. Hypoxia-induced increase in GABA content is essential for restoration of membrane potential and preventing ROS-induced disturbance to ion homeostasis. Plant Commun. 2021, 2, 100188. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Liao, K.; Wang, L.-N.; Shi, L.-L.; Zhang, Y.; Xu, L.-J.; Zhou, Y.; Li, J.-F.; Chen, Y.-Q.; Chen, Q.-F.; et al. Calcium-dependent activation of CPK12 facilitates its cytoplasm-to-nucleus translocation to potentiate plant hypoxia sensing by phosphorylating ERF-VII transcription factors. Mol. Plant 2023, 16, 979–998. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, D.-M.; Yu, W.-W.; Shi, L.-L.; Zhang, Y.; Lai, Y.-X.; Huang, L.-P.; Qi, H.; Chen, Q.-F.; Yao, N.; et al. Phosphatidic acid modulates MPK3- and MPK6-mediated hypoxia signaling in Arabidopsis. Plant Cell 2022, 34, 889–909. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef]
- Pucciariello, C.; Perata, P. The Oxidative Paradox in Low Oxygen Stress in Plants. Antioxidants 2021, 10, 332. [Google Scholar] [CrossRef]
- Hesari, N.; Mirmazloum, I.; Jager, K.; Kolozs, H.; Kiss-Baba, E.; Ramos, M.E.S.; Khan, I.; Babinyec-Czifra, D.; Szego, A.; Papp, I. Nitric oxide mediates nitrate induced alleviation of waterlogging stress in cucumber. Sci. Rep. 2025, 15, 15307. [Google Scholar] [CrossRef]
- Astier, J.; Sanz, L.; Lindermayr, C. S-nitrosylation: An emerging redox-based post-translational modification in plants. Front. Plant Sci. 2018, 9, 1–10. [Google Scholar]
- Mata-Perez, C.; Sanchez-Vicente, I.; Arteaga, N.; Gomez-jimenez, S.; Fuentes-Terron, A.; Oulebsir, C.S.; Calvo-Polanco, M.; Oliver, C.; Lorenzo, O. Functions of nitric oxide-mediated post-tranlastional modifications under abiotic stress. Front. Plant Sci. 2023, 14, 1158184. [Google Scholar] [CrossRef]
- Gupta, K.J.; Shah, J.K.; Brotman, Y.; Jahnke, K.; Willmitzer, L.; Kaiser, W.M.; Bauwe, H.; Igamberdiev, A.U. Inhibition of aconitase by nitric oxide leads to induction of the alternative oxidase and to a shift of metabolism towards biosynthesis of amino acids. J. Exp. Bot. 2012, 63, 1773–1784. [Google Scholar] [CrossRef]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-W.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef]
- Loreti, E.; Perata, P. ERF VII transcription factors and their role in the adaptation to hypoxia in Arabidopsis and crops. Front. Genet. 2023, 14, 1213839. [Google Scholar] [CrossRef]
- Schmidt, R.R.; Weits, D.A.; Feulner, C.F.J.; van Dongen, J.T. Oxygen Sensing and Integrative Stress Signaling in Plants. Plant Physiol. 2018, 176, 1131–1142. [Google Scholar] [CrossRef]
- Carbonare, D.L.; van Veen, H.; Shukla, V.; Perri, M.; Bui, L.; Holdsworth, M.J.; Licausi, F. ERFVIIs as transducers of oxygen-sensing in the evolution of land plant response to hypoxia. Mol. Plant 2025, 18, 1072–1087. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-S.; Javed, T.; Liu, T.-T.; Ali, A.; Gao, S.-J. Mechanisms of Abscisic acid (ABA)-mediated plant defense responses: An updated review. Plant Stress 2025, 15, 100724. [Google Scholar] [CrossRef]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Ni, X.L.; Gui, M.Y.; Tan, L.L.; Zhu, Q.; Liu, W.Z.; Li, C.X. Programmed cell death and aerenchyma formation in water-logged sunflower stems and its promotion by ethylene and ROS. Front. Plant Sci. 2019, 9, 1928. [Google Scholar] [CrossRef]
- Li, J.; Ishii, T.; Yoshioka, M.; Hino, Y.; Nomoto, M.; Tada, Y.; Yoshioka, H.; Takahashi, H.; Yamauchi, T.; Nakazono, M. CDPK5 and CDPK13 play key roles in acclimation to low oxygen through the control of RBOH-mediated ROS production in rice. Plant Physiol. 2024, 197, kiae293. [Google Scholar] [CrossRef]
- Singh, P.; Jaiswal, S.; Kushwaha, A.; Gahlowt, P.; Mishra, V.; Tripathi, D.K.; Singh, S.P.; Gupta, R.; Singh, V.P. Peroxynitrite is essential for aerenchyma formation in rice roots under waterlogging conditions. Planta 2023, 258, 2. [Google Scholar] [CrossRef]
- Li, L.; Dong, X.; He, M.; Huang, M.; Cai, J.; Zhou, Q.; Zhong, Y.; Jiang, D.; Wan, X. Unravelling the role of adventitious roots under priming-induced tolerance to waterlogging stress in wheat. Environ. Exp. Bot. 2023, 216, 105516. [Google Scholar] [CrossRef]
- Tong, B.; Liu, Y.; Wang, Y.; Li, Q. PagMYB180 regulates adventitious rooting via a ROS/PCD-dependent pathway in poplar. Plant Sci. 2024, 346, 112115. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Song, J.; Sohail, H.; Sharif, R.; Yan, W.; Hu, Q.; Qi, X.; Yang, X.; Xu, X.; Chen, X. RNA-seq-based comparative transcriptome analysis reveals the role of CsPrx73 in waterlogging-triggered adventitious root formation in cucumber. Hortic. Res. 2024, 11, uhae062. [Google Scholar] [CrossRef]
- Shiono, K.; Yoshikawa, M.; Kreszies, T.; Yamada, S.; Hojo, Y.; Matsuura, T.; Mori, I.C.; Schreiber, L.; Yoshioka, T. Abscisic acid is required for exodermal suberization to form a barrier to radial oxygen loss in the adventitious roots of rice (Oryza sativa). New Phytol. 2022, 233, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Paradiso, A.; Caretto, S.; Leone, A.; Bove, A.; Nisi, R.; De Gara, L. ROS production and scavenging under Anoxia and Re-Oxygenation in Arabidopsis Cells: A Balance between Redox Signaling and Impairment. Front. Plant Sci. 2016, 7, 1803. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Espinosa, J.; Shabala, S.; Leon, J.; Romero-Puertas, M.C. Reactive oxygen species- and nitric oxide-dependent regulation of ion and metal homeostasis in plants. J. Exp. Bot. 2023, 74, 5970–5986. [Google Scholar] [CrossRef] [PubMed]
- Saini, N.; Joshi, G.; Sangwan, N.S. The role of nitric oxide in plants under salt stress: A review. J. Plant Biol. Crop Res. 2024, 7, 1098. [Google Scholar]
- Timilsina, A.; Dong, W.; Hasanuzzaman, M.; Liu, B.; Hu, C. Nitrate–Nitrite–Nitric Oxide pathway: A mechanism of hypoxia and anoxia tolerance in plants. Int. J. Mol. Sci. 2022, 23, 11522. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Nitric oxide synthase-like activity in higher plants. Nitric Oxide 2017, 68, 5–6. [Google Scholar] [CrossRef]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154. [Google Scholar] [CrossRef]
- Romero-Puertas, M.C.; Sandalio, L.M. Nitric oxide level is self-regulating and also regulates its ROS partners. Front. Plant Sci. 2016, 7, 316. [Google Scholar] [CrossRef]
- Duhan, S.; Kumari, A.; Bala, S.; Sharma, N.; Sheokand, S. Effects of waterlogging, salinity and their combination on stress indices and yield attributes in pigeon pea (Cajanus cajan L. Millsp.) genotypes. Indian J. Plant Physiol. 2018, 23, 65–76. [Google Scholar] [CrossRef]
- Haddadi, B.S.; Hassanpour, H.; Niknam, V. Effect of salinity and waterlogging on growth, anatomical and antioxidative responses in Mentha aquatica L. Acta Physiol. Plant. 2016, 38, 119. [Google Scholar]
- Zheng, C.; Jiang, D.; Liu, F.; Dai, T.; Jing, Q.; Cao, W. Effects of salt and waterlogging stresses and their combination on leaf photosynthesis, chloroplast ATP synthesis, and antioxidant capacity in wheat. Plant Sci. 2009, 176, 575–582. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Kumari, R.; Rakhra, G.; Alsahli, A.A.; Bhat, J.A.; Ahmad, P. Exploring the potential of signalling molecules hydrogen sulfide and nitric oxide in augmenting salt stress resilience in bitter gourd. BMC Plant Biol. 2025, 25, 1008. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Zhong, C.; Li, W.; Dinesh-Kumar, S.P.; Zhang, Y. Orchestrating ROS regulation: Coordinated post-translational modification switches in NADPH oxidases. New Phytol. 2025, 245, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Jindal, A.; Seth, C.S. Nitric oxide mediated post-translational modifications and its significance in plants under abiotic stress. In Nitric Oxide in Developing Plant Stress Resilience; Khan, M.I.R., Iqbal, N., Poor, P., Ferrante, A., Singh, V.P., Tripathi, D.K., Fotopoulos, V., Eds.; Academic Press: New York, NY, USA, 2023; pp. 233–250. [Google Scholar]
- Hu, C.-H.; Wang, P.-Q.; Zhang, P.-P.; Nie, X.-M.; Li, B.-B.; Tai, L.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 2020, 9, 437. [Google Scholar] [CrossRef]
- Kumar, J.; Ramlal, A.; Kumar, K.; Rani, A.; Mishra, V. Signaling pathways and downstream effectors of host innate immunity in plants. Int. J. Mol. Sci. 2021, 22, 9022. [Google Scholar] [CrossRef] [PubMed]
- Bali, S.; Gautam, A.; Dhiman, A.; Michael, R.; Dogra, V. Salicylate and jasmonate intertwine in ROS-triggered chloroplast-to-nucleus retrograde signaling. Physiol. Plant. 2023, 175, e14041. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Selinski, J.; Mao, C.; Zhu, Y.; Berkowitz, O.; Whelan, J. Linking mitochondrial and chloroplast retrograde signalling in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190410. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anee, T.I.; Sewelam, N.A.; Bautista, N.S.; Hirayama, T.; Suzuki, N. Roles of ROS and NO in Plant Responses to Individual and Combined Salt Stress and Waterlogging. Antioxidants 2025, 14, 1455. https://doi.org/10.3390/antiox14121455
Anee TI, Sewelam NA, Bautista NS, Hirayama T, Suzuki N. Roles of ROS and NO in Plant Responses to Individual and Combined Salt Stress and Waterlogging. Antioxidants. 2025; 14(12):1455. https://doi.org/10.3390/antiox14121455
Chicago/Turabian StyleAnee, Taufika Islam, Nasser A. Sewelam, Nonnatus S. Bautista, Takashi Hirayama, and Nobuhiro Suzuki. 2025. "Roles of ROS and NO in Plant Responses to Individual and Combined Salt Stress and Waterlogging" Antioxidants 14, no. 12: 1455. https://doi.org/10.3390/antiox14121455
APA StyleAnee, T. I., Sewelam, N. A., Bautista, N. S., Hirayama, T., & Suzuki, N. (2025). Roles of ROS and NO in Plant Responses to Individual and Combined Salt Stress and Waterlogging. Antioxidants, 14(12), 1455. https://doi.org/10.3390/antiox14121455








