Harnessing γ-TMT Genetic Variations and Haplotypes for Vitamin E Diversity in the Korean Rice Collection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Resequencing Data Processing and Variant Calling for γ-TMT Gene
2.3. Population Structure Analysis
2.4. Nucleotide Diversity, Tajima’s D Analysis, and Fixation Index (FST)
2.5. Haplotype Analysis
2.6. Determination of Vitamin E Isomers by Gas Chromatography (GC)
2.7. Statistical Analysis
3. Results
3.1. Genetic Polymorphisms in the γ-TMT of KRICE
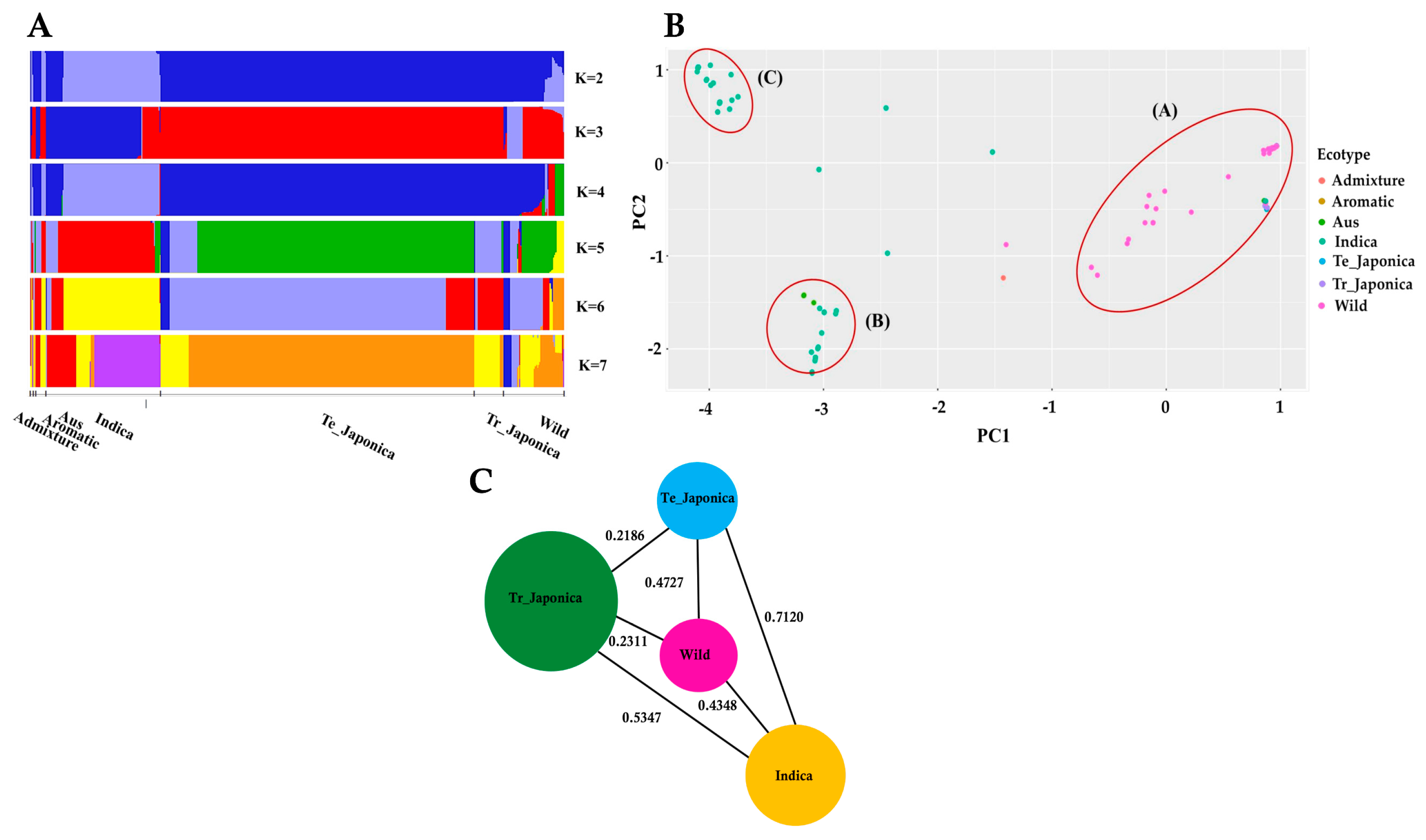
3.2. Population Structure of KRICE Based on the γ-TMT
3.3. Assessment of Population Differentiation via γ-TMT Gene Variations
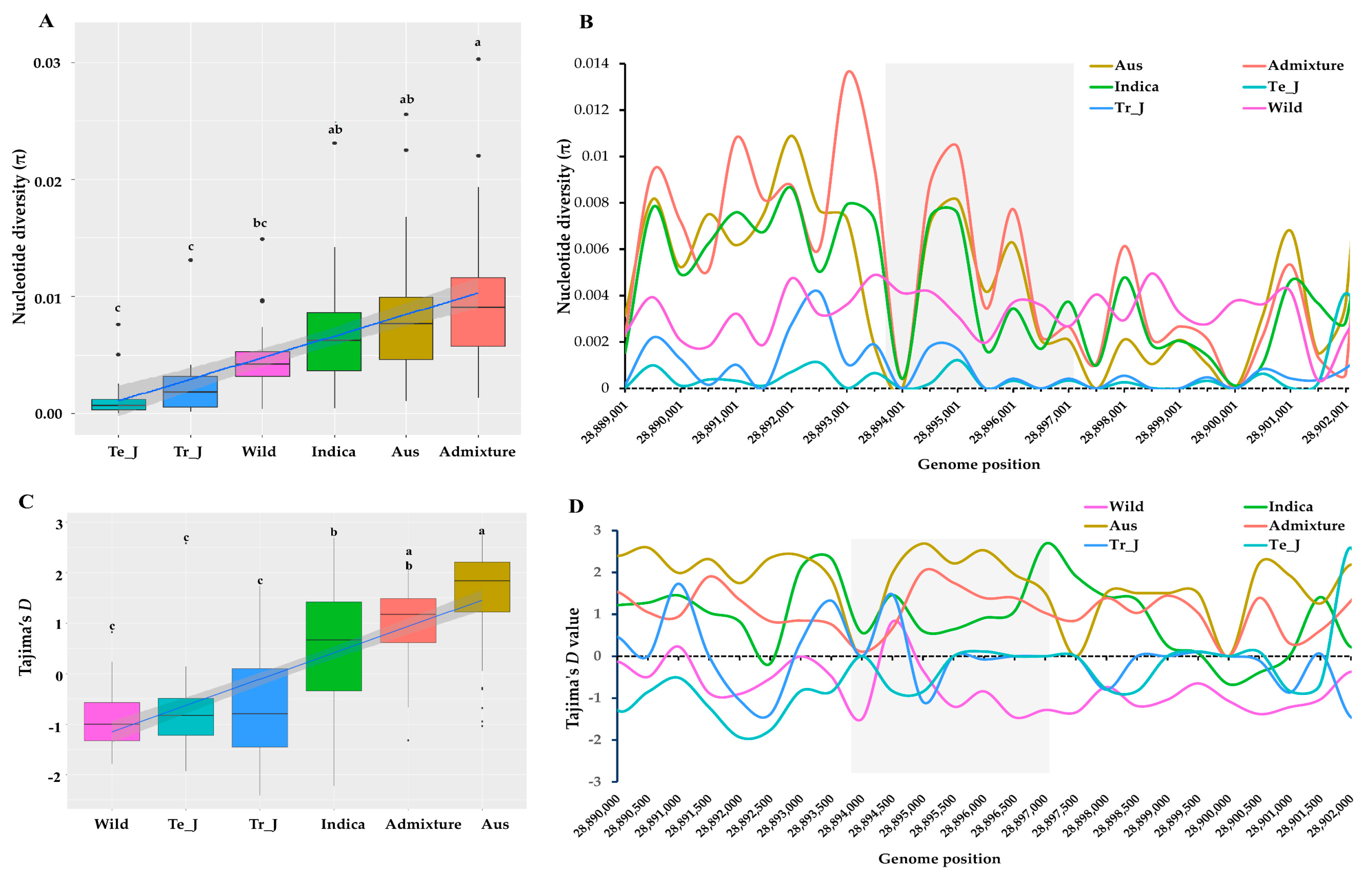
3.4. Polymorphism Assessment in KRICE Groups Based on the γ-TMT Gene
3.5. Insights into Selection Patterns within the γ-TMT Gene
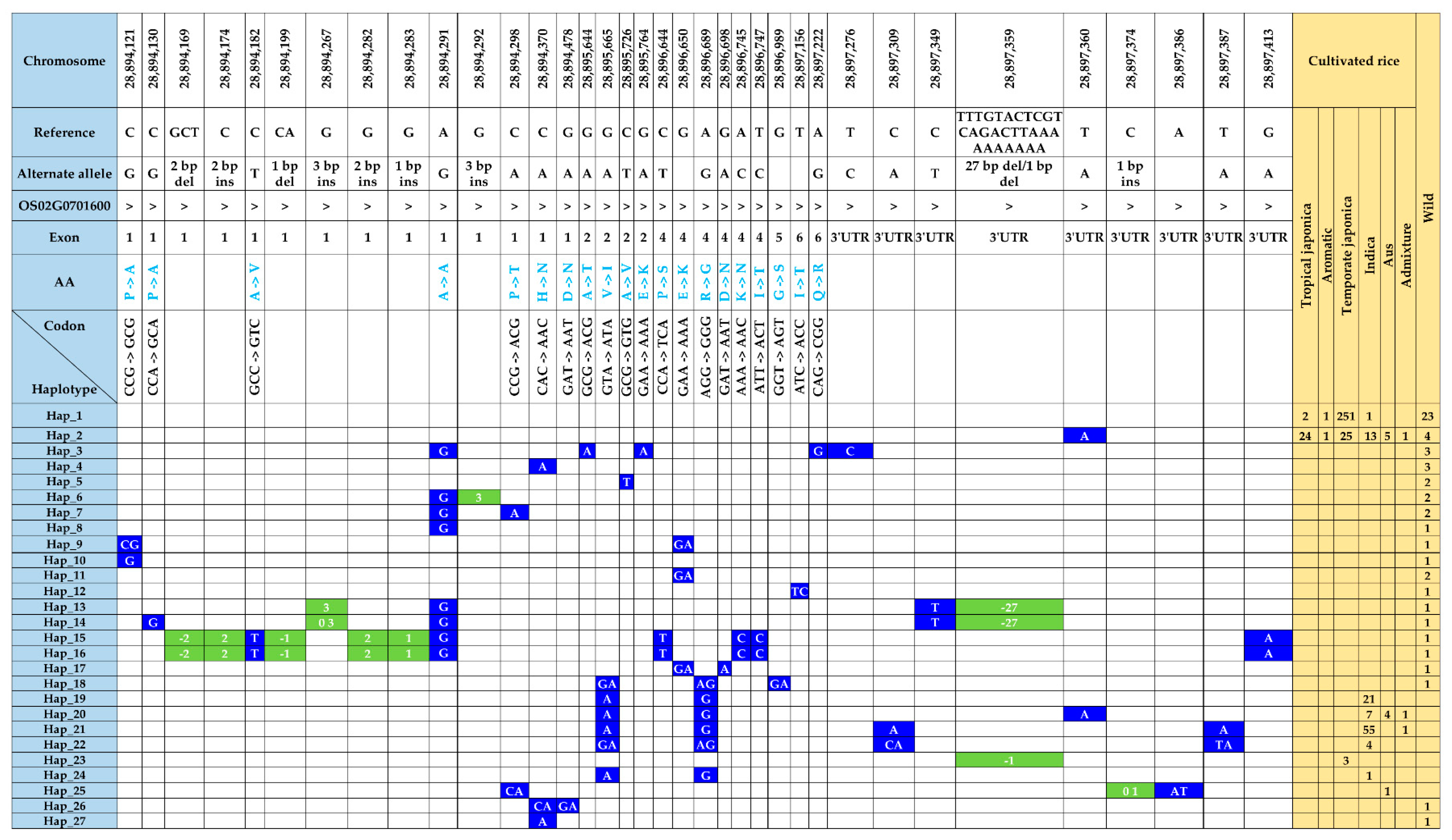
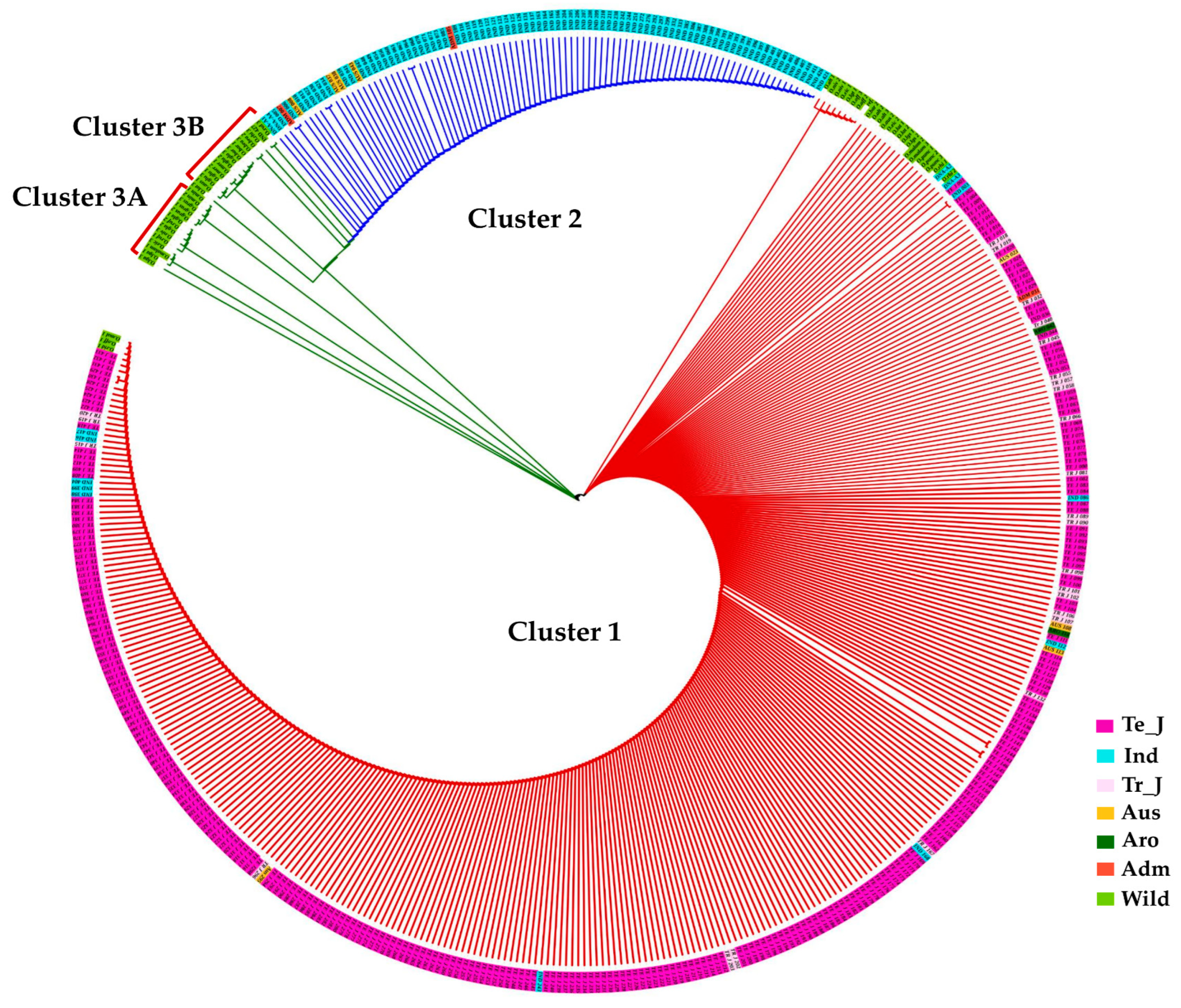
3.6. Functional Polymorphisms in the γ-TMT and Phylogenetic Analysis
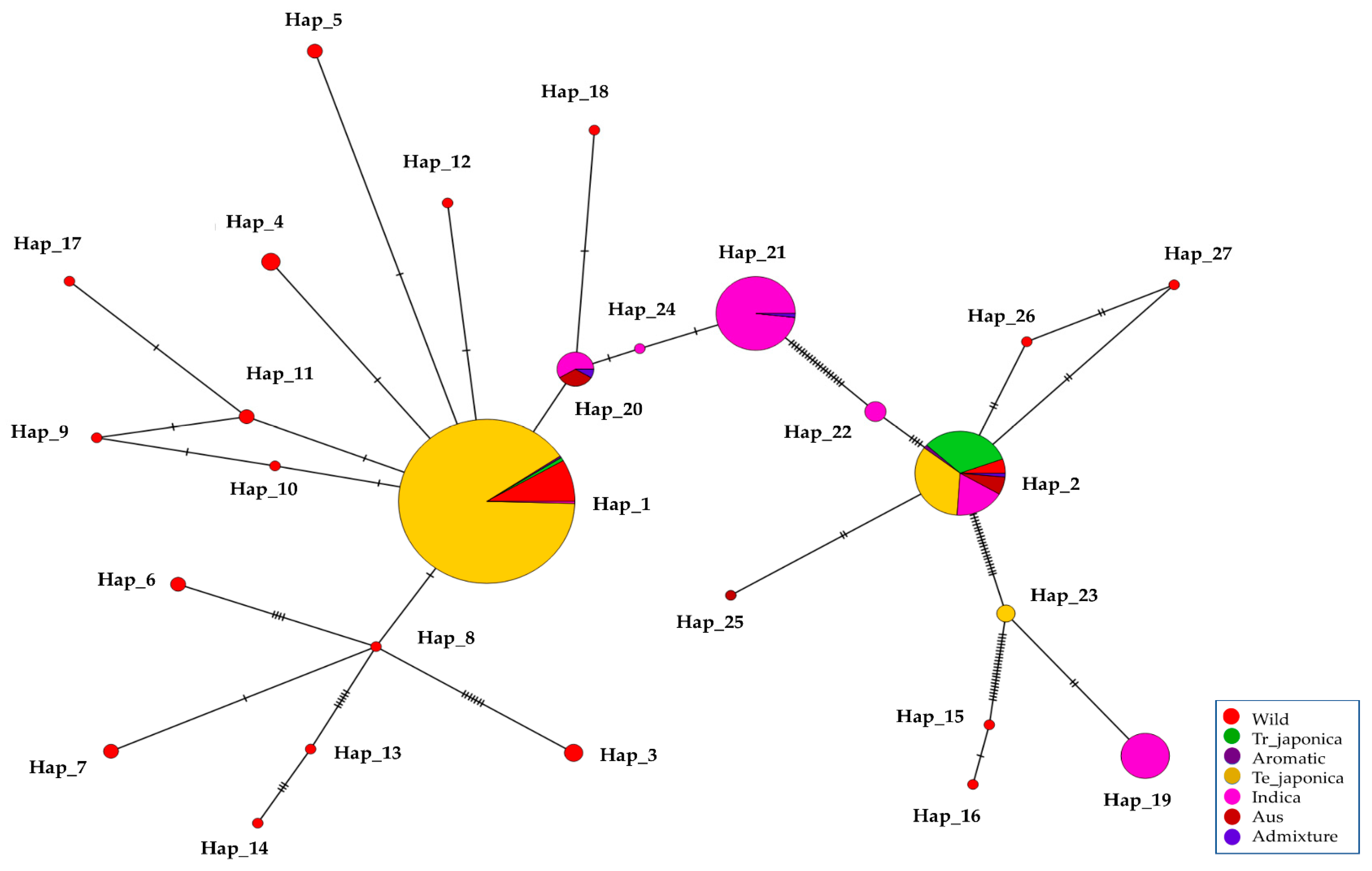
3.7. Haplotype Diversity in the γ-TMT Gene Region
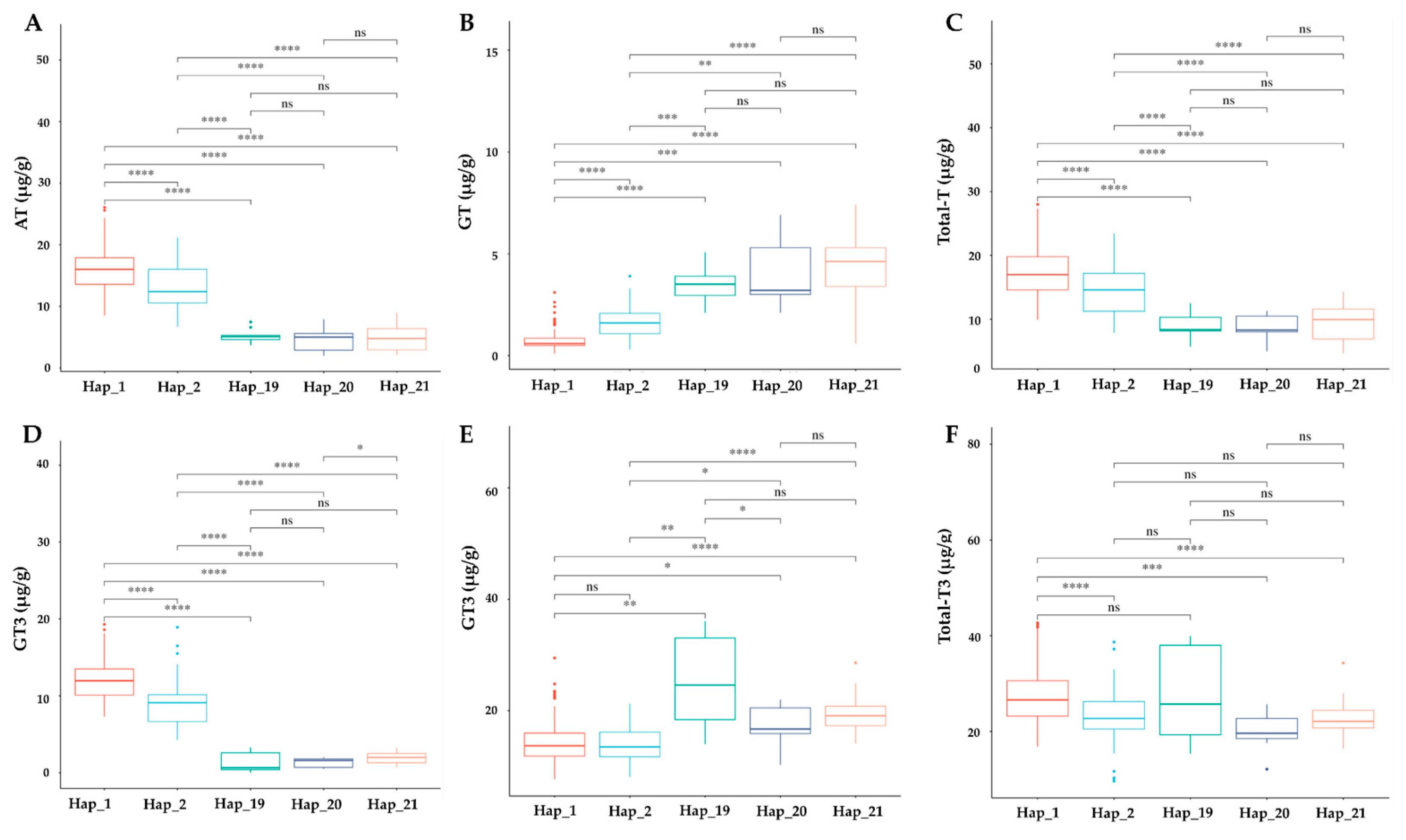
3.8. Phenotypic Effect of TMT Haplotypes on Tocochromanol Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mène-Saffrané, L.; Jones, A.D.; DellaPenna, D. Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 17815–17820. [Google Scholar] [CrossRef]
- Maeda, H.; Sakuragi, Y.; Bryant, D.A.; DellaPenna, D. Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiol. 2005, 138, 1422–1435. [Google Scholar] [CrossRef]
- Muñoz, P.; Briones, M.; Munné-Bosch, S. Photoinhibition and photoprotection during flower opening in lilies. Plant Sci. 2018, 272, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Stacey, M.G.; Cahoon, R.E.; Nguyen, H.T.; Cui, Y.; Sato, S.; Nguyen, C.T.; Phoka, N.; Clark, K.M.; Liang, Y.; Forrester, J.; et al. Identification of homogentisate dioxygenase as a target for vitamin E biofortification in oilseeds. Plant Physiol. 2016, 172, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, K.; Zhu, X.; Wu, Y.; Zhang, S.; Chen, H.; Ling, J.; Wang, Y.; Fang, X. Rice tocopherol deficiency 1 encodes a homogentisate phytyltransferase essential for tocopherol biosynthesis and plant development in rice. Plant Cell Rep. 2018, 37, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Cahoon, R.E.; Hunter, S.C.; Zhang, C.; Han, J.; Borgschulte, T.; Cahoon, E.B. Vitamin E biosynthesis: Functional characterization of the monocot homogentisate geranylgeranyl transferase. Plant J. 2011, 65, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Q.; Yoon, M.-Y.; He, Q.; Kim, T.-S.; Tong, W.; Choi, B.-W.; Lee, Y.-S.; Park, Y.-J. Natural variations in OsγTMT contribute to diversity of the α-tocopherol content in rice. Mol. Genet. Genom. 2015, 290, 2121–2135. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Muthusamy, V.; Zunjare, R.U.; Baveja, A.; Chauhan, H.S.; Bhat, J.S.; Guleria, S.K.; Kumar, B.; Saha, S.; Hossain, F. Genetic variability for kernel tocopherols and haplotype analysis of γ-tocopherol methyl transferase (vte4) gene among exotic- and indigenous- maize inbreds. J. Food Compos. Anal. 2020, 88, 103446. [Google Scholar] [CrossRef]
- Wong, R.S.; Radhakrishnan, A.K. Tocotrienol research: Past into present. Nutr. Rev. 2012, 70, 483–490. [Google Scholar] [CrossRef]
- Tanaka, H.; Yabuta, Y.; Tamoi, M.; Tanabe, N.; Shigeoka, S. Generation of transgenic tobacco plants with enhanced tocotrienol levels through the ectopic expression of rice homogentisate geranylgeranyl transferase. Plant Biotechnol. 2015, 32, 233–238. [Google Scholar] [CrossRef]
- Lushchak, V.I.; Semchuk, N.M. Tocopherol biosynthesis: Chemistry, regulation and effects of environmental factors. Acta Physiol. Plant. 2012, 34, 1607–1628. [Google Scholar] [CrossRef]
- Chaudhary, N.; Khurana, P. Vitamin E biosynthesis genes in rice: Molecular characterization, expression profiling and comparative phylogenetic analysis. Plant Sci. 2009, 177, 479–491. [Google Scholar] [CrossRef]
- Dwiyanti, M.S.; Yamada, T.; Sato, M.; Abe, J.; Kitamura, K. Genetic variation of γ-tocopherol methyltransferase gene contributes to elevated α-tocopherol content in soybean seeds. BMC Plant Biol. 2011, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Mène-Saffrané, L. Vitamin E biosynthesis and its regulation in plants. Antioxidants 2017, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Van Eenennaam, A.L.; Lincoln, K.; Durrett, T.P.; Valentin, H.E.; Shewmaker, C.K.; Thorne, G.M.; Jiang, J.; Baszis, S.R.; Levering, C.K.; Aasen, E.D.; et al. Engineering vitamin E content: From Arabidopsis mutant to soy oil. Plant Cell 2003, 15, 3007–3019. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Quadrana, L.; Asís, R.; Setta, N.; De Godoy, F.; Bermúdez, L.; Otaiza, S.N.; Corrêa Da Silva, J.V.; Fernie, A.R.; Carrari, F.; et al. Genetic dissection of vitamin E biosynthesis in tomato. J. Exp. Bot. 2011, 62, 3781–3798. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.E.; Ahn, J.-W.; Kwon, S.-J.; Kim, J.-B.; Kim, S.H.; Kang, S.-Y.; Kim, D.S. Selection and molecular characterization of a high tocopherol accumulation rice mutant line induced by gamma irradiation. Mol. Biol. Rep. 2014, 41, 7671–7681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lin, Y.; Chen, H. Improving nutritional quality of rice for human health. Theor. Appl. Genet. 2020, 133, 1397–1413. [Google Scholar] [CrossRef]
- Londo, J.P.; Chiang, Y.-C.; Hung, K.-H.; Chiang, T.-Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza Sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar] [CrossRef]
- Hoang, A.T.P.; Prinpreecha, N.; Kim, K.-W. Influence of mining activities on arsenic concentration in rice in Asia: A Review. Minerals 2021, 11, 472. [Google Scholar] [CrossRef]
- Sajid, M.; Khan, S.A.; Khurshid, H.; Iqbal, J.; Muhammad, A.; Saleem, N.; Shah, S.M.A. Characterization of rice (Oryza Sativa L.) germplasm through various agro-morphological traits. Sci. Agric. 2015, 9, 83–88. [Google Scholar] [CrossRef]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza Sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef]
- Oka, H.I. Origin of Cultivated Rice; Elsevier: Amsterdam, The Netherlands, 2012; Volume 14, ISBN 0-444-98919-6. [Google Scholar]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Bhandari, A.; Sandhu, N.; Bartholome, J.; Cao-Hamadoun, T.-V.; Ahmadi, N.; Kumari, N.; Kumar, A. Genome-wide association study for yield and yield related traits under reproductive stage drought in a diverse indica-aus rice panel. Rice 2020, 13, 53. [Google Scholar] [CrossRef]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Shammugasamy, B.; Ramakrishnan, Y.; Ghazali, H.M.; Muhammad, K. Tocopherol and tocotrienol contents of different varieties of rice in Malaysia. J. Sci. Food Agric. 2015, 95, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, D.; Liu, T.; Liao, M.; Li, Y.; Zhang, W.; Liu, Z.; Chen, M. Effect of overexpression of γ-tocopherol methyltransferase on α-tocopherol and fatty acid accumulation and tolerance to salt stress during seed germination in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 15933. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, X.; Xu, S.; Cai, Y.; Zhang, D.; Han, Y.; Li, L.; Zhang, Z.; Gao, S.; Li, J.; et al. Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 2012, 7, e36807. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Yu, Y.; Mao, J.; Liu, H.; Hu, J.; Li, T.; Guo, X.; Liu, R. Evaluation of biosynthesis, accumulation and antioxidant activity of vitamin E in sweet corn (Zea Mays L.) during kernel development. Int. J. Mol. Sci. 2017, 18, 2780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-Y.; Liu, R.-R.; Zhang, P.; Xu, Y.; Zhu, J.; Gu, M.-H.; Liang, G.-H.; Liu, Q.-Q. Variation and distribution of vitamin E and composition in seeds among different rice varieties. Acta Agron. Sin. 2012, 38, 55–61. [Google Scholar] [CrossRef]
- Shintani, D.; DellaPenna, D. Elevating the vitamin E content of plants through metabolic engineering. Science 1998, 282, 2098–2100. [Google Scholar] [CrossRef]
- Zhang, G.-Y.; Liu, R.-R.; Xu, G.; Zhang, P.; Li, Y.; Tang, K.-X.; Liang, G.-H.; Liu, Q.-Q. Increased α-tocotrienol content in seeds of transgenic rice overexpressing Arabidopsis γ-Tocopherol Methyltransferase. Transgenic. Res. 2013, 22, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-W.; Chung, H.-K.; Cho, G.-T.; Ma, K.-H.; Chandrabalan, D.; Gwag, J.-G.; Kim, T.-S.; Cho, E.-G.; Park, Y.-J. PowerCore: A program applying the advanced m strategy with a heuristic search for establishing core sets. Bioinformatics 2007, 23, 2155–2162. [Google Scholar] [CrossRef]
- Phitaktansakul, R.; Kim, K.-W.; Aung, K.M.; Maung, T.Z.; Min, M.-H.; Somsri, A.; Lee, W.; Lee, S.-B.; Nam, J.; Kim, S.-H.; et al. Multi-omics analysis reveals the genetic basis of rice fragrance mediated by betaine aldehyde dehydrogenase 2. J. Adv. Res. 2022, 42, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef]
- Francis, R.M. POPHELPER: An R package and web App to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.R.; Basnet, S.; Chung, K.H.; Ryu, K.-H.; Lee, Y.-S. Comparisons of nutritional and phytochemical property of genetically modified cmv-resistant red pepper and its parental cultivar. Hortic. Environ. Biotechnol. 2012, 53, 151–157. [Google Scholar] [CrossRef]
- Bergman, C.J.; Xu, Z. Genotype and environment effects on tocopherol, tocotrienol, and γ-Oryzanol contents of southern U.S. rice. Cereal Chem. 2003, 80, 446–449. [Google Scholar] [CrossRef]
- Dror, D.K.; Allen, L.H. Vitamin E deficiency in developing countries. Food Nutr. Bull. 2011, 32, 124–143. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, T.; Burdeos, G.C.; Itaya, M.; Nakagawa, K.; Miyazawa, T. Vitamin E: Regulatory redox interactions. IUBMB Life 2019, 71, 430–441. [Google Scholar] [CrossRef]
- Fritsche, S.; Wang, X.; Jung, C. Recent advances in our understanding of tocopherol biosynthesis in plants: An overview of key genes, functions, and breeding of vitamin E improved crops. Antioxidants 2017, 6, 99. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, K.; Xia, H.; Chen, L.; Chen, K. Comparative proteomic analysis of indica and japonica rice varieties. Genet. Mol. Biol. 2014, 37, 652–661. [Google Scholar] [CrossRef]
- Palanisamy, S. Genetic analysis of biofortification of micronutrient breeding in rice (Oryza Sativa L.). In Rice Crop—Current Developments; Shah, F., Khan, Z.H., Iqbal, A., Eds.; InTech: London, UK, 2018; ISBN 978-1-78923-600-2. [Google Scholar]
- Reig-Valiente, J.L.; Viruel, J.; Sales, E.; Marqués, L.; Terol, J.; Gut, M.; Derdak, S.; Talón, M.; Domingo, C. Genetic diversity and population structure of rice varieties cultivated in temperate regions. Rice 2016, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Min, M.-H.; Maung, T.Z.; Cao, Y.; Phitaktansakul, R.; Lee, G.-S.; Chu, S.-H.; Kim, K.-W.; Park, Y.-J. Haplotype analysis of Badh1 by next-generation sequencing reveals association with salt tolerance in rice during domestication. Int. J. Mol. Sci. 2021, 22, 7578. [Google Scholar] [CrossRef] [PubMed]
- Zhai, T.; Yang, H.-Q.; Zhang, R.-C.; Fang, L.-M.; Zhong, G.-H.; Fang, S.-G. Effects of population bottleneck and balancing selection on the chinese alligator are revealed by locus-specific characterization of MHC genes. Sci. Rep. 2017, 7, 5549. [Google Scholar] [CrossRef]
- Carlson, C.S.; Thomas, D.J.; Eberle, M.A.; Swanson, J.E.; Livingston, R.J.; Rieder, M.J.; Nickerson, D.A. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005, 15, 1553–1565. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.A.; Yu, D.; Bohra, A.; Ganie, S.A.; Varshney, R.K. Features and applications of haplotypes in crop breeding. Commun. Biol. 2021, 4, 1266. [Google Scholar] [CrossRef]
- Zhao, H.; Li, J.; Yang, L.; Qin, G.; Xia, C.; Xu, X.; Su, Y.; Liu, Y.; Ming, L.; Chen, L.-L.; et al. An inferred functional impact map of genetic variants in rice. Mol. Plant 2021, 14, 1584–1599. [Google Scholar] [CrossRef]
- Mignone, F. UTRdb and UTRsite: A Collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2004, 33, D141–D146. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Peng, B.; Kong, H.; Li, Y.; Wang, L.; Zhong, M.; Sun, L.; Gao, G.; Zhang, Q.; Luo, L.; Wang, G.; et al. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 2014, 5, 4847. [Google Scholar] [CrossRef]
- Gong, D.; Zhang, X.; He, F.; Chen, Y.; Li, R.; Yao, J.; Zhang, M.; Zheng, W.; Yu, G. Genetic improvements in rice grain quality: A review of elite genes and their applications in molecular breeding. Agronomy 2023, 13, 1375. [Google Scholar] [CrossRef]
- Cai, X.; Wang, Z.; Xing, Y.; Zhang, J.; Hong, M. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of Waxy gene in rice cultivars of intermediate amylose content. Plant J. 1998, 14, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, N.; Cai, Y.; Yang, Q.; Yu, L.; Chen, Z.; Shi, W.; Liu, J.; Pan, C.; Li, Y.; et al. CRISPR-Cas9-mediated editing of the OsHPPD 3′ UTR confers enhanced resistance to HPPD-inhibiting herbicides in rice. Plant Commun. 2023, 4, 100605. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somsri, A.; Chu, S.-H.; Nawade, B.; Lee, C.-Y.; Park, Y.-J. Harnessing γ-TMT Genetic Variations and Haplotypes for Vitamin E Diversity in the Korean Rice Collection. Antioxidants 2024, 13, 234. https://doi.org/10.3390/antiox13020234
Somsri A, Chu S-H, Nawade B, Lee C-Y, Park Y-J. Harnessing γ-TMT Genetic Variations and Haplotypes for Vitamin E Diversity in the Korean Rice Collection. Antioxidants. 2024; 13(2):234. https://doi.org/10.3390/antiox13020234
Chicago/Turabian StyleSomsri, Aueangporn, Sang-Ho Chu, Bhagwat Nawade, Chang-Yong Lee, and Yong-Jin Park. 2024. "Harnessing γ-TMT Genetic Variations and Haplotypes for Vitamin E Diversity in the Korean Rice Collection" Antioxidants 13, no. 2: 234. https://doi.org/10.3390/antiox13020234
APA StyleSomsri, A., Chu, S.-H., Nawade, B., Lee, C.-Y., & Park, Y.-J. (2024). Harnessing γ-TMT Genetic Variations and Haplotypes for Vitamin E Diversity in the Korean Rice Collection. Antioxidants, 13(2), 234. https://doi.org/10.3390/antiox13020234





