First Characterization of a Cyanobacterial Xi-Class Glutathione S-Transferase in Synechocystis PCC 6803
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions and Gene-Transfer Procedures
2.2. Construction of ΔsufR::KmR Deletion Cassette for Targeted Deletion of sufR Gene
2.3. Construction of Δslr0605::KmR Deletion Cassette for Targeted Deletion of slr0605 Gene
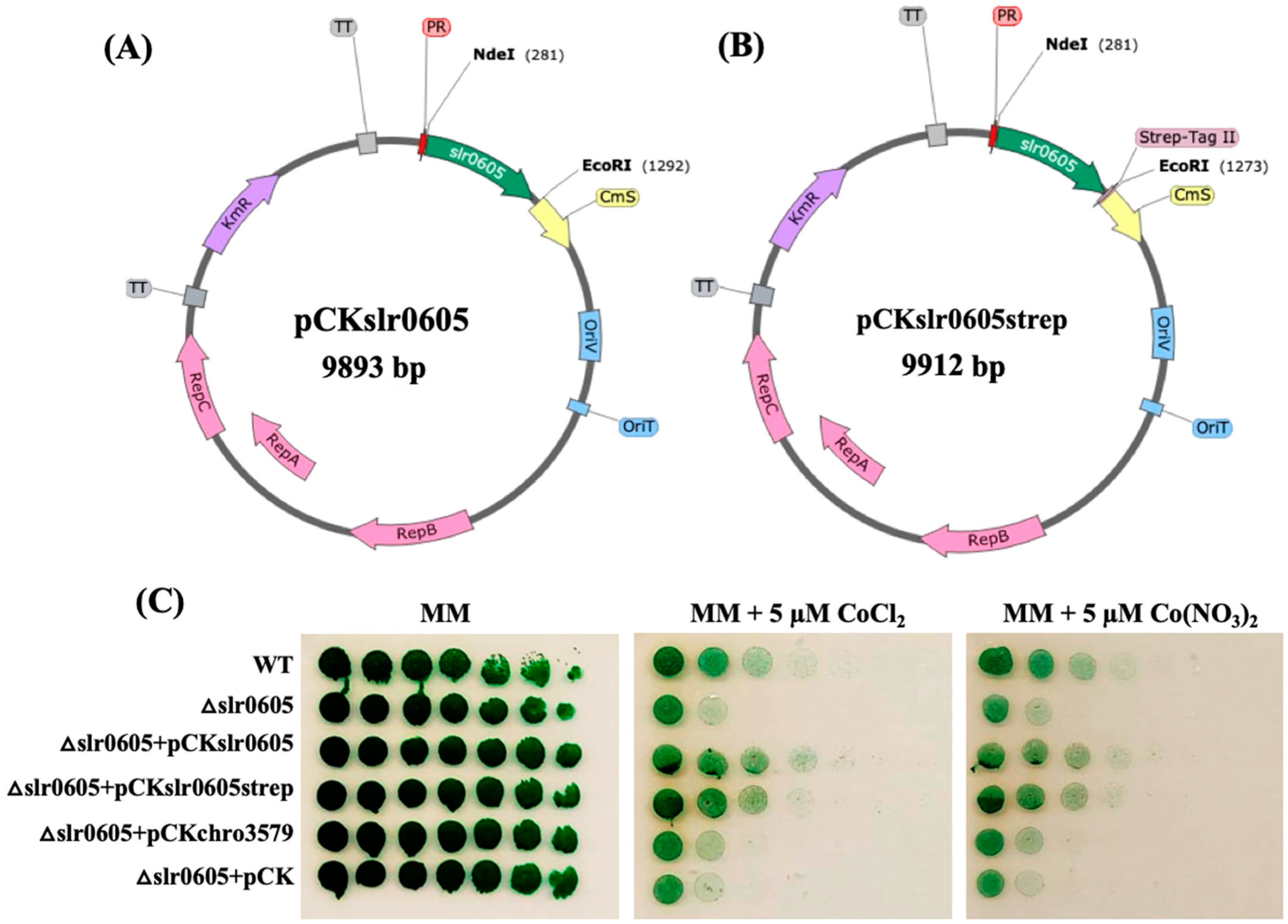
2.4. Construction of pCKslr0605 and pCKslr0605 Plasmids Expressing slr0605 Gene, or Its Strep-Tagged Derivative, Respectively
2.5. Pigment Extraction and Quantification
2.6. Reactive Oxygen Species (ROS) Assay
2.7. Protein Extraction and Purification
2.8. Western Blot Analysis
2.9. Catalase Assay
2.10. Superoxide Dismutase Assay
2.11. Measurement of Glutathionyl–Hydroquinone Reductase Activity
2.12. Statistical Analyses
3. Results and Discussion
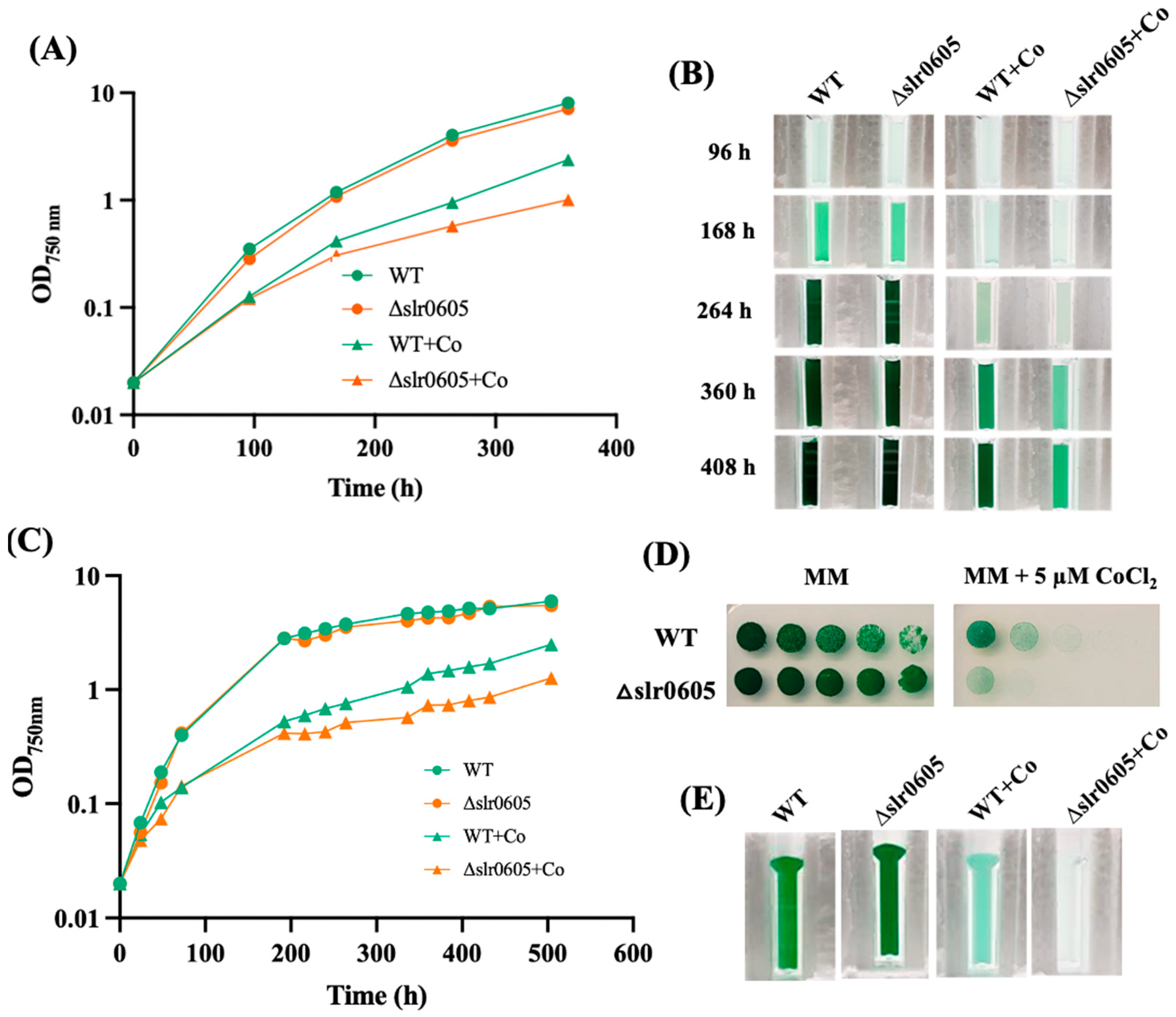
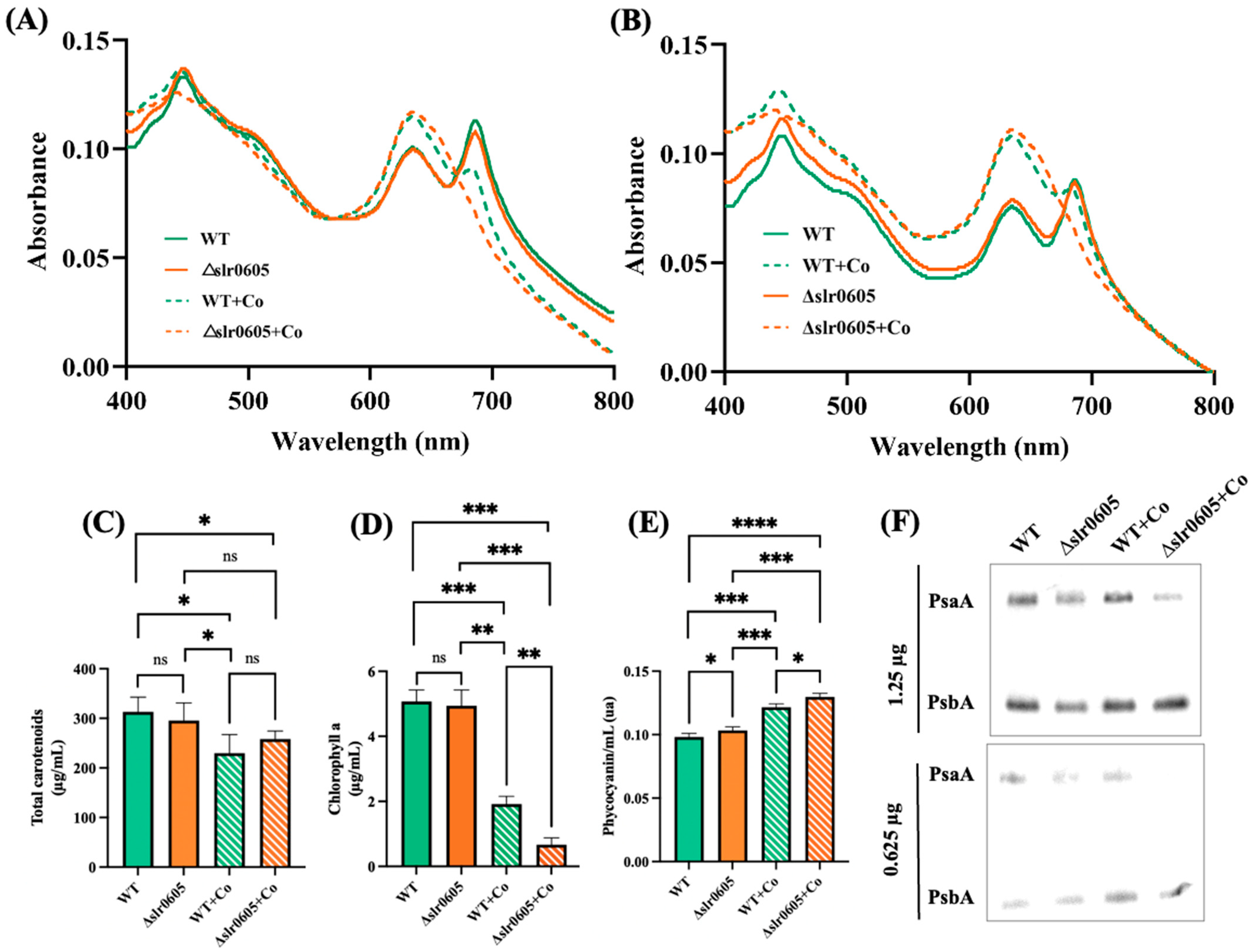
3.1. Synechocystis PCC 6803 Is Particularly Sensitive to Cobalt That Reduces Chlorophyll Content
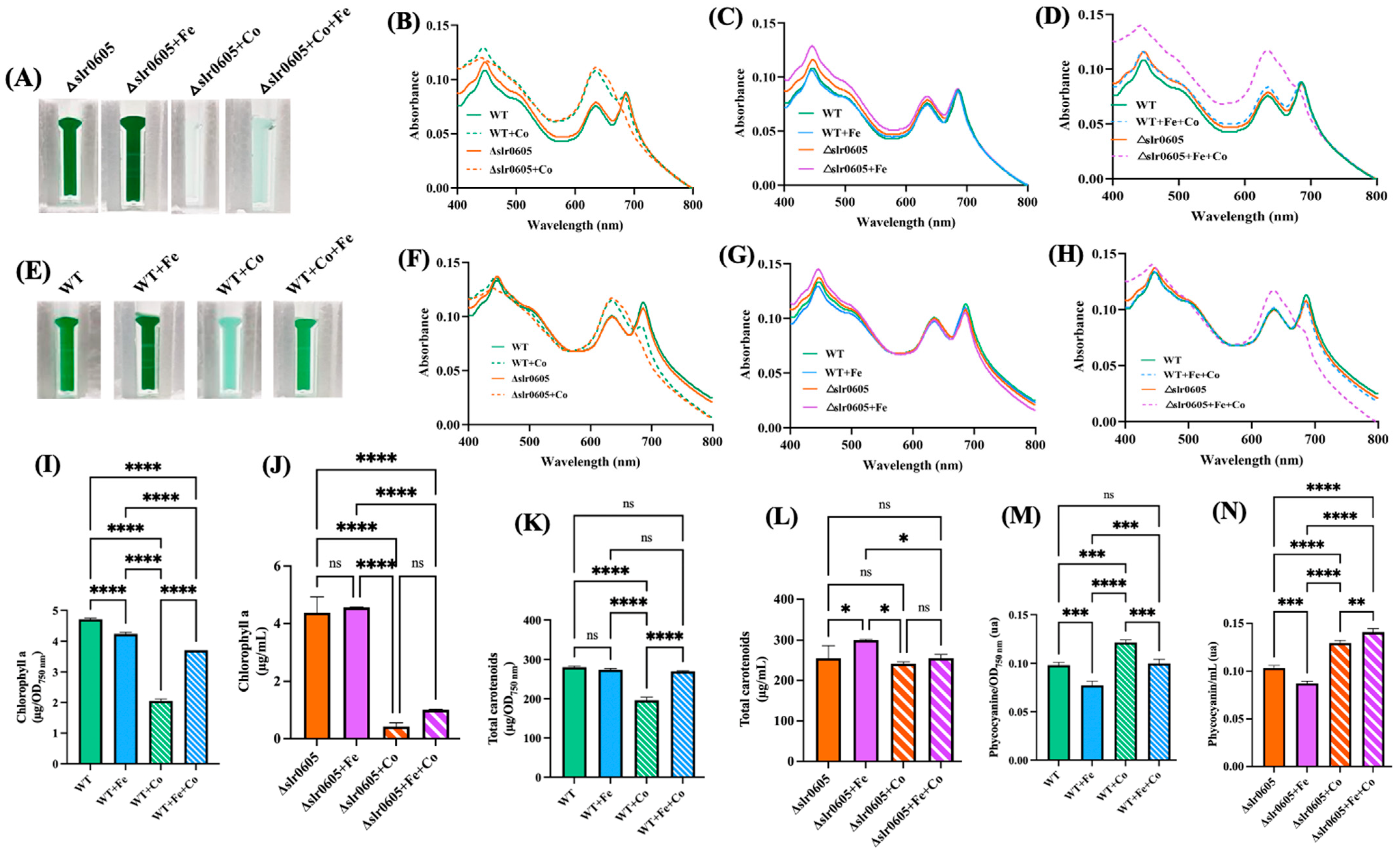
3.2. Iron Supplementation Protects Synechocystis Against Cobalt-Elicited Declines of Growth and Pigments
3.3. Activation of Iron–Sulfur Cluster Repair Genes Alleviates Cobalt Toxicity
3.4. Slr0605 GST Contributes to Cobalt Resistance in Synechocystis
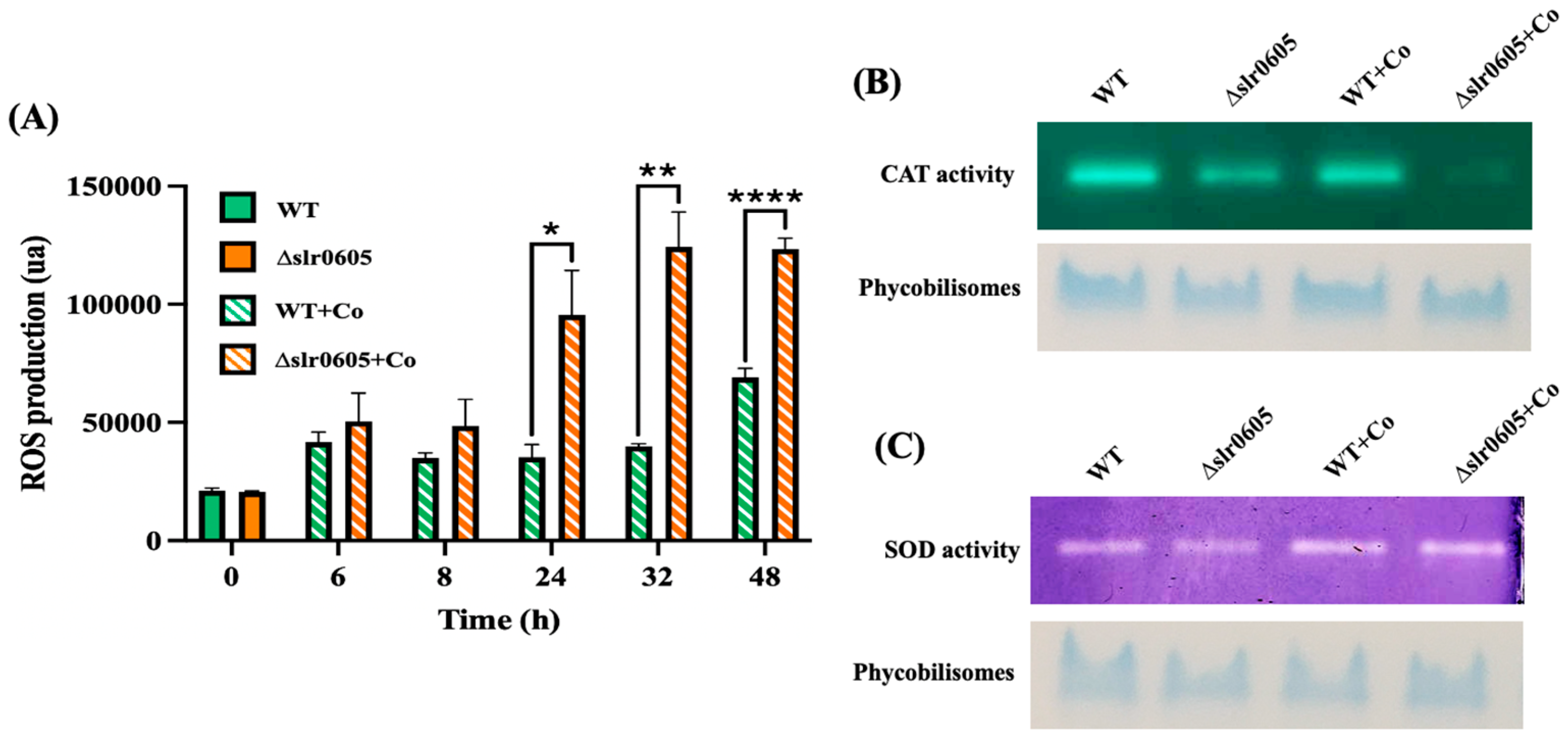
3.5. Cobalt-Elicited Decline of Catalase Activity and Accumulation of ROS Are Exacerbated in the Δslr0605 Mutant as Compared to WT Strain
3.6. Slr0605 Contributes to Iron Mitigation of Cobalt Toxicity
3.7. The Restoration of Cobalt Tolerance in the Δslr0605 Mutant by Expressing the slr0605 Gene, or Its Strep-Tagged Derivative, from a Replicative Plasmid
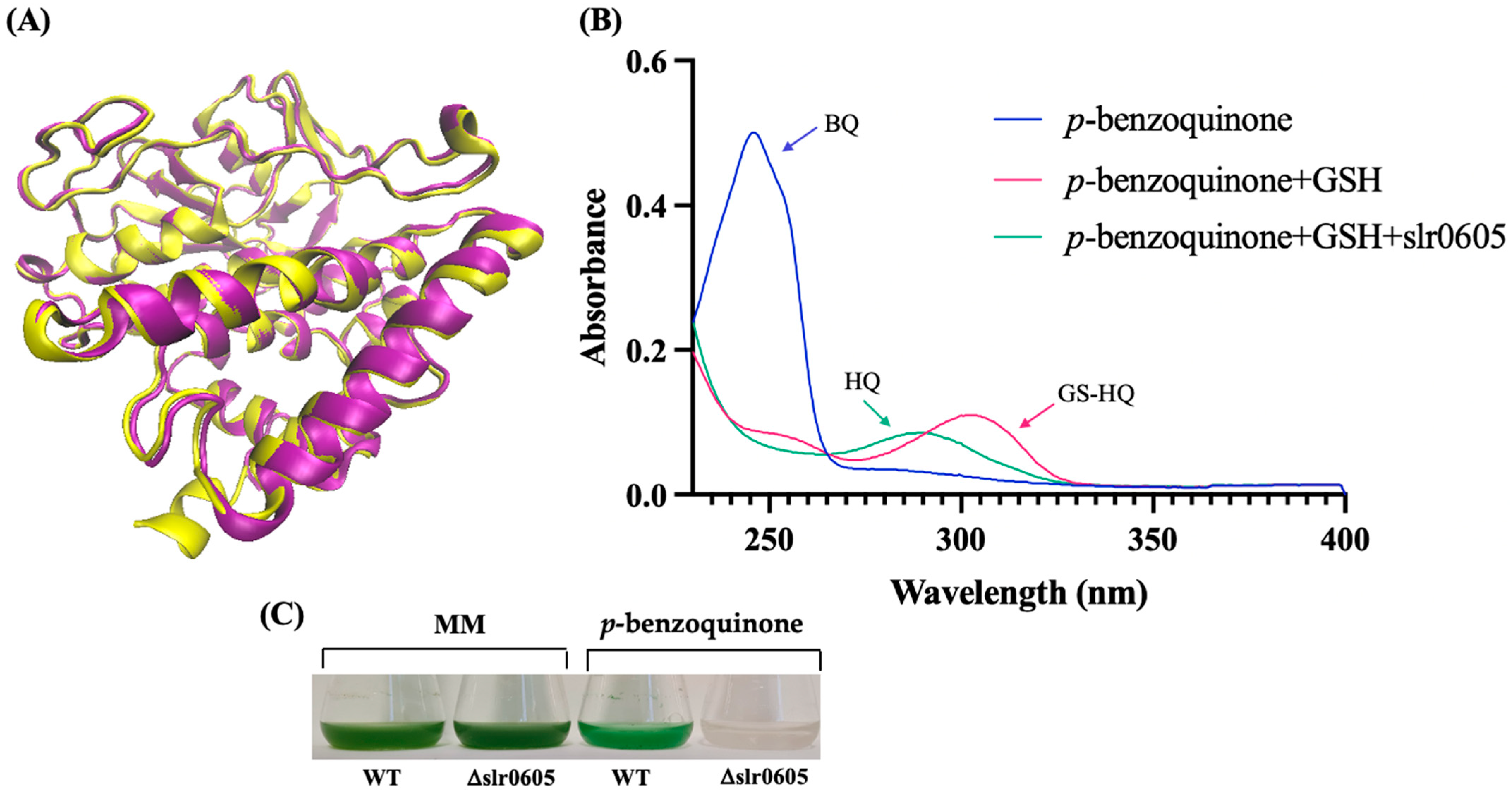
3.8. Slr0605 Is a Genuine Glutathionyl–Hydroquinone Reductase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cassier-Chauvat, C.; Marceau, F.; Farci, S.; Ouchane, S.; Chauvat, F. The Glutathione System: A Journey from Cyanobacteria to Higher Eukaryotes. Antioxidants 2023, 12, 1199. [Google Scholar] [CrossRef] [PubMed]
- Josephy, P.D. Genetic Variations in Human Glutathione Transferase Enzymes: Significance for Pharmacology and Toxicology. Hum. Genom. Proteom. 2010, 2010, 876940. [Google Scholar] [CrossRef] [PubMed]
- Sing, R.R.; Reindi, K.M. Glutathione S-Transferases in Cancer. Antioxidants 2021, 10, 701. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Kiraly, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Gallé, Á.; Czékus, Z.; Bela, K.; Horváth, E.; Ördög, A.; Csiszár, J.; Poór, P. Plant Glutathione Transferases and Light. Front. Plant Sci. 2019, 9, 1944. [Google Scholar] [CrossRef] [PubMed]
- Kammerscheit, X.; Hecker, A.; Rouhier, N.; Chauvat, F.; Cassier-Chauvat, C. Methylglyoxal Detoxification Revisited: Role of Glutathione Transferase in Model Cyanobacterium Synechocystis sp. Strain PCC 6803. mBio 2020, 11, e00882-20. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; Park, E.-H.; Fuchs, J.A.; Lim, C.-J. Characterization, Expression and Regulation of a Third Gene Encoding Glutathione S-Transferase from the Fission Yeast. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 2002, 1577, 164–170. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef]
- Schwartz, M.; Perrot, T.; Aubert, E.; Dumarçay, S.; Favier, F.; Gérardin, P.; Morel-Rouhier, M.; Mulliert, G.; Saiag, F.; Didierjean, C.; et al. Molecular Recognition of Wood Polyphenols by Phase II Detoxification Enzymes of the White Rot Trametes versicolor. Sci. Rep. 2018, 8, 8472. [Google Scholar] [CrossRef]
- Kalinina, E.; Novichkova, M. Glutathione in Protein Redox Modulation through S-Glutathionylation and S-Nitrosylation. Molecules 2021, 26, 435. [Google Scholar] [CrossRef] [PubMed]
- Musaogullari, A.; Chai, Y.C. Redox Regulation by Protein S-Glutathionylation: From Molecular Mechanisms to Implications in Health and Disease. Int. J. Mol. Sci. 2020, 21, 8113. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, A.; Federici, L.; Masuli, M.; Carletti, E.; Santorelli, D.; Paradisi, P.; Di Ilio, C.; Allocati, N. Structural Characterization of the Xi Class Glutathione Transferase From the Haloalkaliphilic Archaeon Natrialba magadii. Front. Microbiol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Green, A.R.; Hayes, R.P.; Xun, L.; Kang, C. Structural Understanding of the Glutathione-Dependent Reduction Mechanism of Glutathionyl-Hydroquinone Reductases. J. Biol. Chem. 2012, 287, 35838–35848. [Google Scholar] [CrossRef] [PubMed]
- Lallement, P.-A.; Meux, E.; Gualberto, J.M.; Dumarcay, S.; Favier, F.; Didierjean, C.; Saul, F.; Haouz, A.; Morel-Rouhier, M.; Gelhaye, E.; et al. Glutathionyl-Hydroquinone Reductases from Poplar Are Plastidial Proteins That Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones. FEBS Lett. 2015, 539, 37–44. [Google Scholar] [CrossRef]
- Fahey, R.C.; Buschbacher, R.M.; Newton, G.L. The Evolution of Glutathione Metabolism in Phototrophic Microorganisms. J. Mol. Evol. 1987, 25, 81–88. [Google Scholar] [CrossRef]
- ShylajaNaciyar, M.; Karthick, L.; Prakasam, P.A.; Deviram, G.; Uma, L.; Prabaharan, D.; Saha, S.K. Diversity of Glutathione S-Transferases (GSTs) in Cyanobacteria with Reference to Their Structures, Substrate Recognition and Catalytic Functions. Microorganisms 2020, 8, 712. [Google Scholar] [CrossRef] [PubMed]
- Partensky, F.; Hess, W.R.; Vaulot, D. Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance. Microbiol. Mol. Biol. Rev. 1999, 63, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Cassier-Chauvat, C.; Chauvat, F. Responses to Oxidative and Heavy Metal Stresses in Cyanobacteria: Recent Advances. Int. J. Mol. Sci. 2015, 16, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef]
- Victoria, A.J.; Asrbury, M.J.; McCormick, A.J. Engineering Highly Productive Cyanobacteria towards Carbon Negative Emissions Technologies. Curr. Opin. Biotechnol. 2024, 87, 103141. [Google Scholar] [CrossRef] [PubMed]
- Kammerscheit, X.; Chauvat, F.; Cassier-Chauvat, C. First In Vivo Evidence That Glutathione-S-Transferase Operates in Photo-Oxidative Stress in Cyanobacteria. Front. Microbiol. 2019, 10, 1899. [Google Scholar] [CrossRef]
- Pandey, T.; Chhetri, G.; Chinta, R.; Kumar, B.; Singh, D.B.; Tripathi, T.; Singh, A.K. Functional Classification and Biochemical Characterization of a Novel Rho Class Glutathione S-Transferase in Synechocystis PCC 6803. FEBS Open Bio 2015, 5, 1–7. [Google Scholar] [CrossRef]
- Pandey, T.; Shukla, R.; Shukla, H.; Sonkar, A.; Tripathi, T.; Singh, A.K. A Combined Biochemical and Computational Studies of the Rho-Class Glutathione s-Transferase Sll1545 of Synechocystis PCC 6803. Int. J. Biol. Macromol. 2017, 94, 378–385. [Google Scholar] [CrossRef]
- Kammerscheit, X.; Chauvat, F.; Cassier-Chauvat, C. From Cyanobacteria to Human, MAPEG-Type Glutathione-S-Transferases Operate in Cell Tolerance to Heat, Cold, and Lipid Peroxidation. Front. Microbiol. 2019, 10, 2248. [Google Scholar] [CrossRef]
- Pandey, T.; Singh, S.K.; Chhetri, G.; Tripathi, T.; Singh, A.K. Characterization of a Highly pH Stable Chi-Class Glutathione S-Transferase from Synechocystis PCC 6803. PLoS ONE 2015, 10, e0126811. [Google Scholar] [CrossRef] [PubMed]
- Mocchetti, E.; Morette, L.; Mulliert, G.; Mathiot, S.; Guillot, B.; Dehez, F.; Chauvat, F.; Cassier-Chauvat, C.; Brochier-Armanet, C.; Didierjean, C.; et al. Biochemical and Structural Characterization of Chi-Class Glutathione Transferases: A Snapshot on the Glutathione Transferase Encoded by Sll0067 Gene in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Biomolecules 2022, 12, 1466. [Google Scholar] [CrossRef]
- Thornalley, P.J. Protein and Nucleotide Damage by Glyoxal and Methylglyoxal in Physiological Systems-Role in Ageing and Disease. Drug Metab. Drug Interact. 2008, 23, 125–150. [Google Scholar] [CrossRef] [PubMed]
- Hawco, N.J.; Mcilvin, M.M.; Bundy, R.M.; TagliabueSaito, A.; Goepfert, T.J.; Moran, D.M.; Valentin-Alvarado, L.; DiTullio, G.R.; Saito, M.A. Minimal Cobalt Metabolism in the Marine Cyanobacterium Prochlorococcus. Proc. Natl. Acad. Sci. USA 2020, 117, 15740–15747. [Google Scholar] [CrossRef]
- Gleason, F.K.; Wood, J.M. Ribonucleotide Reductase in Blue-Green Algae: Dependence on Adenosylcobalamin. Science 1976, 192, 1343–1344. [Google Scholar] [CrossRef] [PubMed]
- Barras, F.; Fontecave, M. Cobalt Stress in Escherichia Coli and Salmonella Enterica: Molecular Bases for Toxicity and Resistance. Metallomics 2011, 3, 1130–1134. [Google Scholar] [CrossRef]
- García-Domínguez, M.; Lopez-Maury, L.; Florencio, F.J. A Gene Cluster Involved in Metal Homeostasis in the Cyanobacterium Synechocystis. J. Bacteriol. 2000, 182, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and Properties of Unicellular Blue-Green Algae (Order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef] [PubMed]
- Labarre, J.; Chauvat, F.; Thuriaux, P. Insertional Mutagenesis by Random Cloning of Antibiotic Resistance Genes into the Genome of the Cyanobacterium Synechocystis Strain PCC 6803. J. Bacteriol. 1989, 171, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
- Mermet-Bouvier, P.; Chauvat, F. A Conditional Expression Vector for the Cyanobacteria Synechocystis sp. Strains PCC6803 and PCC6714 or Synechococcus sp. Strains PCC7942 and PCC6301. Curr. Microbiol. 1994, 28, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, R.J. Consistent Sets of Spectrophotometric Chlorophyll Equations for Acetone, Methanol and Ethanol Solvents. Photosynth. Res. 2006, 89, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R. The Spectral Determination of Chlorophylls a and b, as Well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. Chlorosis Induced by Nutrient Deprivation in Synechococcus sp. Strain PCC 7942: Not All Bleaching Is the Same. J. Bacteriol. 1992, 174, 4718–4726. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Fernandes, E.; Lima, J. Fluorescence Probes Used for Detection of Reactive Oxygen Species. J. Biochem. Biophys. Methods 2005, 65, 45–80. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; Volume 112, pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Haan, C.; Behrmann, I. A Cost Effective Non-Commercial ECL-Solution for Western Blot Detections Yielding Strong Signals and Low Background. J. Immunol. Methods 2007, 118, 11–19. [Google Scholar] [CrossRef]
- Lorusso, C.; Calisi, A.; Sarà, G.; Dondero, F. In-Gel Assay to Evaluate Antioxidant Enzyme Response to Silver Nitrate and Silver Nanoparticles in Marine Bivalve Tissues. Appl. Sci. 2022, 12, 2760. [Google Scholar] [CrossRef]
- Meux, E.; Prosper, P.; Ngadin, A.; Didierjean, C.; Morel, M.; Dumarçay, S.; Lamant, T.; Jacquot, J.P.; Favier, F.; Gelhaye, E. Glutathione Transferases of Phanerochaete chrysosporium: S-Glutathionyl-p-Hydroquinone Reductase Belongs to a New Structural Class. J. Biol. Chem. 2011, 286, 9162–9173. [Google Scholar] [CrossRef] [PubMed]
- West, R.M. Best Practice in Statistics: Use the Welch t-Test When Testing the Difference between Two Groups. Ann. Clin. Biochem. 2021, 58, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Willson, V.L. One-Way Anova. In Basic and Advanced Statistical Tests; SensePublishers: Rotterdam, The Netherlands, 2017; pp. 21–24. ISBN 978-94-6351-086-8. [Google Scholar]
- Huertas, M.J.; López-Maury, L.; Giner-Lamia, J.; Sánchez-Riego, A.M.; Florencio, F.J. Metals in Cyanobacteria: Analysis of the Copper, Nickel, Cobalt and Arsenic Homeostasis Mechanisms. Life 2014, 9, 865–886. [Google Scholar] [CrossRef]
- Babu, T.S.; Sabat, S.C.; Mohanty, P. Alterations in Photosystem II Organization by Cobalt Treatment in the Cyanobacterium Spirulina platensis. J. Plant Biochem. Biotechnol. 1992, 1, 61–63. [Google Scholar] [CrossRef]
- Lee, L.H.; Lustigman, B.; Chu, I.-Y.; Hsu, S. Effect of Lead and Cobalt on the Growth of Anacystis nidulans. Bull. Environ. Contam. Toxicol. 1992, 48, 230–236. [Google Scholar] [CrossRef]
- Martín-Betancor, K.; Aguado, S.; Rodea-Palomares, I.; Tamayo-Belda, M.; Leganés, F.; Rosal, R.; Fernández-Piñas, F. Co, Zn and Ag-MOFs Evaluation as Biocidal Materials towards Photosynthetic Organisms. Sci. Total Environ. 2017, 595, 547–555. [Google Scholar] [CrossRef]
- Golden, S.S. The International Journeys and Aliases of Synechococcus elongatus. N. Z. J. Bot. 2018, 57, 70–75. [Google Scholar] [CrossRef]
- Mettert, E.; Kiley, P. How Is Fe-S Cluster Formation Regulated? Annu. Rev. Microbiol. 2015, 69, 505–526. [Google Scholar] [CrossRef] [PubMed]
- Gao, F. Iron–Sulfur Cluster Biogenesis and Iron Homeostasis in Cyanobacteria. Front. Microbiol. 2020, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Prieto, M.A.; Schön, V.; Georg, J.; Barreira, L.; Varela, J.; Hess, W.R.; Futschik, M.E. Iron Deprivation in Synechocystis: Inference of Pathways, Non-Coding RNAs, and Regulatory Elements from Comprehensive Expression Profiling. G3 Genes Genomes Genet. 2012, 2, 1475–1495. [Google Scholar] [CrossRef] [PubMed]
- Vuorijoki, L.; Tiwari, A.; Kallio, P.; Aro, E.M. Inactivation of Iron-Sulfur Cluster Biogenesis Regulator SufR in Synechocystis sp. PCC 6803 Induces Unique Iron-Dependent Protein-Level Responses. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Ranquet, C.; Ollagnier-de-Choudens, S.; Loiseau, L.; Barras, F.; Fontecave, M. Cobalt Stress in Escherichia coli. The Effect on the Iron-Sulfur Proteins. J. Biol. Chem. 2007, 282, 30442–30451. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Shen, G.; Balasubramanian, R.; McIntosh, L.; Bryant, D.; Golbeck, J. The sufR Gene (Sll0088 in Synechocystis sp. Strain PCC 6803) Functions as a Repressor of the sufBCDS Operon in Iron-Sulfur Cluster Biogenesis in Cyanobacteria. J. Bacteriol. 2004, 186, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.-M.; Agalarov, R.; Brettel, K.; Carmeli, C. Control of Electron Transport in Photosystem I by the Iron-Sulfur Cluster FX in Response to Intra- and Intersubunit Interactions. J. Biol. Chem. 2003, 278, 19141–19150. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Graziantonio, L.; Catalano, A.; Carocci, A.; Sinicropi, M.S. Prevalence of Cobalt in the Environment and Its Role in Biological Processes. Biology 2023, 12, 1335. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marceau, F.; Lamothe-Sibold, M.; Farci, S.; Ouchane, S.; Cassier-Chauvat, C.; Chauvat, F. First Characterization of a Cyanobacterial Xi-Class Glutathione S-Transferase in Synechocystis PCC 6803. Antioxidants 2024, 13, 1577. https://doi.org/10.3390/antiox13121577
Marceau F, Lamothe-Sibold M, Farci S, Ouchane S, Cassier-Chauvat C, Chauvat F. First Characterization of a Cyanobacterial Xi-Class Glutathione S-Transferase in Synechocystis PCC 6803. Antioxidants. 2024; 13(12):1577. https://doi.org/10.3390/antiox13121577
Chicago/Turabian StyleMarceau, Fanny, Marlène Lamothe-Sibold, Sandrine Farci, Soufian Ouchane, Corinne Cassier-Chauvat, and Franck Chauvat. 2024. "First Characterization of a Cyanobacterial Xi-Class Glutathione S-Transferase in Synechocystis PCC 6803" Antioxidants 13, no. 12: 1577. https://doi.org/10.3390/antiox13121577
APA StyleMarceau, F., Lamothe-Sibold, M., Farci, S., Ouchane, S., Cassier-Chauvat, C., & Chauvat, F. (2024). First Characterization of a Cyanobacterial Xi-Class Glutathione S-Transferase in Synechocystis PCC 6803. Antioxidants, 13(12), 1577. https://doi.org/10.3390/antiox13121577






