Antioxidants Improve the Proliferation and Efficacy of hUC-MSCs against H2O2-Induced Senescence
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Immunophenotypical Analysis of hUC-MSCs
2.3. Antioxidants Treatment
2.4. Cell Growth Curve Assay
2.5. Cell ROS Level Measurement
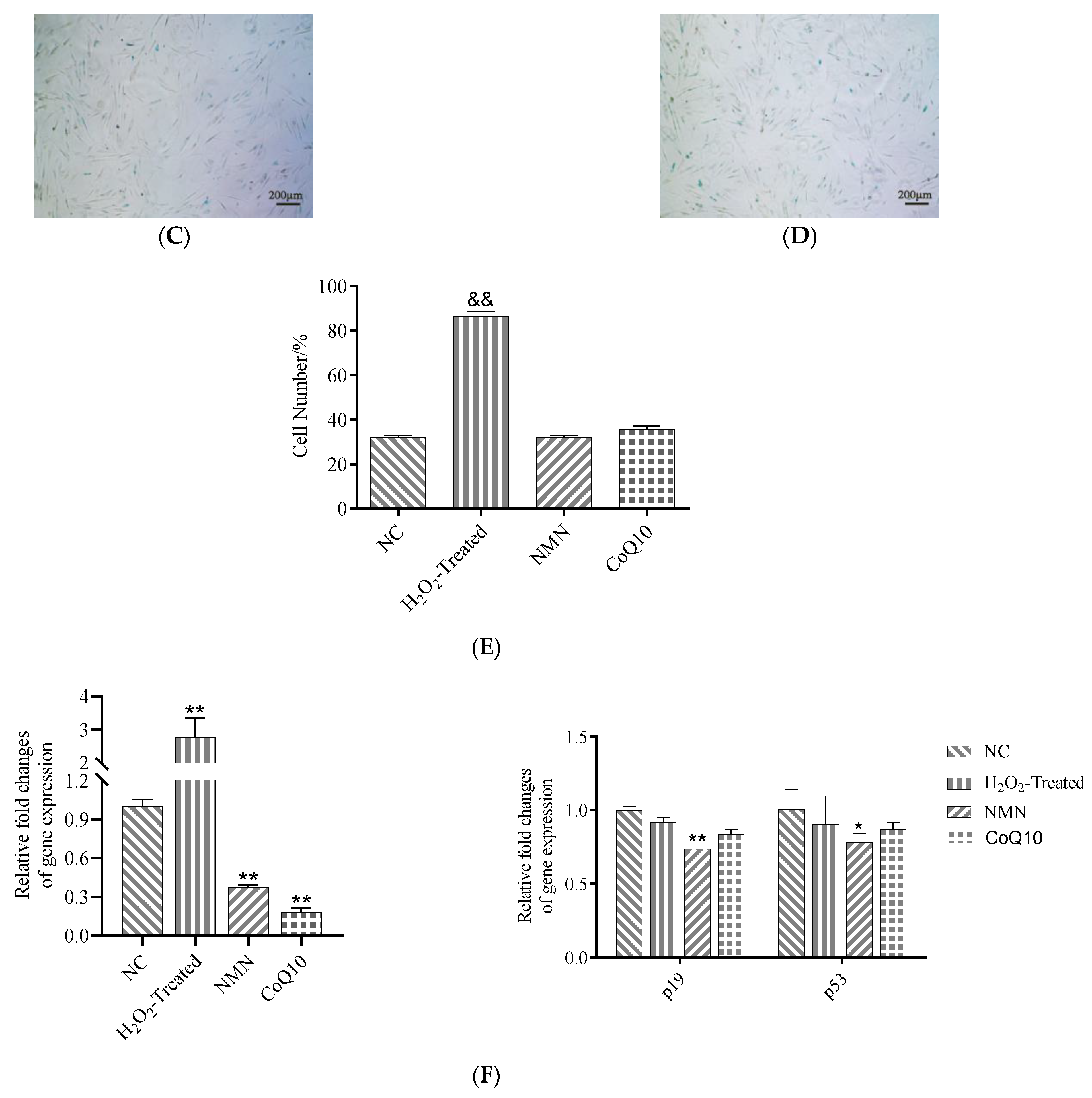
2.6. Senescence-Associated β-Galactosidase (SA-β-Gal) Assay
2.7. qRT-PCR Analysis
2.8. Differentiation Ability of the hUC-MSCs
2.9. Migration Assay
- RS: Relative area ratio
- St36/St0: growth area of cells in the antioxidant treatment group at 36 h or 0 h
- Sn36/Sn0: growth area of cells in the control group at 36 h or 0 h
2.10. RNA Sequencing
2.11. RNAseq Data Analysis
2.12. Statistical Analysis
3. Results
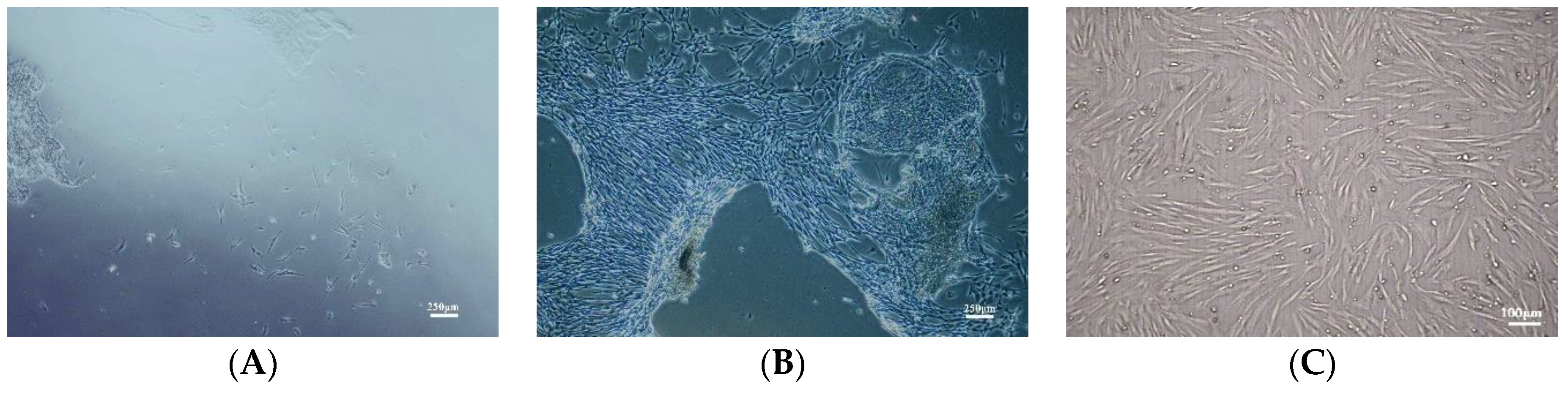
3.1. Characterization of Primary Culture of hUC-MSCs
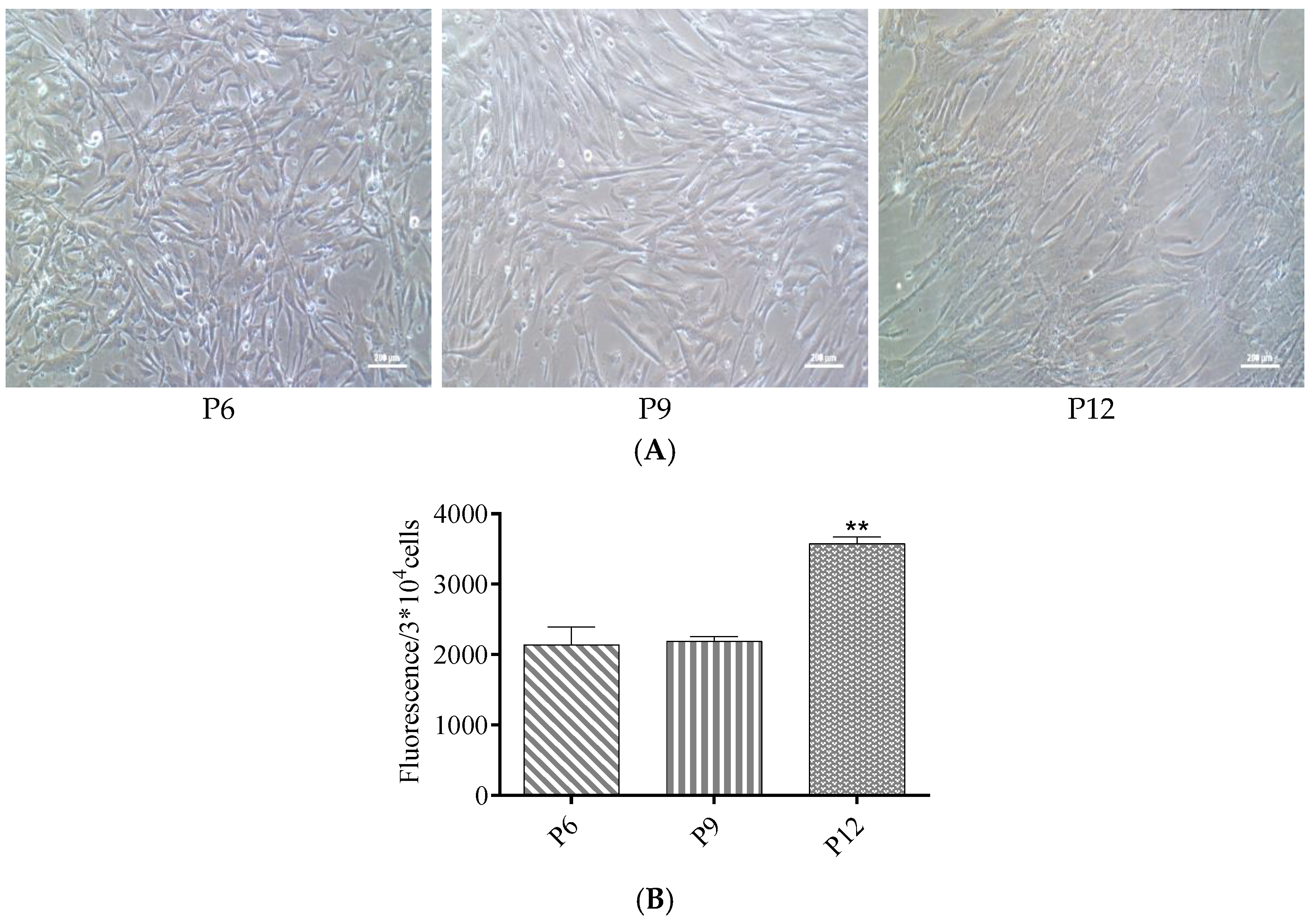
3.2. ROS Levels Increased in Late Passages of hUC-MSCs Cultured in a Serum-Free Medium
3.3. The Proliferative Capacity Increased in Late Passages of hUC-MSCs after Treatment with NMN and CoQ10
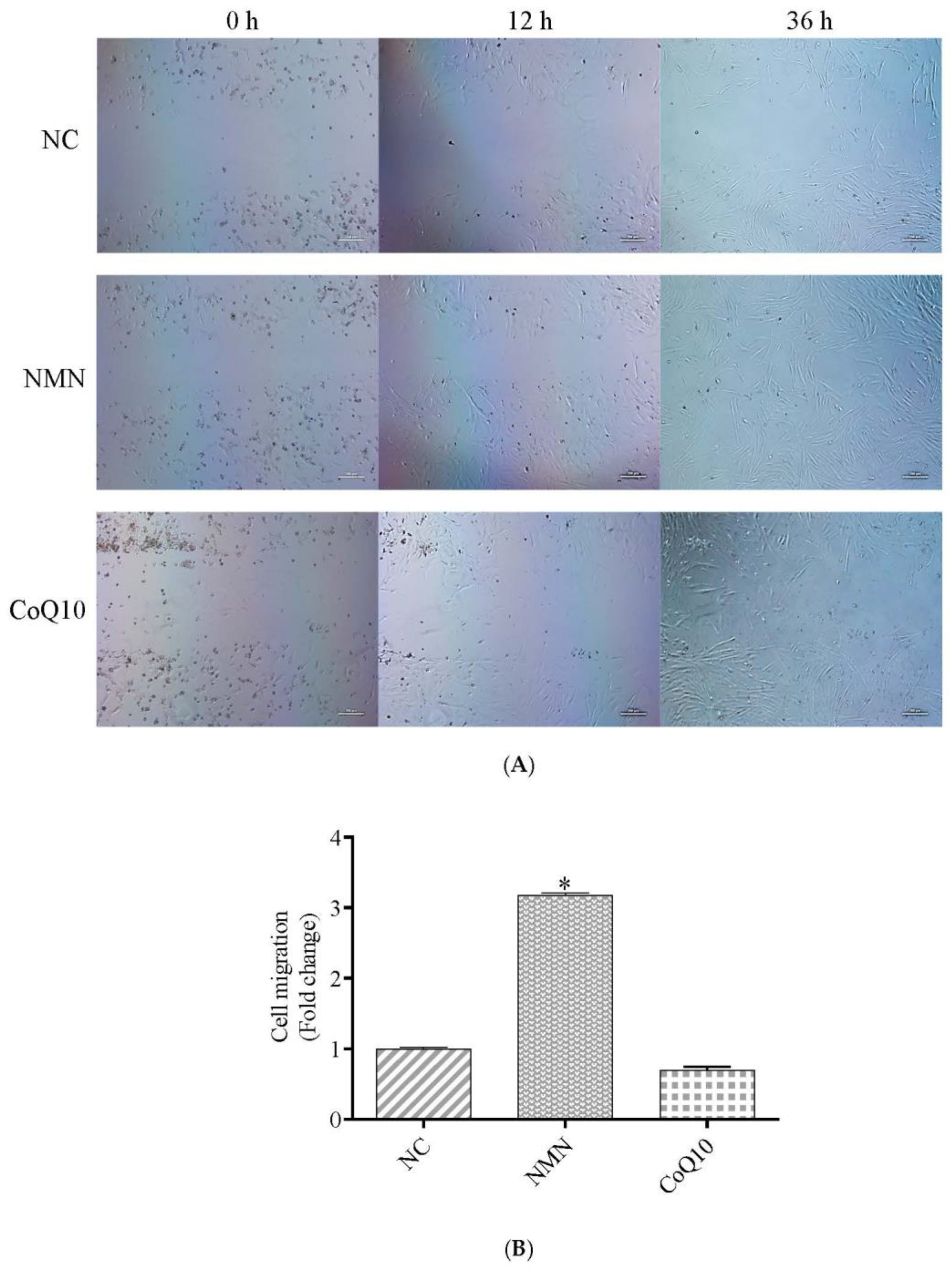
3.4. Cell Migration Ability Increase after Treatment with NMN
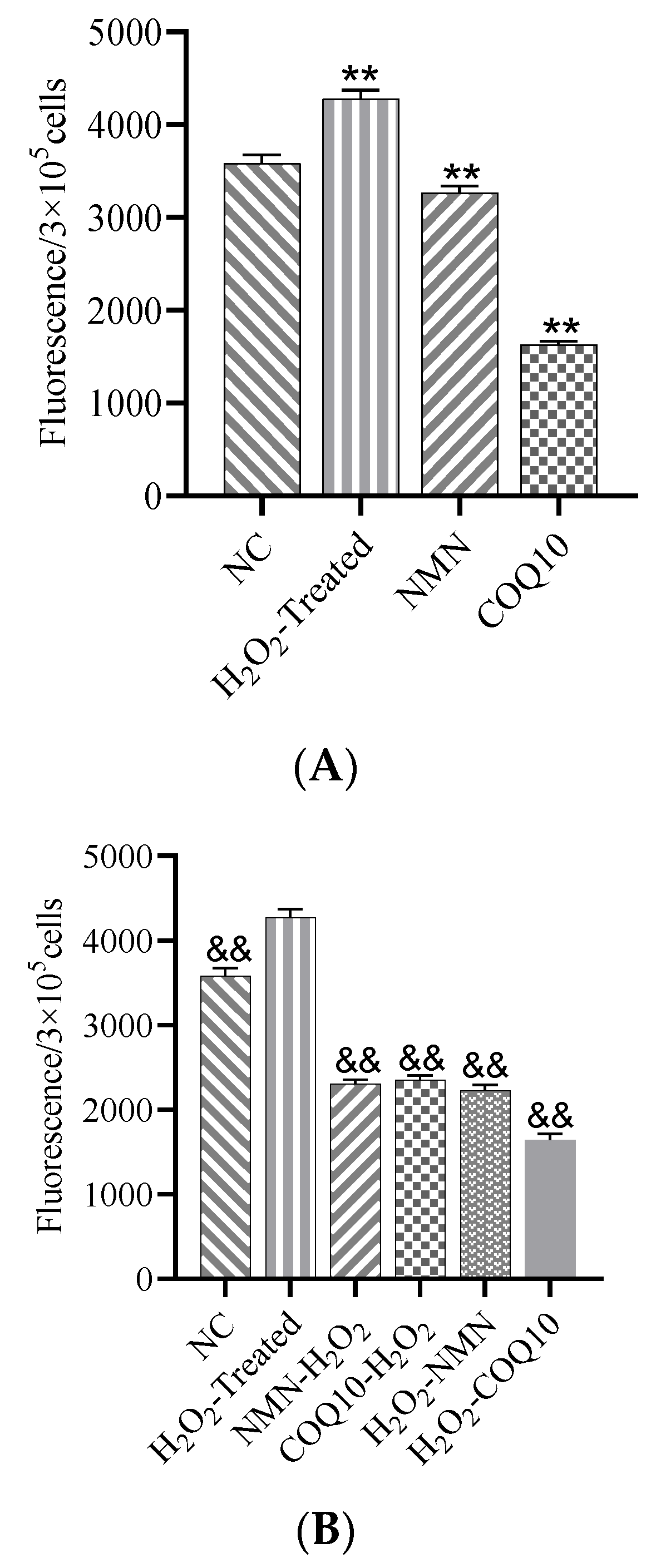
3.5. ROS Levels Decreased after Treatment with NMN and CoQ10
3.6. SA-β-Gal Activity and Senescence-Related Gene Expression Difference after Treatment with NMN and CoQ10
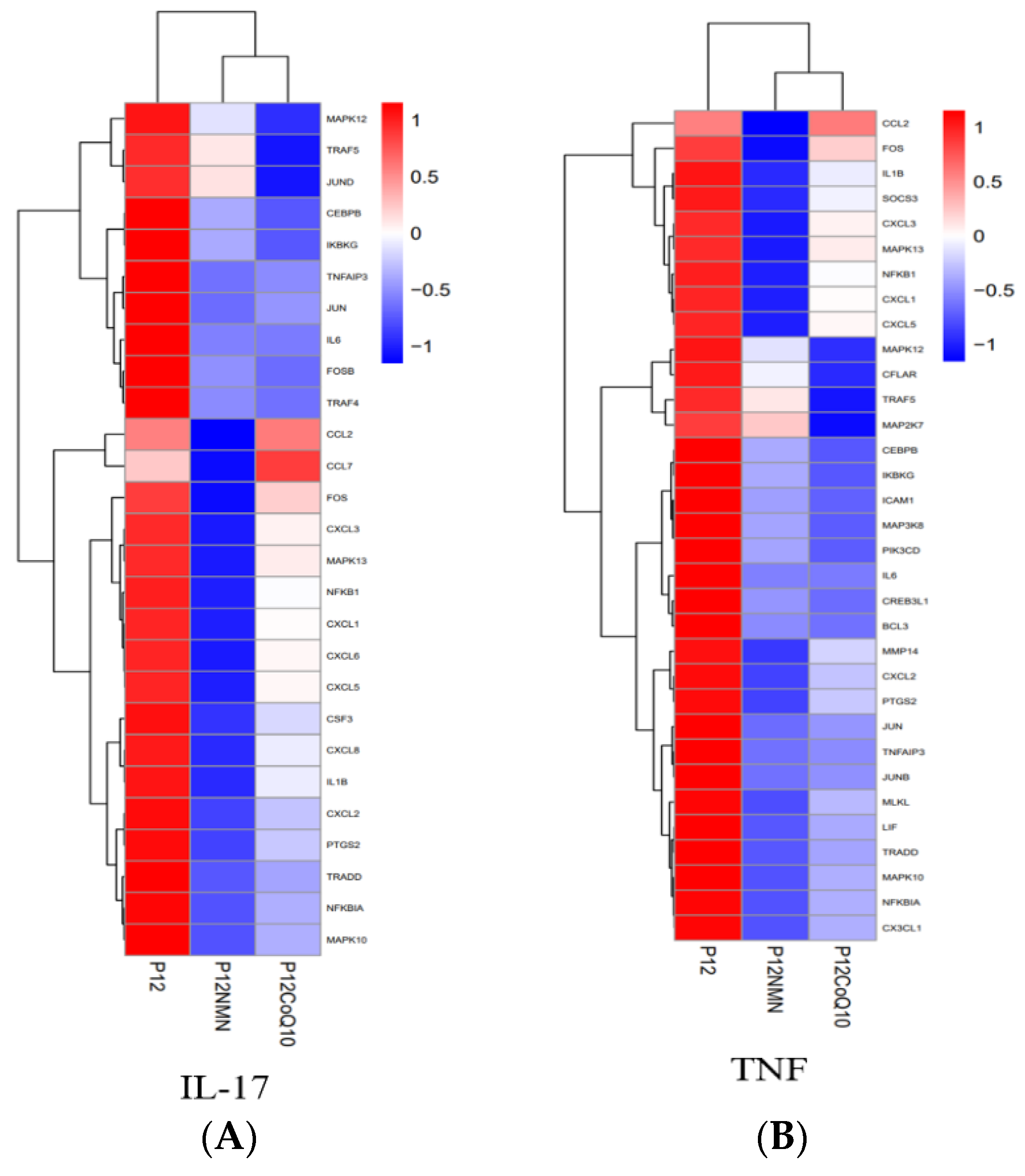
3.7. Increase of TNFα Gene Expression due to H2O2 Induction was Significantly Reduced by NMN and CoQ10
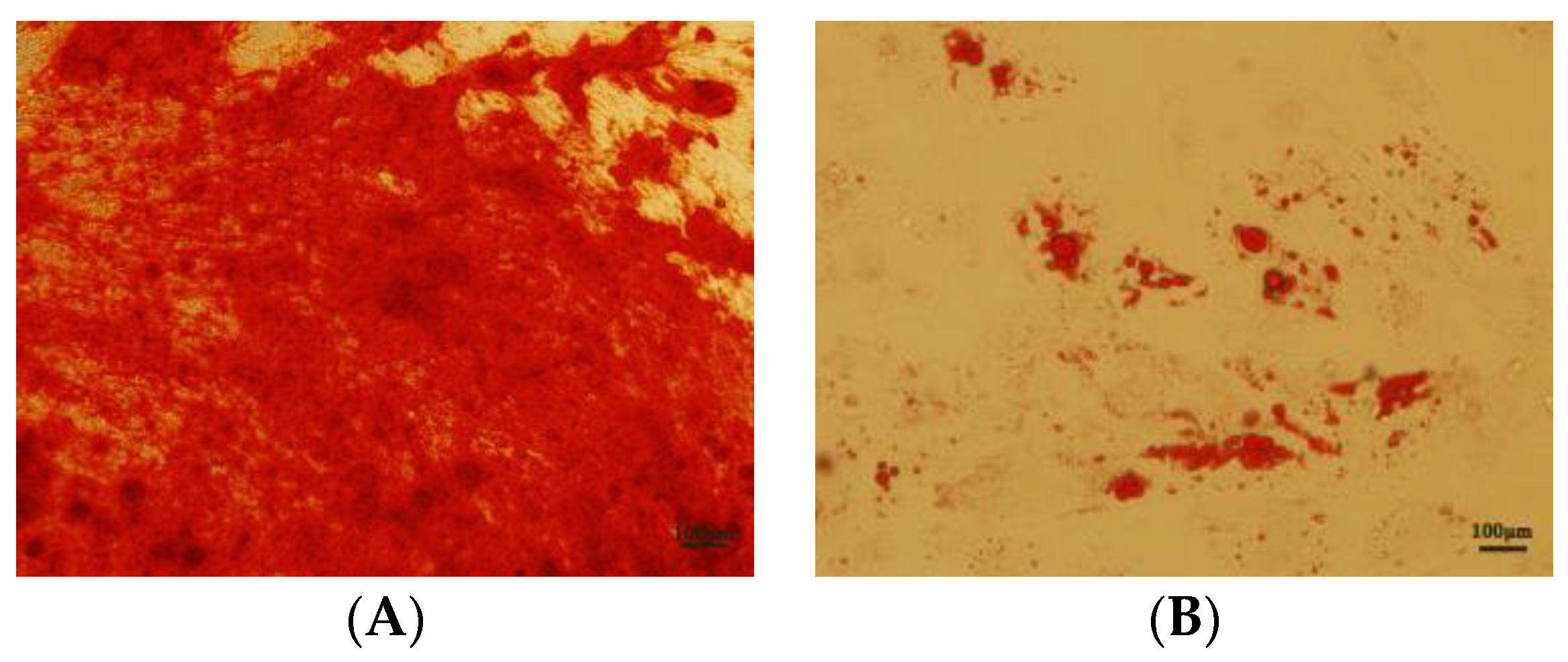
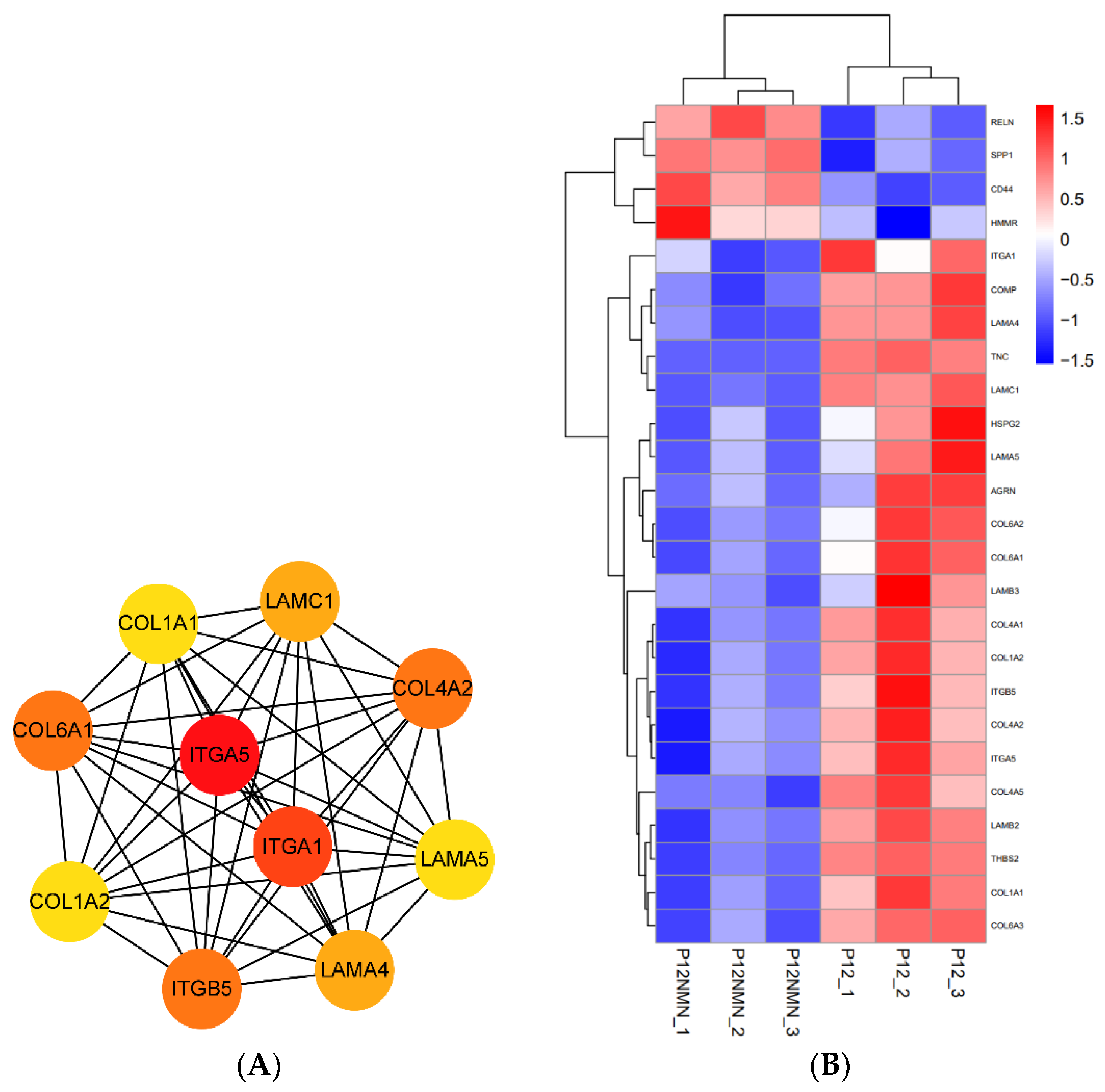
3.8. Effects of NMN and CoQ10 Treatment on Cell Tri-Lineage Differentiation Ability and Expression Genes Related to Stemness and Differentiation
3.9. Transcriptomic Profiles of hUC-MSCs after NMN/CoQ10 Treatment
3.10. Functional Enrichment Analysis of Differentially Expressed Genes in hUC-MSCs in the Antioxidants-Treated Group
3.11. KEGG Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parsons Ronald, F.; Baquerizo, A.; Kirchner Varvara, A.; Malek, S.; Desai Chirag, S. Challenges, highlights, and opportunities in cellular transplantation: A white paper of the current landscape. American journal of transplantation. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2021, 21, 3225–3238. [Google Scholar] [CrossRef] [PubMed]
- Cassano Jennifer, M.; Schnabel Lauren, V.; Goodale Margaret, B.; Fortier Lisa, A. Inflammatory licensed equine MSCs are chondroprotective and exhibit enhanced immunomodulation in an inflammatory environment. Stem Cell Res. Ther. 2018, 9, 82. [Google Scholar] [CrossRef]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical Trials with Mesenchymal Stem Cells: An Update. Cell Transplant. 2015, 25, 829–848. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wehner, R.; Bornhäuser, M.; Wassmuth, R.; Bachmann, M. Immunomodulatory properties of mesenchymal stromal cells and their therapeutic consequences for immune-mediated disorders. Stem Cells Dev. 2010, 19, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Nauta, A.J.; Fibbe, W.E. Immunomodulatory properties of mesenchymal stromal cells. Blood 2007, 110, 3499–3506. [Google Scholar] [CrossRef] [PubMed]
- Plock, J.A.; Schnider, J.T.; Zhang, W.; Schweizer, R.; Tsuji, W. Adipose- and Bone Marrow-Derived Mesenchymal Stem Cells Prolong Graft Survival in Vascularized Composite Allotransplantation. Transplantation 2015, 99, 1765–1773. [Google Scholar] [CrossRef]
- Garcia, J.; Garcia, J.; Wright, K.; Wright, K.; Roberts, S. Characterisation of synovial fluid and infrapatellar fat pad derived mesenchymal stromal cells: The influence of tissue source and inflammatory stimulus. Sci. Rep. 2016, 6, 24295. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Cao, W.; Shi, Y. Plasticity of mesenchymal stem cells in immunomodulation: Pathological and therapeutic implications. Nat. Immunol. 2014, 15, 1009–1016. [Google Scholar] [CrossRef]
- Chen, X.; Gan, Y.; Li, W.; Su, J.; Zhang, Y. The interaction between mesenchymal stem cells and steroids during inflammation. Cell Death Dis. 2014, 5, e1009. [Google Scholar] [CrossRef]
- Chen, L.; Liu, G.; Wu, J.; Zhou, X.; Zhao, Y.; Chen, Z.; Lin, Z.; Xiao, Y. Multi-faceted effects of mesenchymal stem cells (MSCs) determined by immune microenvironment and their implications on MSC/biomaterial-based inflammatory disease therapy. Appl. Mater. Today 2020, 18, 100485. [Google Scholar] [CrossRef]
- Effros, R.B. Replicative senescence in the immune system: Impact of the Hayflick limit on T-cell function in the elderly. Am. J. Hum. Genet. 1998, 62, 1003–1007. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, H.; Zhuang, Y.; Liu, H.; Shi, Q. The role of N-acetyltransferase 8 in mesenchymal stem cell-based therapy for liver ischemia/reperfusion injury in rats. PLoS ONE 2014, 9, e103355. [Google Scholar] [CrossRef]
- Abdul-Muneer, P.M.; Chandra, N.; Haorah, J. Interactions of oxidative stress and neurovascular inflammation in the pathogenesis of traumatic brain injury. Mol. Neurobiol. 2015, 51, 966–979. [Google Scholar] [CrossRef]
- Toygar, M.; Aydin, I.; Agilli, M.; Aydin, F.N.; Oztosun, M. The relation between oxidative stress, inflammation, and neopterin in the paraquat-induced lung toxicity. Hum. Exp. Toxicol. 2014, 15, 1009–1016. [Google Scholar] [CrossRef]
- Guzik, T.J.; Touyz, R.M. Oxidative Stress, Inflammation, and Vascular Aging in Hypertension. Hypertension 2017, 70, 660–667. [Google Scholar] [CrossRef]
- Cannizzo, E.S.; Clement, C.C.; Sahu, R.; Follo, C.; Santambrogio, L. Oxidative stress, inflamm-aging and immunosenescence. J. Proteom. 2011, 74, 2313–2323. [Google Scholar] [CrossRef]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef]
- Kawabori, M.; Kuroda, S.; Ito, M.; Shichinohe, H.; Houkin, K. Timing and cell dose determine therapeutic effects of bone marrow stromal cell transplantation in rat model of cerebral infarct. Neuropathology 2013, 33, 140–148. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, A.; Xu, D.; Yao, K.; Huang, Z. Impact of timing on efficacy and safetyof intracoronary autologous bone marrow stem cells transplantation in acute myocardial infarction: A pooled subgroup analysis of randomized controlled trials. Clin. Cardiol. 2009, 32, 458–466. [Google Scholar] [CrossRef]
- Hajam, Y.A.; Rani, R.; Ganie, S.Y.; Sheikh, T.A.; Javaid, D. Oxidative Stress in Human Pathology and Aging: Molecular Mechanisms and Perspectives. Cells 2022, 11, 552. [Google Scholar] [CrossRef]
- Zhu, X.-H.; Lu, M.; Lee, B.-Y.; Ugurbil, K.; Chen, W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA 2015, 112, 2876–2881. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.; Campisi, J. NAD+ controls neural stem cell fate in the aging brain. EMBO J. 2014, 33, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, J.; Yang, F.; Ning, G.; Zhen, L. Nicotinamide mononucleotide promotes osteogenesis and reduces adipogenesis by regulating mesenchymal stromal cells via the SIRT1 pathway in aged bone marrow. Cell Death Dis. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maraver, J.; Cordero, M.D.; Oropesa-Avila, M.; Vega, A.F.; de la Mata, M. Clinical applications of coenzyme Q10. Front. Biosci. 2014, 19, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Huo, J.; Xu, Z.; Xu, Z.; Hosoe, K. Coenzyme Q10 Prevents Senescence and Dysfunction Caused by Oxidative Stress in Vascular Endothelial Cells. Oxidative Med. Cell. Longev. 2018, 2018, 3181759. [Google Scholar] [CrossRef]
- Mantle, D.; Hargreaves, I. Coenzyme Q10 and Degenerative Disorders Affecting Longevity: An Overview. Antioxidants 2019, 8, 44. [Google Scholar] [CrossRef]
- Soma, M.; Lalam, S.K. The role of nicotinamide mononucleotide (NMN) in anti-aging, longevity, and its potential for treating chronic conditions. Mol. Biol. Rep. 2022, 49, 9737–9748. [Google Scholar] [CrossRef]
- Kabat, M.; Bobkov, I.; Kumar, S.; Grumet, M. Trends in mesenchymal stem cell clinical trials 2004–2018: Is efficacy optimal in a narrow dose range? Stem Cells Transl. Med. 2020, 9, 12–27. [Google Scholar] [CrossRef]
- Ranjbaran, H.; Abediankenari, S.; Mohammadi, M.; Jafari, N.; Khalilian, A. Wharton’s Jelly Derived-Mesenchymal Stem Cells: Isolation and Characterization. Acta Med. Iran. 2018, 56, 28–33. [Google Scholar]
- Sorrell, J.M.; Caplan, A.I. Topical delivery of mesenchymal stem cells and their function in wounds. Stem Cell Res. Ther. 2010, 1, 1–6. [Google Scholar] [CrossRef]
- Wang, Y.; Yago, T.; Zhang, N.; Abdisalaam, S.; Alexandrakis, G. Cytoskeletal regulation of CD44 membrane organization and interactions with E-selectin. J. Biol. Chem. 2014, 289, 35159–35171. [Google Scholar] [CrossRef]
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e10. [Google Scholar] [CrossRef]
- Medeiros, T.M.; Joana, C.; Cornélio, D.A.; Nogueira, S.V.; Ducati, L.A.; de Souza, S. Identification of new genes associated to senescent and tumorigenic phenotypes in mesenchymal stem cells. Sci. Rep. 2017, 7, 17837. [Google Scholar] [CrossRef]
- Yue, Z.; Rong, J.; Ping, W.; Bing, Y.; Xin, Y. Gene expression of the p16(INK4a)-Rb and p19(Arf)-p53-p21(Cip/Waf1) signaling pathways in the regulation of hematopoietic stem cell aging by ginsenoside Rg1. Genet. Mol. Res. GMR 2014, 13, 10086–10096. [Google Scholar] [CrossRef]
- Parsch, D.; Fellenberg, J.; Brümmendorf, T.H.; Eschlbeck, A.-M.; Richter, W. Telomere length and telomerase activity during expansion and differentiation of human mesenchymal stem cells and chondrocytes. J. Mol. Med. 2003, 82, 49–55. [Google Scholar]
- Hall, J.A.; Dominy, J.E.; Lee, Y.; Puigserver, P. The sirtuin family’s role in aging and age-associated pathologies. J. Clin. Invest. 2013, 123, 973–979. [Google Scholar] [CrossRef]
- Hargreaves, I.P.; Mantle, D. Coenzyme Q10 Supplementation in Fibrosis and Aging. Adv. Exp. Med. Biol. 2019, 1178, 103–112. [Google Scholar]
- Ko, H.J.; Kim, J.; Ahn, M.; Kim, J.H.; Lee, G.S. Ergothioneine alleviates senescence of fibroblasts induced by UVB damage of keratinocytes via activation of the Nrf2/HO-1 pathway and HSP70 in keratinocytes. Exp. Cell Res. 2021, 400, 112516. [Google Scholar] [CrossRef]
- Hernández-Pérez, O.R.; Juárez-Navarro, K.J.; Diaz, N.F.; Padilla-Camberos, E.; Beltran-Garcia, M.J. Biomolecules resveratrol + coenzyme Q10 recover the cell state of human mesenchymal stem cells after 1-methyl-4-phenylpyridinium-induced damage and improve proliferation and neural differentiation. Front. Neurosci. 2022, 31, 929590. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Xu, E.; Musich, P.R.; Lin, F. Mitochondrial dysfunction in neurodegenerative diseases and the potential countermeasure. CNS Neurosci. Ther. 2019, 25, 816–824. [Google Scholar] [CrossRef]
- Lei, Q.; Liu, T.; Gao, F.; Xie, H.; Sun, L. Microvesicles as Potential Biomarkers for the Identification of Senescence in Human Mesenchymal Stem Cells. Theranostics 2017, 7, 2673–2689. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Z.; Wang, F.; Gao, P.; Pei, J.-F.; Liu, Y. Age-Associated Sirtuin 1 Reduction in Vascular Smooth Muscle Links Vascular Senescence and Inflammation to Abdominal Aortic Aneurysm. Circ. Res. 2016, 119, 1076–1088. [Google Scholar] [CrossRef] [PubMed]
- Scialo, F.; Sanz, A. Coenzyme Q redox signalling and longevity. Free. Radic. Biol. Med. 2021, 164, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Mori, N.; Shibata, K. β-Nicotinamide Mononucleotide, an Anti-Aging Candidate Compound, Is Retained in the Body for Longer than Nicotinamide in Rats. J. Nutr. Sci. Vitaminol. 2016, 62, 272–276. [Google Scholar] [CrossRef]







| Gene Name | Primer | Sequence (5′-3′) | Accension Number |
|---|---|---|---|
| foxe1 | Forward Reverse | GGAGTGTTCAGATGGACTGTTT CAGCCCTTCCCTGTTAAGTTTAT | NM_004473 |
| p19 | Forward | AGTCCAGTCCATGACGCAG | NM_001800.4 |
| Reverse | ATCAGGCACGTTGACATCAGC | ||
| p53 | Forward | CGCTTCGAGATGTTCCGAGA | NC_000017.11 |
| Reverse | CTGGGACCCAATGAGATGGG | ||
| pparg | Forward Reverse | GAAGACGGAGACAGACATGAG GCAACTGGAAGAAGGGAAATG | NM_001354666 |
| runx2 | Forward Reverse | ATGTCCGCCACCACTCACTACC TGGAGTGCTGCTGGTCTGGAAG | NM_001024630 |
| col2a1 | Forward Reverse | TGAACCTGGACGAGAGGGAAGC ACAGCACCAGTCTCACCACGAT | X16711 |
| oct4 | Forward | CTTGCTGCAGAAGTGGGTGGAGGAA | NM_001285986.2 |
| Reverse | CTGCAGTGTGGGTTTCGGGCA | ||
| sox2 | Forward | TGGACAGTTACGCGCACAT | NM_003106.4 |
| Reverse | CGAGTAGGACATGCTGTAGGT | ||
| nanog | Forward | AATACCTCAGCCTCCAGCAGAT | NM_024865.4 |
| Reverse | TGCGTCACACCATTGCTATTCTT | ||
| IL10 | Forward Reverse | CTGGGAGCCCACACTATTTATT GATGCACCACTACCTTTCTACC | NM_001382624 |
| TNFα | Forward Reverse | GGAGTGTTCAGATGGACTGTTT CAGCCCTTCCCTGTTAAGTTTAT | NM_000594 |
| alas1 | Forward Reverse | CCTGGATGGATGAGTGGCTTCT AATGGGCAGCGGCGAACAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Z.; Wang, X.; Ouyang, L.; Chen, W.; Zhang, L.; Cao, Y. Antioxidants Improve the Proliferation and Efficacy of hUC-MSCs against H2O2-Induced Senescence. Antioxidants 2023, 12, 1334. https://doi.org/10.3390/antiox12071334
Zheng Z, Wang X, Ouyang L, Chen W, Zhang L, Cao Y. Antioxidants Improve the Proliferation and Efficacy of hUC-MSCs against H2O2-Induced Senescence. Antioxidants. 2023; 12(7):1334. https://doi.org/10.3390/antiox12071334
Chicago/Turabian StyleZheng, Zhaojuan, Xia Wang, Liming Ouyang, Wenxia Chen, Lixin Zhang, and Yulin Cao. 2023. "Antioxidants Improve the Proliferation and Efficacy of hUC-MSCs against H2O2-Induced Senescence" Antioxidants 12, no. 7: 1334. https://doi.org/10.3390/antiox12071334
APA StyleZheng, Z., Wang, X., Ouyang, L., Chen, W., Zhang, L., & Cao, Y. (2023). Antioxidants Improve the Proliferation and Efficacy of hUC-MSCs against H2O2-Induced Senescence. Antioxidants, 12(7), 1334. https://doi.org/10.3390/antiox12071334







