The Fate of Oxidative Strand Breaks in Mitochondrial DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Measurement of H2O2 Decay
2.3. Total DNA Isolation
2.4. Isolation of mtDNA
2.5. Southern Blot
2.6. Assessment of the Number of mtDNA Breaks by qPCR
2.7. BrdU Incorporation
2.8. Short-Read (Illumina) Sequencing of Linker-Ligated Isolated mtDNA
2.9. PacBio Single-Molecule Long-Read Sequencing of Isolated mtDNA
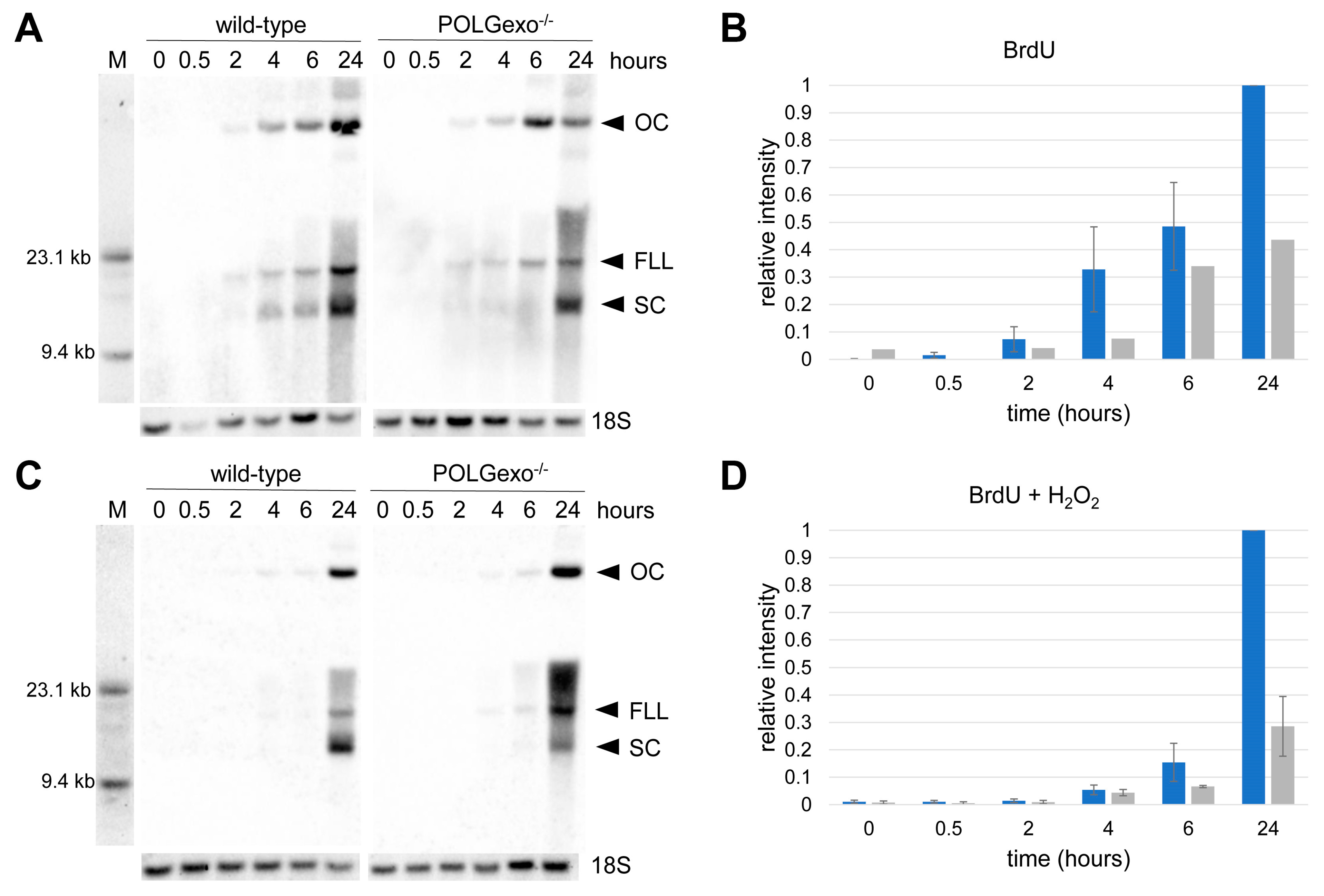
3. Results
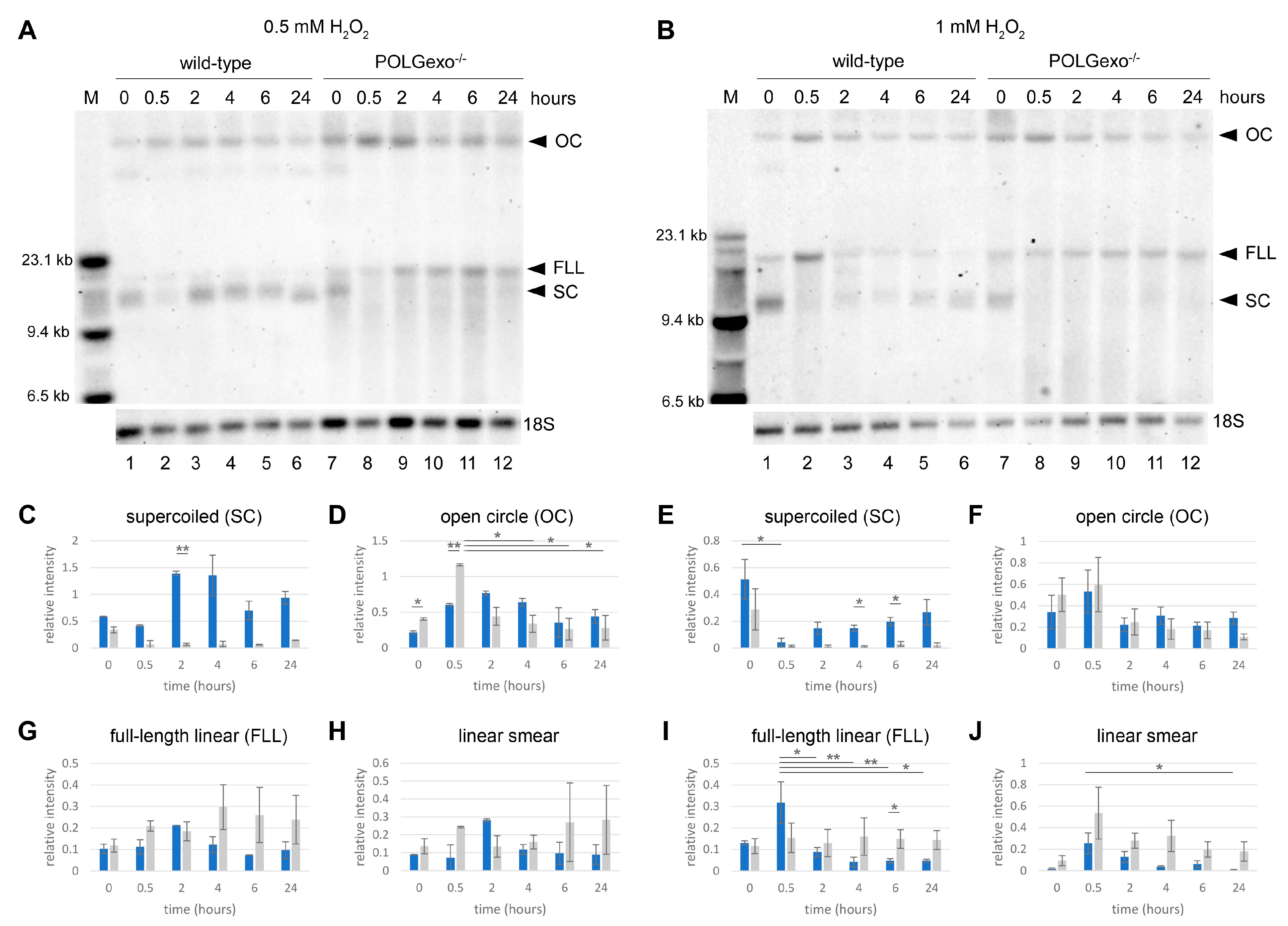
3.1. Transient Hydrogen Peroxide Treatment of HEK 293 Cells Causes Reversible Oxidative Damage of Mitochondrial DNA
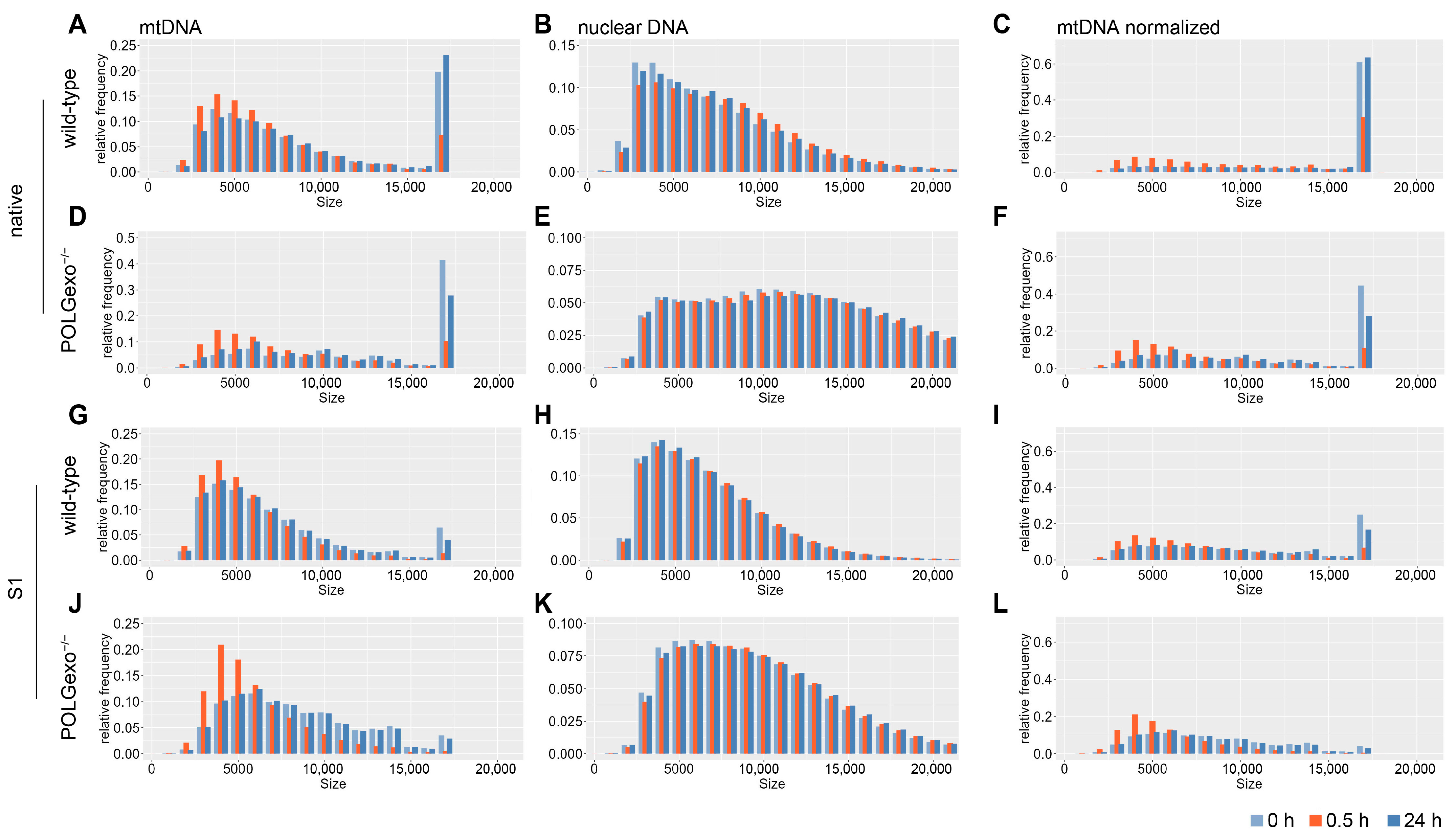
3.2. Differential Impact of Hydrogen Peroxide Pulse on Mitochondrial and Nuclear DNA Integrity
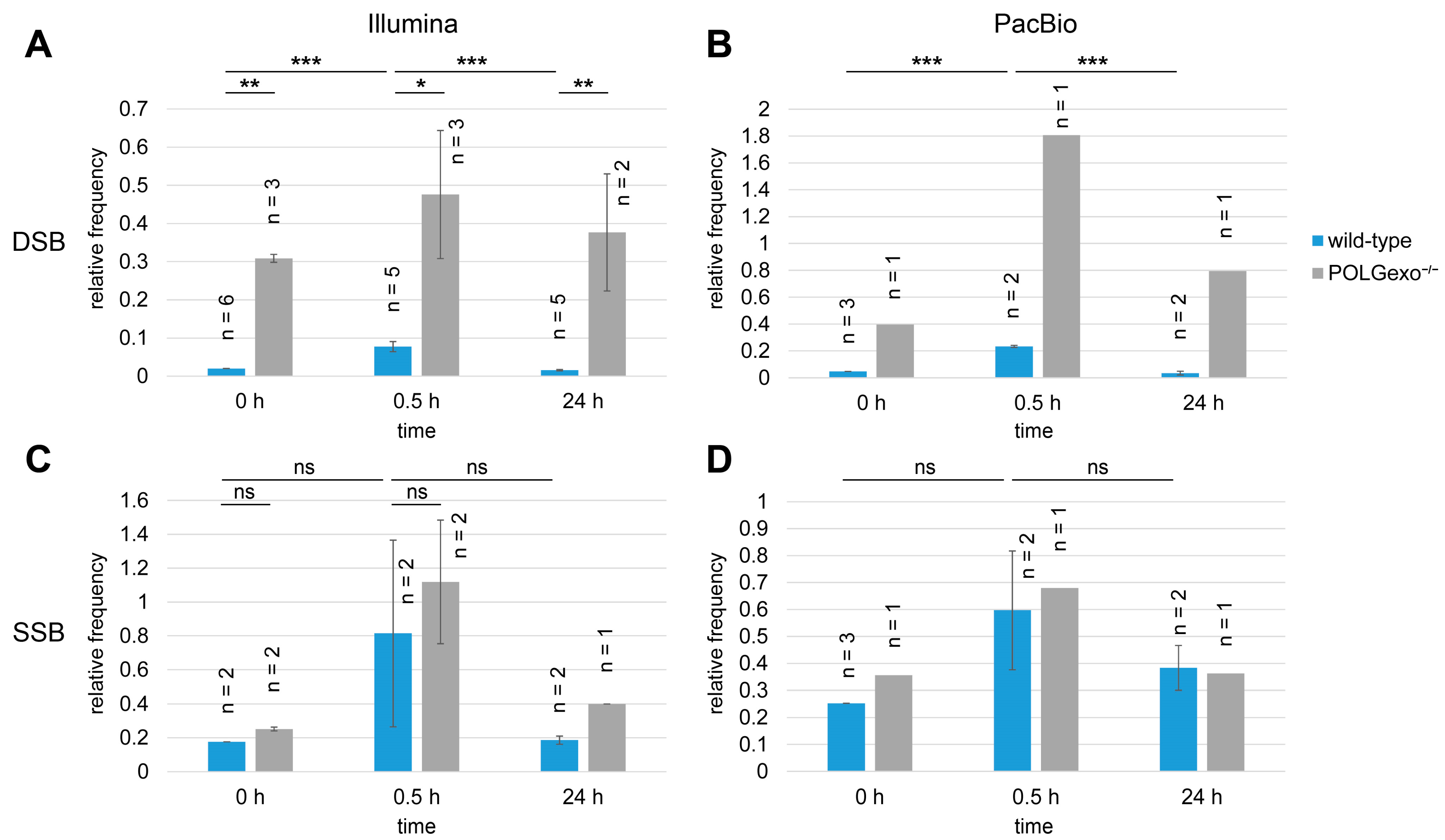
3.3. Increased Amount of DSBs but Not SSBs in POLGexo-/- Cells
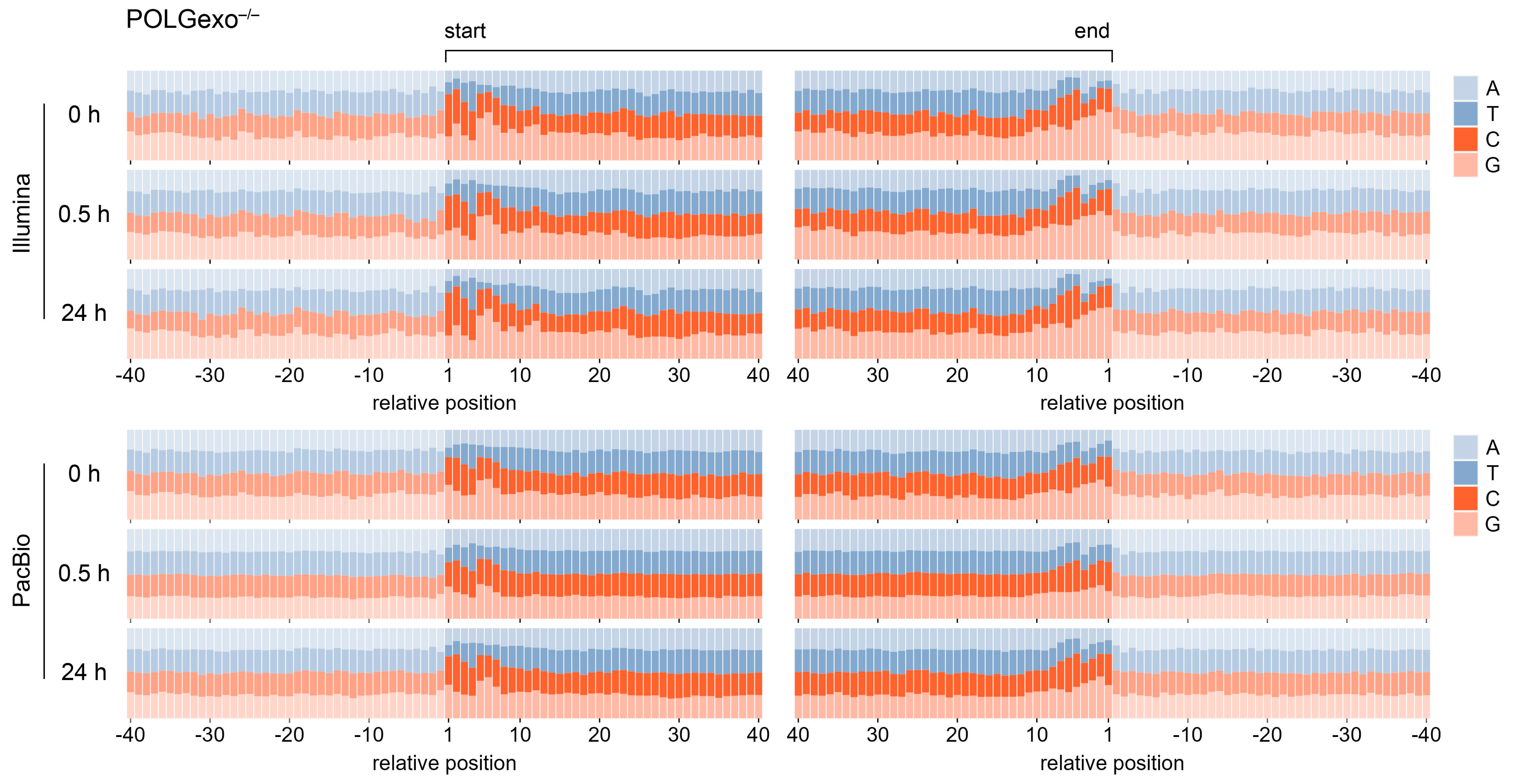
3.4. Degradation-Specific Sequence Motifs Are Detectable in POLGexo-/- Cells and upon H2O2 Damage
3.5. De Novo mtDNA Synthesis Does Not Play a Crucial Role in Short-Term Recovery after H2O2 Damage
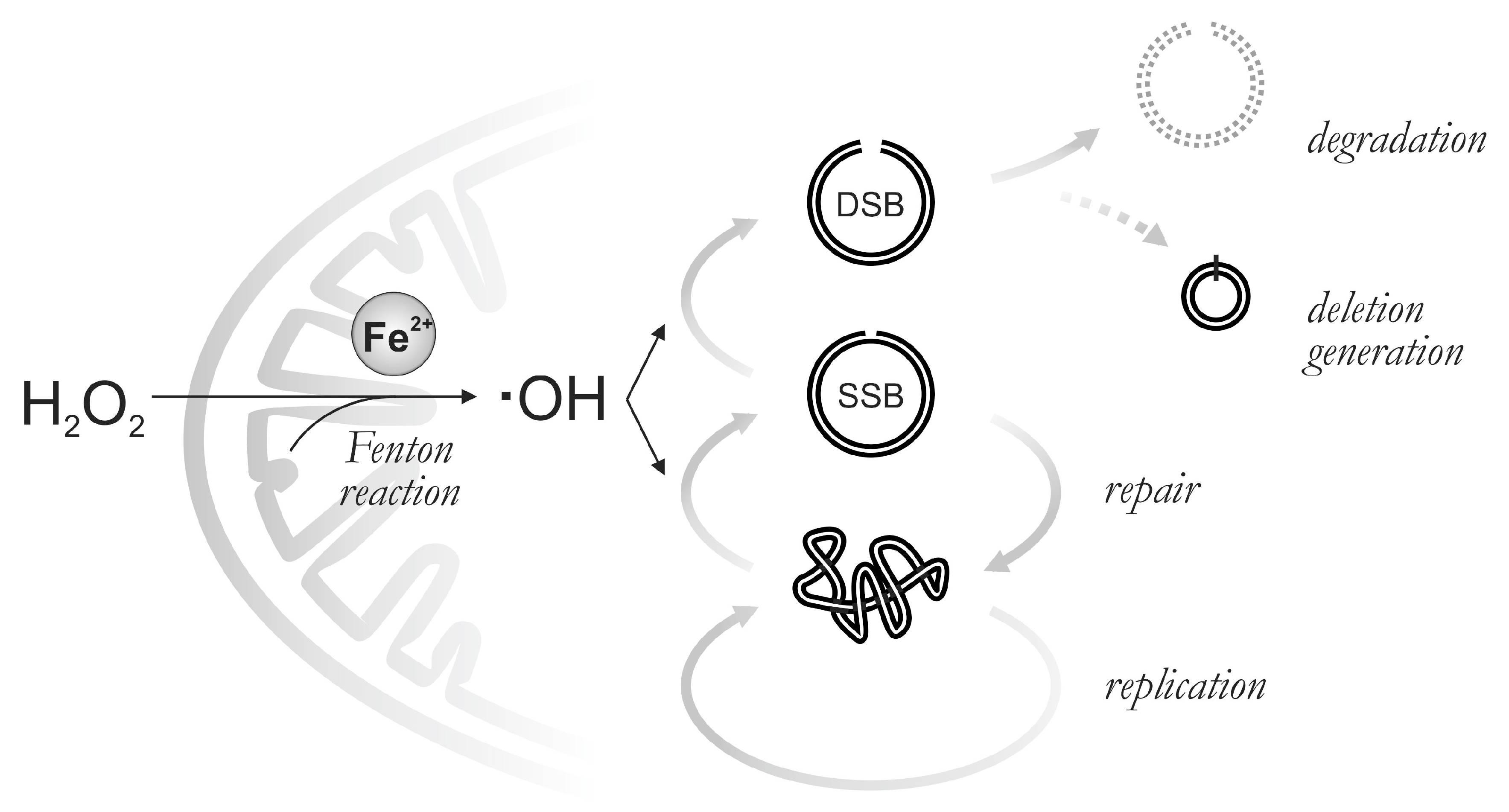
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexeyev, M.F. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009, 276, 5768–5787. [Google Scholar] [CrossRef]
- Levi, S.; Rovida, E. The role of iron in mitochondrial function. Biochim. Biophys. Acta 2009, 1790, 629–636. [Google Scholar] [CrossRef]
- Chance, B.; Sies, H.; Boveris, A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979, 59, 527–605. [Google Scholar] [CrossRef]
- Itsara, L.S.; Kennedy, S.R.; Fox, E.J.; Yu, S.; Hewitt, J.J.; Sanchez-Contreras, M.; Cardozo-Pelaez, F.; Pallanck, L.J. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014, 10, e1003974. [Google Scholar] [CrossRef]
- Szczepanowska, K.; Trifunovic, A. Origins of mtDNA mutations in ageing. Essays Biochem. 2017, 61, 325–337. [Google Scholar]
- Kauppila, T.E.S.; Kauppila, J.H.K.; Larsson, N.G. Mammalian mitochondria and aging: An update. Cell Metab. 2017, 25, 57–71. [Google Scholar] [CrossRef]
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.G. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 2016, 85, 133–160. [Google Scholar] [CrossRef]
- Sanchez-Contreras, M.; Sweetwyne, M.T.; Kohrn, B.F.; Tsantilas, K.A.; Hipp, M.J.; Schmidt, E.K.; Fredrickson, J.; Whitson, J.A.; Campbell, M.D.; Rabinovitch, P.S.; et al. A replication-linked mutational gradient drives somatic mutation accumulation and influences germline polymorphisms and genome composition in mitochondrial DNA. Nucleic Acids Res. 2021, 49, 11103–11118. [Google Scholar] [CrossRef]
- Zsurka, G.; Peeva, V.; Kotlyar, A.; Kunz, W.S. Is There Still Any Role for Oxidative Stress in Mitochondrial DNA-Dependent Aging? Genes 2018, 9, 175. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef]
- Alencar, R.R.; Batalha, C.M.P.F.; Freire, T.S.; de Souza-Pinto, N.C. Enzymology of mitochondrial DNA repair. Enzymes 2019, 45, 257–287. [Google Scholar]
- Peeva, V.; Blei, D.; Trombly, G.; Corsi, S.; Szukszto, M.J.; Rebelo-Guiomar, P.; Gammage, P.A.; Kudin, A.P.; Becker, C.; Altmüller, J.; et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat. Commun. 2018, 9, 1727. [Google Scholar] [CrossRef]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef]
- Shokolenko, I.N.; Alexeyev, M.F. Mitochondrial DNA: A disposable genome? Biochim. Biophys. Acta 2015, 1852, 1805–1809. [Google Scholar] [CrossRef]
- Malinska, D.; Kudin, A.P.; Bejtka, M.; Kunz, W.S. Changes in mitochondrial reactive oxygen species synthesis during differentiation of skeletal muscle cells. Mitochondrion 2012, 12, 144–148. [Google Scholar] [CrossRef]
- Trounce, I.A.; Kim, Y.L.; Jun, A.S.; Wallace, D.C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996, 264, 484–509. [Google Scholar] [CrossRef]
- Hussain, M.; Mohammed, A.; Saifi, S.; Khan, A.; Kaur, E.; Priya, S.; Agarwal, H.; Sengupta, S. MITOL-dependent ubiquitylation negatively regulates the entry of PolγA into mitochondria. PLoS Biol. 2021, 19, e3001139. [Google Scholar] [CrossRef]
- Davis, A.F.; Clayton, D.A. In situ localization of mitochondrial DNA replication in intact mammalian cells. J. Cell Biol. 1996, 135, 883–893. [Google Scholar] [CrossRef]
- Magnusson, J.; Orth, M.; Lestienne, P.; Taanman, J.W. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp. Cell Res. 2003, 289, 133–142. [Google Scholar] [CrossRef]
- Tullman, J.; Guntas, G.; Dumont, M.; Ostermeier, M. Protein switches identified from diverse insertion libraries created using S1 nuclease digestion of supercoiled-form plasmid DNA. Biotechnol. Bioeng. 2011, 108, 2535–2543. [Google Scholar] [CrossRef]
- Guarino, V.A.; Oldham, W.M.; Loscalzo, J.; Zhang, Y.Y. Reaction rate of pyruvate and hydrogen peroxide: Assessing antioxidant capacity of pyruvate under biological conditions. Sci. Rep. 2019, 9, 19568. [Google Scholar] [CrossRef]
- Trifunovic, A.; Wredenberg, A.; Falkenberg, M.; Spelbrink, J.N.; Rovio, A.T.; Bruder, C.E.; Bohlooly, Y.M.; Gidlöf, S.; Oldfors, A.; Wibom, R.; et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 2004, 429, 417–423. [Google Scholar] [CrossRef]
- Hämäläinen, R.H.; Landoni, J.C.; Ahlqvist, K.J.; Goffart, S.; Ryytty, S.; Rahman, M.O.; Brilhante, V.; Icay, K.; Hautaniemi, S.; Wang, L.; et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nat. Metab. 2019, 1, 958–965. [Google Scholar] [CrossRef]
- Kudin, A.P.; Bimpong-Buta, N.Y.; Vielhaber, S.; Elger, C.E.; Kunz, W.S. Characterization of superoxide-producing sites in isolated brain mitochondria. J. Biol. Chem. 2004, 279, 4127–4135. [Google Scholar] [CrossRef]
- Kudin, A.P.; Malinska, D.; Kunz, W.S. Sites of generation of reactive oxygen species in homogenates of brain tissue determined with the use of respiratory substrates and inhibitors. Biochim. Biophys. Acta 2008, 1777, 689–695. [Google Scholar] [CrossRef]
- Petrat, F.; Weisheit, D.; Lensen, M.; de Groot, H.; Sustmann, R.; Rauen, U. Selective determination of mitochondrial chelatable iron in viable cells with a new fluorescent sensor. Biochem. J. 2002, 362, 137–147. [Google Scholar] [CrossRef]
- Kudin, A.P.; Augustynek, B.; Lehmann, A.K.; Kovács, R.; Kunz, W.S. The contribution of thioredoxin-2 reductase and glutathione peroxidase to H(2)O(2) detoxification of rat brain mitochondria. Biochim. Biophys. Acta 2012, 1817, 1901–1906. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Role of Free Radicals and Catalytic Metal Ions in Human Disease. Methods Enzymol. 1990, 186, 1–85. [Google Scholar]
- Henle, E.S.; Han, Z.; Tang, N.; Rai, P.; Luo, Y.; Linn, S. Sequence-specific DNA cleavage by Fe2+-mediated fenton reactions has possible biological implications. J. Biol. Chem. 1999, 274, 962–971. [Google Scholar] [CrossRef]
- Henzler, T.; Steudle, E. Transport and metabolic degradation of hydrogen peroxide in Chara corallina: Model calculations and measurements with the pressure probe suggest transport of H2O2 across water channels. J. Exp. Bot. 2000, 51, 2053–2066. [Google Scholar] [CrossRef]
- Kaneko, M.; Inoue, F. The sensitivity to DNA single strand breakage in mitochondria, but not in nuclei, of Chinese hamster V79 and variant cells correlates with their cellular sensitivity to hydrogen peroxide. Toxicol. Lett. 1998, 99, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Goffart, S.; Tikkanen, P.; Michell, C.; Wilson, T.; Pohjoismäki, J. The Type and Source of Reactive Oxygen Species Influences the Outcome of Oxidative Stress in Cultured Cells. Cells. 2021, 10, 1075. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, P.A.; Zhang, Z.; Pearson, D.V.; Phebus, L.A. Measurement of striatal H2O2 by microdialysis following global forebrain ischemia and reperfusion in the rat: Correlation with the cytotoxic potential of H2O2 in vitro. Brain Res. 1995, 671, 181–186. [Google Scholar] [CrossRef]
- Claude, J.; Linnartz-Gerlach, B.; Kudin, A.P.; Kunz, W.S.; Neumann, H. Microglial CD33-related Siglec-E inhibits neurotoxicity by preventing the phagocytosis-associated oxidative burst. J. Neurosci. 2013, 33, 18270–18276. [Google Scholar] [CrossRef]
- Simpson, D.S.A.; Oliver, P.L. ROS Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef] [PubMed]
- Nissanka, N.; Bacman, S.R.; Plastini, M.J.; Moraes, C.T. The mitochondrial DNA polymerase gamma degrades linear DNA fragments precluding the formation of deletions. Nat. Commun. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Medeiros, T.C.; Thomas, R.L.; Ghillebert, R.; Graef, M. Autophagy balances mtDNA synthesis and degradation by DNA polymerase POLG during starvation. J. Cell Biol. 2018, 217, 1601–1611. [Google Scholar] [CrossRef]
- Volmering, E.; Niehusmann, P.; Peeva, V.; Grote, A.; Zsurka, G.; Altmüller, J.; Nürnberg, P.; Becker, A.J.; Schoch, S.; Elger, C.E.; et al. Neuropathological signs of inflammation correlate with mitochondrial DNA deletions in mesial temporal lobe epilepsy. Acta Neuropathol. 2016, 132, 277–288. [Google Scholar] [CrossRef]
- Campbell, G.R.; Kraytsberg, Y.; Krishnan, K.J.; Ohno, N.; Ziabreva, I.; Reeve, A.; Trapp, B.D.; Newcombe, J.; Reynolds, R.; Lassmann, H.; et al. Clonally expanded mitochondrial DNA deletions within the choroid plexus in multiple sclerosis. Acta Neuropathol. 2012, 124, 209–220. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trombly, G.; Said, A.M.; Kudin, A.P.; Peeva, V.; Altmüller, J.; Becker, K.; Köhrer, K.; Zsurka, G.; Kunz, W.S. The Fate of Oxidative Strand Breaks in Mitochondrial DNA. Antioxidants 2023, 12, 1087. https://doi.org/10.3390/antiox12051087
Trombly G, Said AM, Kudin AP, Peeva V, Altmüller J, Becker K, Köhrer K, Zsurka G, Kunz WS. The Fate of Oxidative Strand Breaks in Mitochondrial DNA. Antioxidants. 2023; 12(5):1087. https://doi.org/10.3390/antiox12051087
Chicago/Turabian StyleTrombly, Genevieve, Afaf Milad Said, Alexei P. Kudin, Viktoriya Peeva, Janine Altmüller, Kerstin Becker, Karl Köhrer, Gábor Zsurka, and Wolfram S. Kunz. 2023. "The Fate of Oxidative Strand Breaks in Mitochondrial DNA" Antioxidants 12, no. 5: 1087. https://doi.org/10.3390/antiox12051087
APA StyleTrombly, G., Said, A. M., Kudin, A. P., Peeva, V., Altmüller, J., Becker, K., Köhrer, K., Zsurka, G., & Kunz, W. S. (2023). The Fate of Oxidative Strand Breaks in Mitochondrial DNA. Antioxidants, 12(5), 1087. https://doi.org/10.3390/antiox12051087







