Novel Role of CETP in Macrophages: Reduction of Mitochondrial Oxidants Production and Modulation of Cell Immune-Metabolic Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Peritoneal Macrophages
2.3. Bone Marrow Derived Macrophages (BMDM)
2.4. Oxidant Generation
2.5. Oxygen Consumption Rates (OCR) and Extracellular Acidification Rates (ECAR)
2.6. Crystal Violet DNA Staining
2.7. Mitochondrial Network Analysis
2.8. Gene Expression by Real Time PCR
2.9. Cytokine Secretion Assay
2.10. Phagocytosis Activity
2.11. Cell Cholesterol Content
2.12. Unesterified Cholesterol Determination by Filipin Staining
2.13. Cholesterol Efflux Assay
2.14. CETP Knockdown by siRNA
2.15. Statistical Data Analyses
3. Results
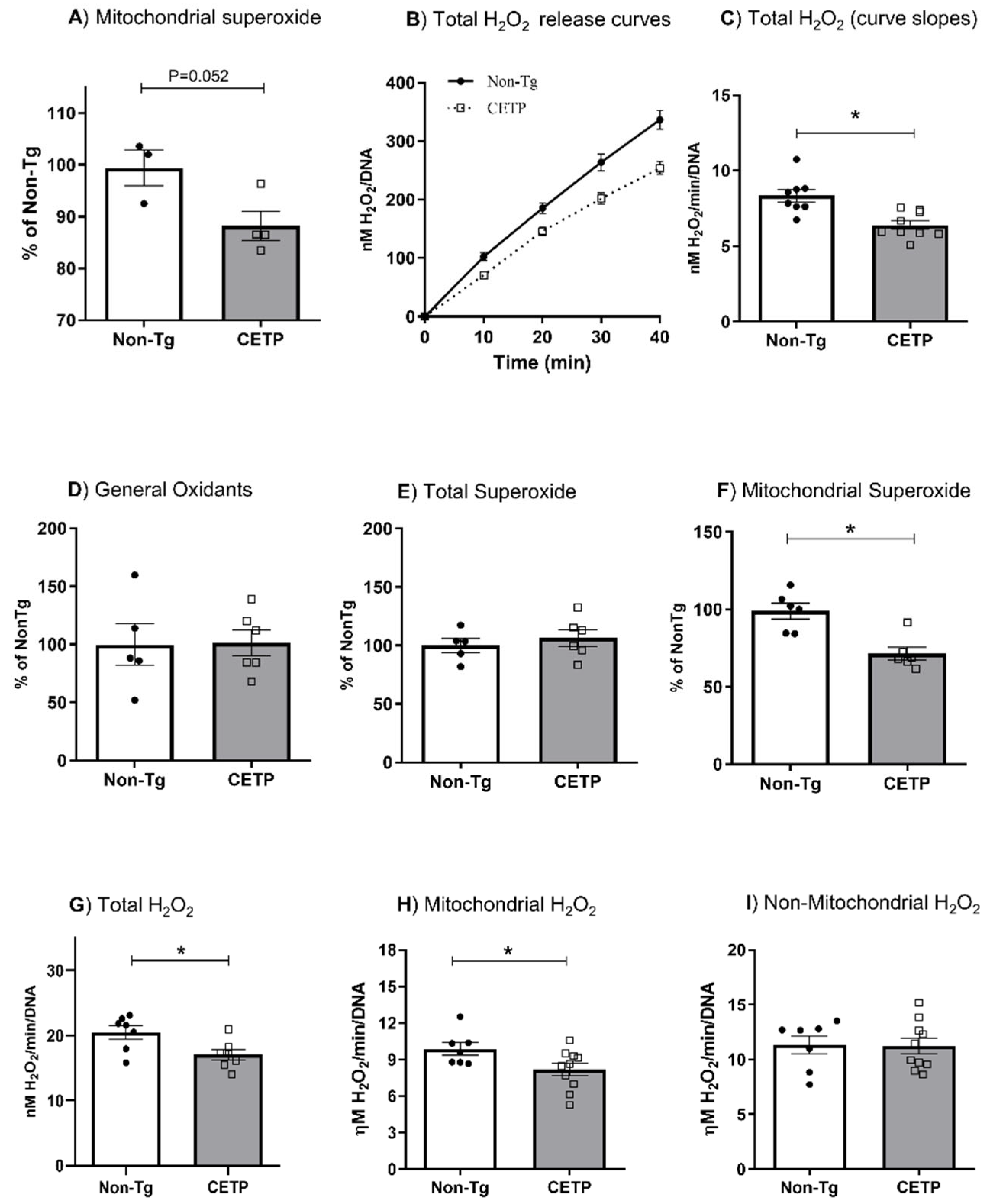
3.1. CETP Expression in Macrophages Decreases Mitochondrial Oxidant Production
3.2. CETP Expression in Macrophages Increases Mitochondrial Maximal Respiration
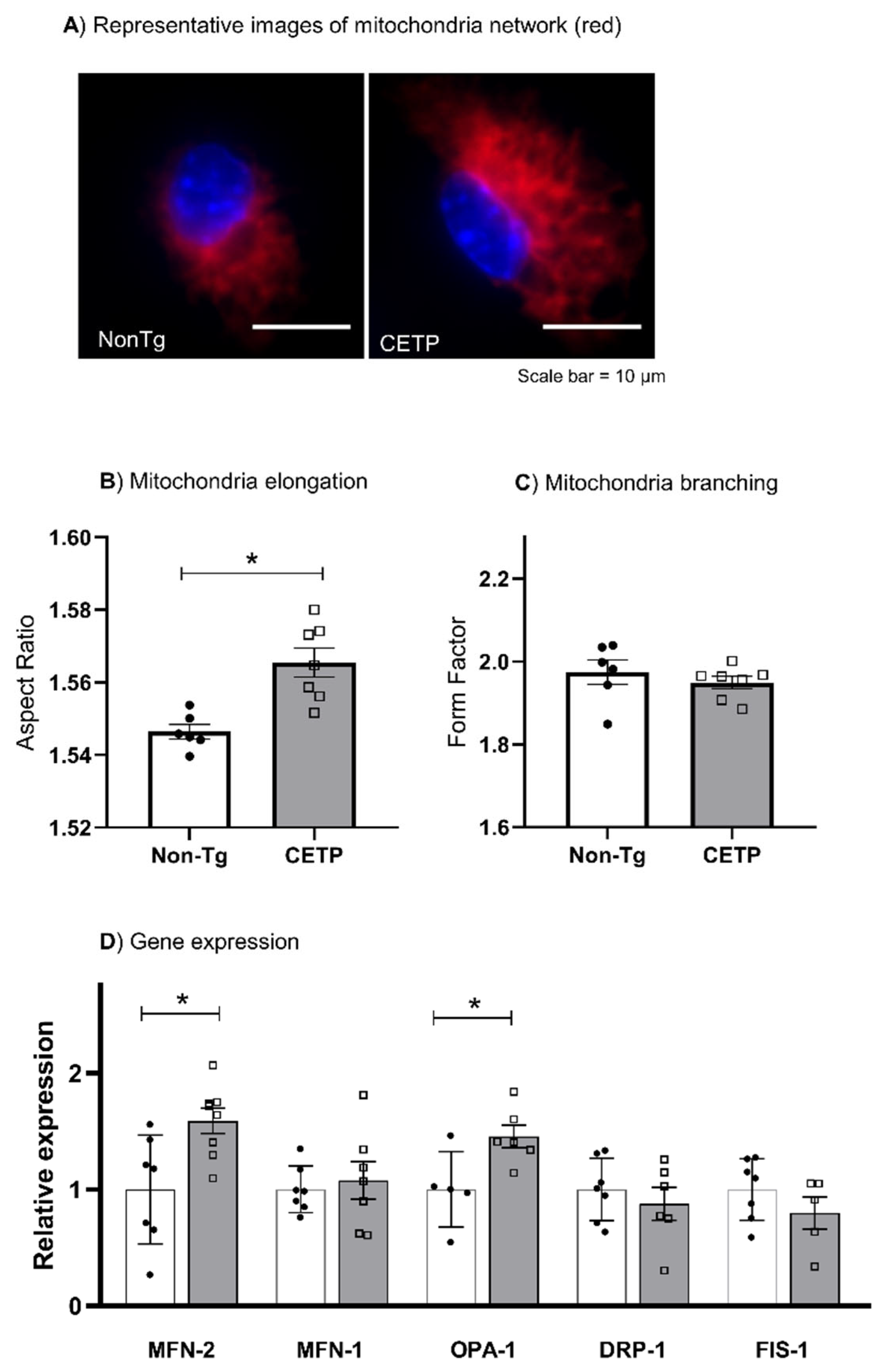
3.3. CETP Expression in Macrophages Increases Mitochondrial Network Elongation
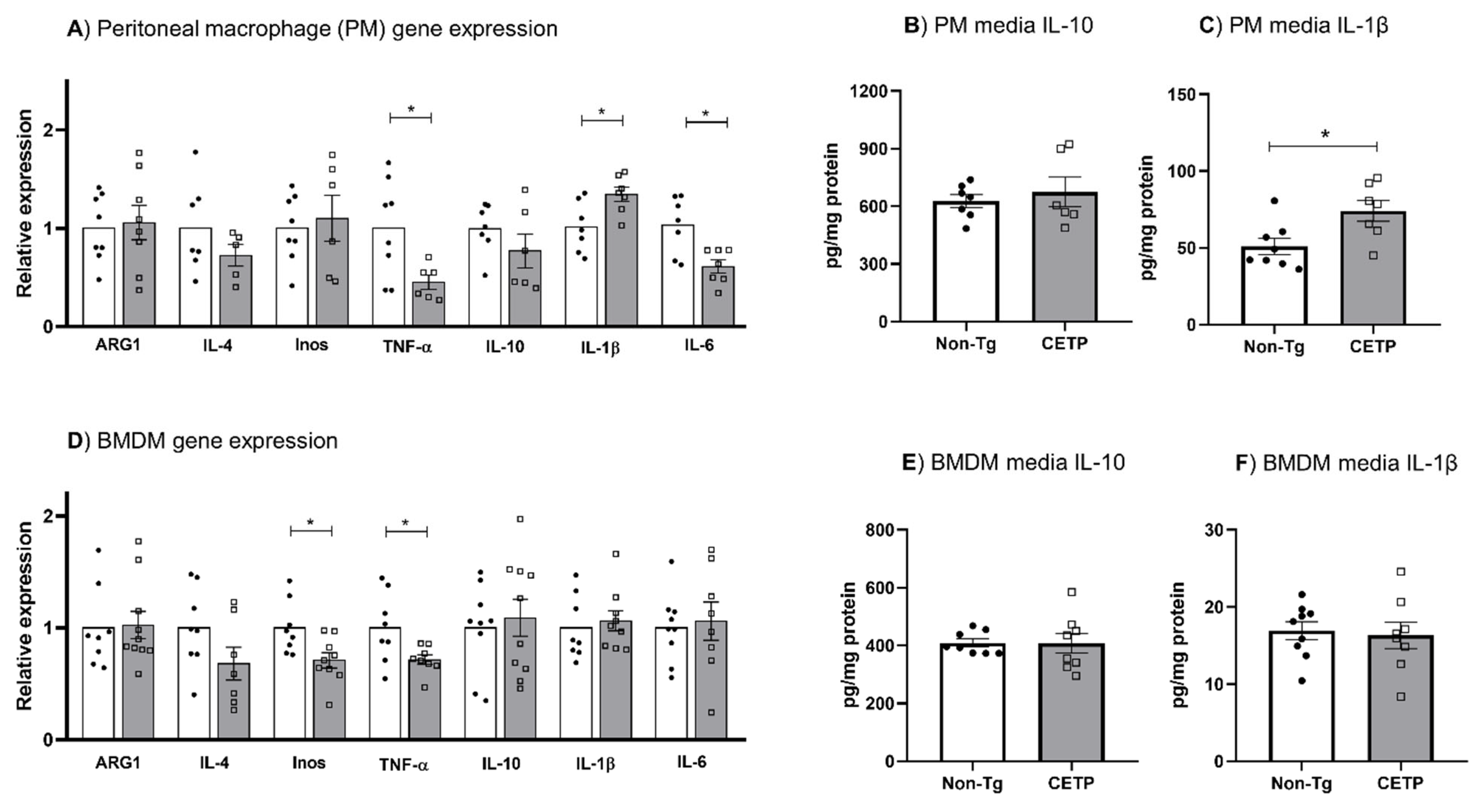
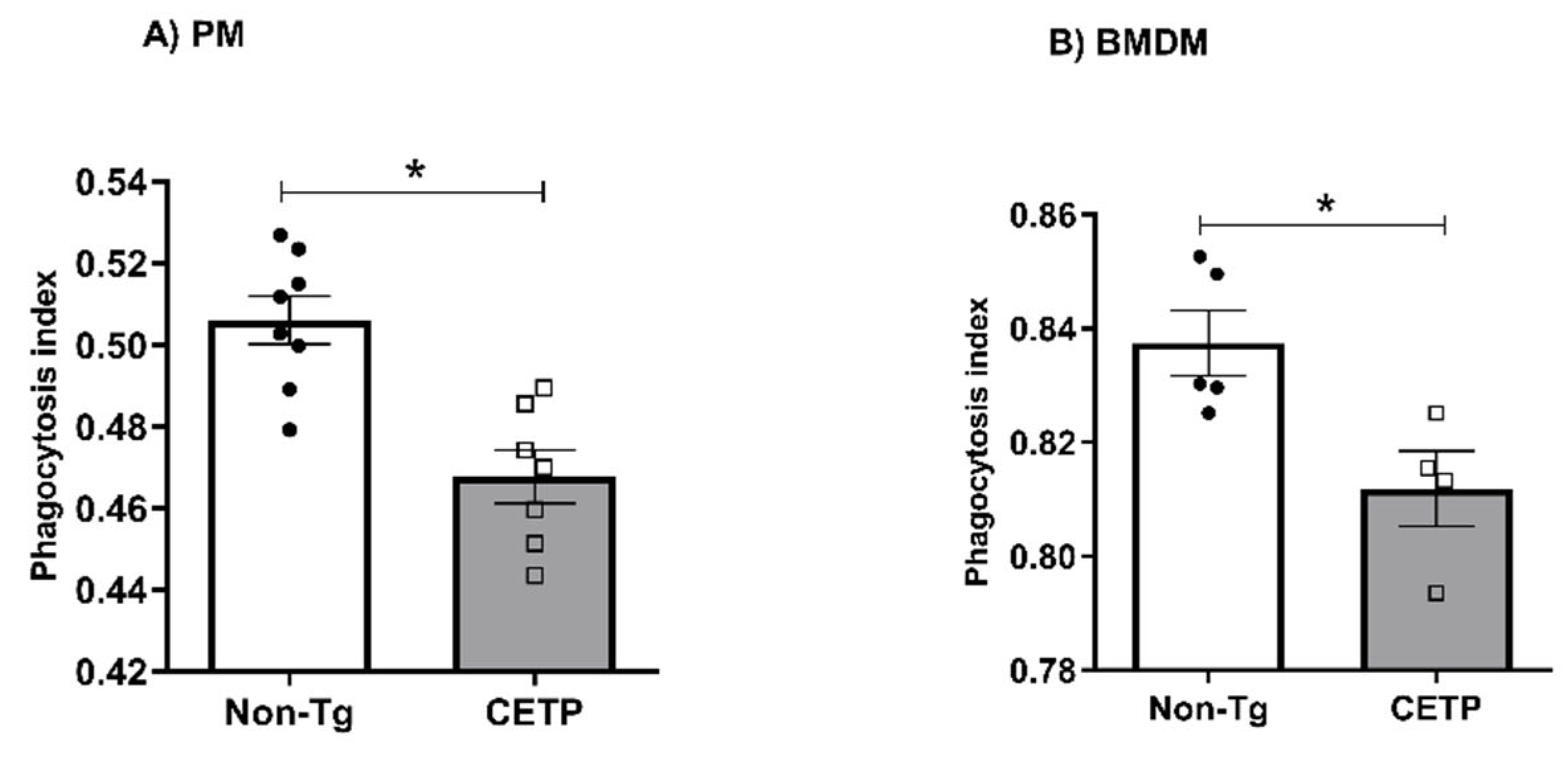
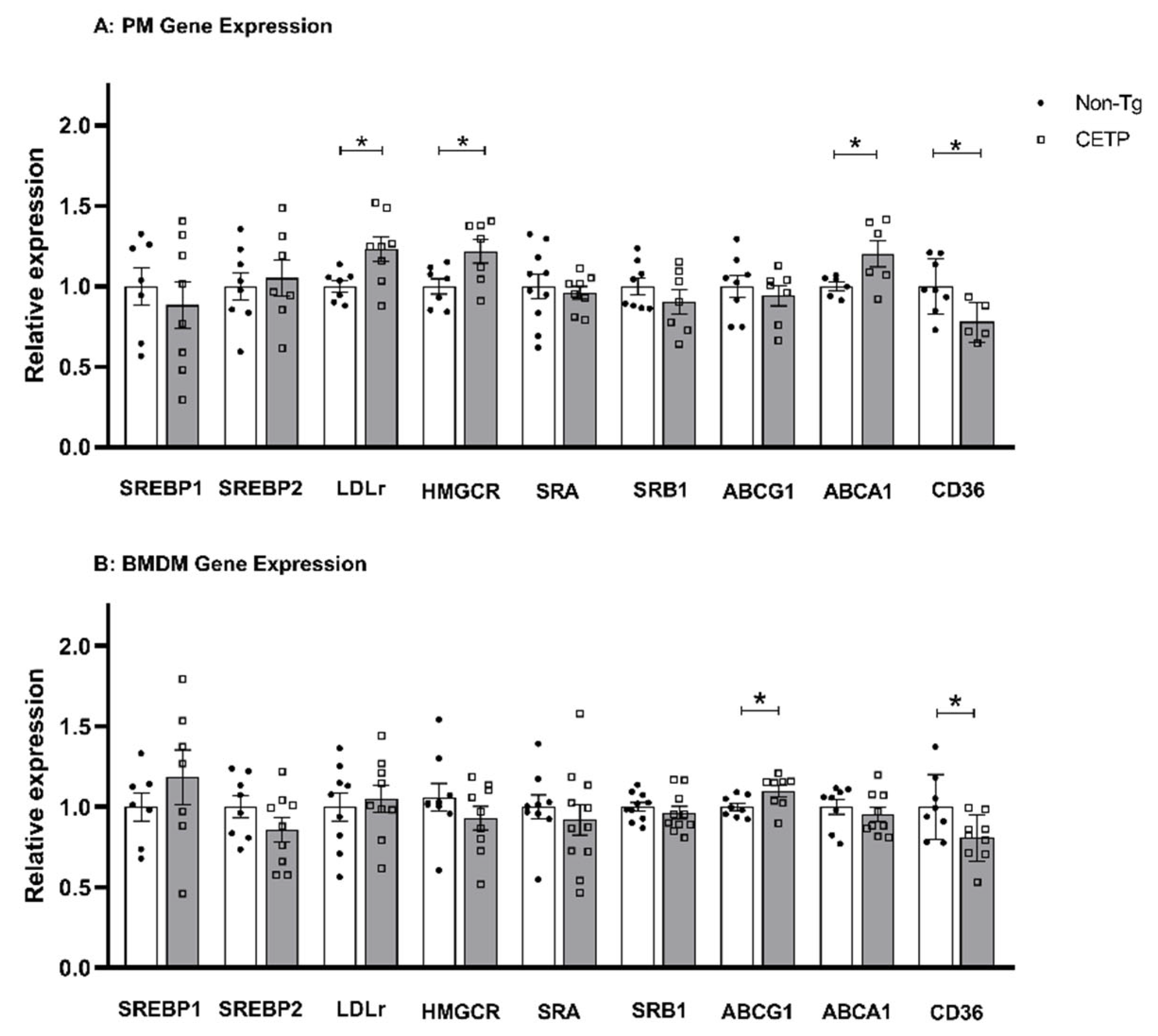
3.4. Effects of CETP Expression on Inflammation Related Genes, Cytokine Secretion and Phagocytosis
3.5. Macrophages Expressing CETP Have Reduced Unesterified Cholesterol Content
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chajek, T.; Fielding, C.J. Isolation and Characterization of a Human Serum Cholesteryl Ester Transfer Protein. Proc. Natl. Acad. Sci. USA 1978, 75, 3445–3449. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.C.F.; Raposo, H.F. Cholesteryl Ester Transfer Protein and Lipid Metabolism and Cardiovascular Diseases. In Lipid Transfer in Lipoprotein Metabolism and Cardiovascular Disease; Springer: Singapore, 2020; pp. 15–25. [Google Scholar]
- Quintão, E.C.R.; Cazita, P.M. Lipid Transfer Proteins: Past, Present and Perspectives. Atherosclerosis 2010, 209, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bowman, L.; Hopewell, J.C.; Chen, F.; Wallendszus, K.; Stevens, W.; Collins, R.; Wiviott, S.D.; Cannon, C.P.; Braunwald, E.; Sammons, E.; et al. Effects of Anacetrapib in Patients with Atherosclerotic Vascular Disease. N. Engl. J. Med. 2017, 377, 1217–1227. [Google Scholar] [CrossRef]
- Morehouse, L.A.; Sugarman, E.D.; Bourassa, P.-A.; Sand, T.M.; Zimetti, F.; Gao, F.; Rothblat, G.H.; Milici, A.J. Inhibition of CETP Activity by Torcetrapib Reduces Susceptibility to Diet-Induced Atherosclerosis in New Zealand White Rabbits. J. Lipid Res. 2007, 48, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Francone, O.L.; Royer, L.; Haghpassand, M. Increased Prebeta-HDL Levels, Cholesterol Efflux, and LCAT-Mediated Esterification in Mice Expressing the Human Cholesteryl Ester Transfer Protein (CETP) and Human Apolipoprotein A-I (ApoA-I) Transgenes. J. Lipid Res. 1996, 37, 1268–1277. [Google Scholar] [CrossRef]
- Collet, X.; Tall, A.R.; Serajuddin, H.; Guendouzi, K.; Royer, L.; Oliveira, H.; Barbaras, R.; Jiang, X.; Francone, O.L. Remodeling of HDL by CETP in Vivo and by CETP and Hepatic Lipase in Vitro Results in Enhanced Uptake of HDL CE by Cells Expressing Scavenger Receptor B-I. J. Lipid Res. 1999, 40, 1185–1193. [Google Scholar] [CrossRef]
- Ishigami, M.; Yamashita, S.; Sakai, N.; Hirano, K.-I.; Arai, T.; Maruyama, T.; Takami, S.; Koyama, M.; Kameda-Takemura, K.; Matsuzawa, Y. High-density Lipoproteins from Probucol-treated Patients Have Increased Capacity to Promote Cholesterol Efflux from Mouse Peritoneal Macrophages Loaded with Acetylated Low-density Lipoproteins. Eur. J. Clin. Investig. 1997, 27, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Villard, E.F.; el Khoury, P.; Duchene, E.; Bonnefont-Rousselot, D.; Clement, K.; Bruckert, E.; Bittar, R.; le Goff, W.; Guerin, M. Elevated CETP Activity Improves Plasma Cholesterol Efflux Capacity from Human Macrophages in Women. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2341–2349. [Google Scholar] [CrossRef]
- Matsuura, F. HDL from CETP-Deficient Subjects Shows Enhanced Ability to Promote Cholesterol Efflux from Macrophages in an ApoE- and ABCG1-Dependent Pathway. J. Clin. Investig. 2006, 116, 1435–1442. [Google Scholar] [CrossRef]
- Venancio, T.M.; Machado, R.M.; Castoldi, A.; Amano, M.T.; Nunes, V.S.; Quintao, E.C.R.; Camara, N.O.S.; Soriano, F.G.; Cazita, P.M. CETP Lowers TLR4 Expression Which Attenuates the Inflammatory Response Induced by LPS and Polymicrobial Sepsis. Mediat. Inflamm. 2016, 2016, 1784014. [Google Scholar] [CrossRef]
- Santana, K.G.; Righetti, R.F.; Breda, C.N.d.S.; Domínguez-Amorocho, O.A.; Ramalho, T.; Dantas, F.E.B.; Nunes, V.S.; Tibério, I.d.F.L.C.; Soriano, F.G.; Câmara, N.O.S.; et al. Cholesterol-Ester Transfer Protein Alters M1 and M2 Macrophage Polarization and Worsens Experimental Elastase-Induced Pulmonary Emphysema. Front. Immunol. 2021, 12, 684076. [Google Scholar] [CrossRef]
- Cazita, P.M.; Barbeiro, D.F.; Moretti, A.I.S.; Quintão, E.C.R.; Soriano, F.G. Human Cholesteryl Ester Transfer Protein Expression Enhances the Mouse Survival Rate in an Experimental Systemic Inflammation Model. Shock 2008, 30, 590–595. [Google Scholar] [CrossRef]
- Grion, C.M.C.; Cardoso, L.T.Q.; Perazolo, T.F.; Garcia, A.S.; Barbosa, D.S.; Morimoto, H.K.; Matsuo, T.; Carrilho, A.J.F. Lipoproteins and CETP Levels as Risk Factors for Severe Sepsis in Hospitalized Patients. Eur. J. Clin. Investig. 2010, 40, 330–338. [Google Scholar] [CrossRef]
- Barter, P.J.; Caulfield, M.; Eriksson, M.; Grundy, S.M.; Kastelein, J.J.P.; Komajda, M.; Lopez-Sendon, J.; Mosca, L.; Tardif, J.-C.; Waters, D.D.; et al. Effects of Torcetrapib in Patients at High Risk for Coronary Events. N. Engl. J. Med. 2007, 357, 2109–2122. [Google Scholar] [CrossRef]
- Martinon, F. Signaling by ROS Drives Inflammasome Activation. Eur. J. Immunol. 2010, 40, 616–619. [Google Scholar] [CrossRef]
- Oliveira, H.C.F.; Vercesi, A.E. Mitochondrial Bioenergetics and Redox Dysfunctions in Hypercholesterolemia and Atherosclerosis. Mol. Asp. Med. 2020, 71, 100840. [Google Scholar] [CrossRef]
- Tschopp, J. Mitochondria: Sovereign of Inflammation? Eur. J. Immunol. 2011, 41, 1196–1202. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in Innate Immune Responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef]
- Emre, Y.; Nübel, T. Uncoupling Protein UCP2: When Mitochondrial Activity Meets Immunity. FEBS Lett. 2010, 584, 1437–1442. [Google Scholar] [CrossRef]
- Rousset, S.; Emre, Y.; Joinlambert, O.; Hurtaud, C.; Ricquier, D.; Cassarddoulcier, A. The Uncoupling Protein 2 Modulates the Cytokine Balance in Innate Immunity. Cytokine 2006, 35, 135–142. [Google Scholar] [CrossRef]
- Assis, L.H.P.; Dorighello, G.G.; Rentz, T.; Souza, J.C.; Vercesi, A.E.; Oliveira, H.C.F. In Vivo Pravastatin Treatment Reverses Hypercholesterolemia Induced Mitochondria-Associated Membranes Contact Sites, Foam Cell Formation, and Phagocytosis in Macrophages. Front. Mol. Biosci. 2022, 9, 839428. [Google Scholar] [CrossRef] [PubMed]
- Trudeau, K.; Molina, A.J.A.; Roy, S. High Glucose Induces Mitochondrial Morphology and Metabolic Changes in Retinal Pericytes. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8657–8664. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lima, C.; Alves, L.E.; Iagher, F.; Machado, A.F.; Bonatto, S.J.; Kuczera, D.; de Souza, C.F.; Pequito, D.C.; Muritiba, A.L.; Nunes, E.A.; et al. Anaerobic Exercise Reduces Tumor Growth, Cancer Cachexia and Increases Macrophage and Lymphocyte Response in Walker 256 Tumor-Bearing Rats. Eur. J. Appl. Physiol. 2008, 104, 957–964. [Google Scholar] [CrossRef]
- Robinet, P.; Wang, Z.; Hazen, S.L.; Smith, J.D. A Simple and Sensitive Enzymatic Method for Cholesterol Quantification in Macrophages and Foam Cells. J. Lipid Res. 2010, 51, 3364–3369. [Google Scholar] [CrossRef]
- Pavlou, S.; Wang, L.; Xu, H.; Chen, M. Higher Phagocytic Activity of Thioglycollate-Elicited Peritoneal Macrophages Is Related to Metabolic Status of the Cells. J. Inflamm. 2017, 14, 4. [Google Scholar] [CrossRef]
- Koshiba, T.; Detmer, S.A.; Kaiser, J.T.; Chen, H.; McCaffery, J.M.; Chan, D.C. Structural Basis of Mitochondrial Tethering by Mitofusin Complexes. Science 2004, 305, 858–862. [Google Scholar] [CrossRef]
- Chinetti-Gbaguidi, G.; Colin, S.; Staels, B. Macrophage Subsets in Atherosclerosis. Nat. Rev. Cardiol. 2015, 12, 10–17. [Google Scholar] [CrossRef]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 Ligands Promote Sterile Inflammation through Assembly of a Toll-like Receptor 4 and 6 Heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef]
- Kowaltowski, A.J.; de Souza-Pinto, N.C.; Castilho, R.F.; Vercesi, A.E. Mitochondria and Reactive Oxygen Species. Free. Radic. Biol. Med. 2009, 47, 333–343. [Google Scholar] [CrossRef]
- van den Bossche, J.; O’Neill, L.A.; Menon, D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 2017, 38, 395–406. [Google Scholar] [CrossRef]
- Flynn, J.M.; Choi, S.W.; Day, N.U.; Gerencser, A.A.; Hubbard, A.; Melov, S. Impaired Spare Respiratory Capacity in Cortical Synaptosomes from Sod2 Null Mice. Free. Radic. Biol. Med. 2011, 50, 866–873. [Google Scholar] [CrossRef][Green Version]
- Connolly, N.M.C.; Theurey, P.; Adam-Vizi, V.; Bazan, N.G.; Bernardi, P.; Bolaños, J.P.; Culmsee, C.; Dawson, V.L.; Deshmukh, M.; Duchen, M.R.; et al. Guidelines on Experimental Methods to Assess Mitochondrial Dysfunction in Cellular Models of Neurodegenerative Diseases. Cell Death Differ. 2018, 25, 542–572. [Google Scholar] [CrossRef]
- Cappel, D.A.; Lantier, L.; Palmisano, B.T.; Wasserman, D.H.; Stafford, J.M. CETP Expression Protects Female Mice from Obesity-Induced Decline in Exercise Capacity. PLoS ONE 2015, 10, e0136915. [Google Scholar] [CrossRef]
- Ježek, J.; Cooper, K.; Strich, R. Reactive Oxygen Species and Mitochondrial Dynamics: The Yin and Yang of Mitochondrial Dysfunction and Cancer Progression. Antioxidants 2018, 7, 13. [Google Scholar] [CrossRef]
- Mills, E.L.; Kelly, B.; O’Neill, L.A.J. Mitochondria Are the Powerhouses of Immunity. Nat. Immunol. 2017, 18, 488–498. [Google Scholar] [CrossRef]
- Erdman, L.K.; Cosio, G.; Helmers, A.J.; Gowda, D.C.; Grinstein, S.; Kain, K.C. CD36 and TLR Interactions in Inflammation and Phagocytosis: Implications for Malaria. J. Immunol. 2009, 183, 6452–6459. [Google Scholar] [CrossRef]
- Ichinohe, T.; Yamazaki, T.; Koshiba, T.; Yanagi, Y. Mitochondrial Protein Mitofusin 2 Is Required for NLRP3 Inflammasome Activation after RNA Virus Infection. Proc. Natl. Acad. Sci. USA 2013, 110, 17963–17968. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Sun, Y.; Ishibashi, M.; Seimon, T.; Lee, M.; Sharma, S.M.; Fitzgerald, K.A.; Samokhin, A.O.; Wang, Y.; Sayers, S.; Aikawa, M.; et al. Free Cholesterol Accumulation in Macrophage Membranes Activates Toll-like Receptors and P38 Mitogen-Activated Protein Kinase and Induces Cathepsin K. Circ. Res. 2009, 104, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Yvan-Charvet, L.; Welch, C.; Pagler, T.A.; Ranalletta, M.; Lamkanfi, M.; Han, S.; Ishibashi, M.; Li, R.; Wang, N.; Tall, A.R. Increased Inflammatory Gene Expression in ABC Transporter–Deficient Macrophages. Circulation 2008, 118, 1837–1847. [Google Scholar] [CrossRef]
- Zhu, X.; Lee, J.-Y.; Timmins, J.M.; Brown, J.M.; Boudyguina, E.; Mulya, A.; Gebre, A.K.; Willingham, M.C.; Hiltbold, E.M.; Mishra, N.; et al. Increased Cellular Free Cholesterol in Macrophage-Specific Abca1 Knock-out Mice Enhances pro-Inflammatory Response of Macrophages. J. Biol. Chem. 2008, 283, 22930–22941. [Google Scholar] [CrossRef]
- Lozhkin, A.; Vendrov, A.E.; Pan, H.; Wickline, S.A.; Madamanchi, N.R.; Runge, M.S. NADPH Oxidase 4 Regulates Vascular Inflammation in Aging and Atherosclerosis. J. Mol. Cell. Cardiol. 2017, 102, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Wang, N.; Tall, A.R.; Tabas, I. Mitochondrial Oxidative Stress Promotes Atherosclerosis and Neutrophil Extracellular Traps in Aged Mice. Arterioscler. Thromb. Vasc. Biol. 2017, 37, e99–e107. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorighello, G.G.; Assis, L.H.P.; Rentz, T.; Morari, J.; Santana, M.F.M.; Passarelli, M.; Ridgway, N.D.; Vercesi, A.E.; Oliveira, H.C.F. Novel Role of CETP in Macrophages: Reduction of Mitochondrial Oxidants Production and Modulation of Cell Immune-Metabolic Profile. Antioxidants 2022, 11, 1734. https://doi.org/10.3390/antiox11091734
Dorighello GG, Assis LHP, Rentz T, Morari J, Santana MFM, Passarelli M, Ridgway ND, Vercesi AE, Oliveira HCF. Novel Role of CETP in Macrophages: Reduction of Mitochondrial Oxidants Production and Modulation of Cell Immune-Metabolic Profile. Antioxidants. 2022; 11(9):1734. https://doi.org/10.3390/antiox11091734
Chicago/Turabian StyleDorighello, Gabriel G., Leandro H. P. Assis, Thiago Rentz, Joseane Morari, Monique F. M. Santana, Marisa Passarelli, Neale D. Ridgway, Anibal E. Vercesi, and Helena C. F. Oliveira. 2022. "Novel Role of CETP in Macrophages: Reduction of Mitochondrial Oxidants Production and Modulation of Cell Immune-Metabolic Profile" Antioxidants 11, no. 9: 1734. https://doi.org/10.3390/antiox11091734
APA StyleDorighello, G. G., Assis, L. H. P., Rentz, T., Morari, J., Santana, M. F. M., Passarelli, M., Ridgway, N. D., Vercesi, A. E., & Oliveira, H. C. F. (2022). Novel Role of CETP in Macrophages: Reduction of Mitochondrial Oxidants Production and Modulation of Cell Immune-Metabolic Profile. Antioxidants, 11(9), 1734. https://doi.org/10.3390/antiox11091734







