Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptomic and qPCR Analysis
2.2. Chemical and Reagents for Metabolic Analysis
2.3. Extraction of Sugarcane Rind Samples
2.4. Chromatography Mass Spectrometry Acquisition Conditions
2.5. Mass Spectrometry Conditions and Metabolic Analysis
2.6. Agrobacterium Mediated Transformation and Phylogenetic Analysis
2.7. Total Amino Acid and Organic Acid Contents and Their Antioxidant Capacity Assays
2.8. Statistical Analysis
3. Results
3.1. Diversity of Major Amino Acids and Organic Acids from the Sugarcane Rind
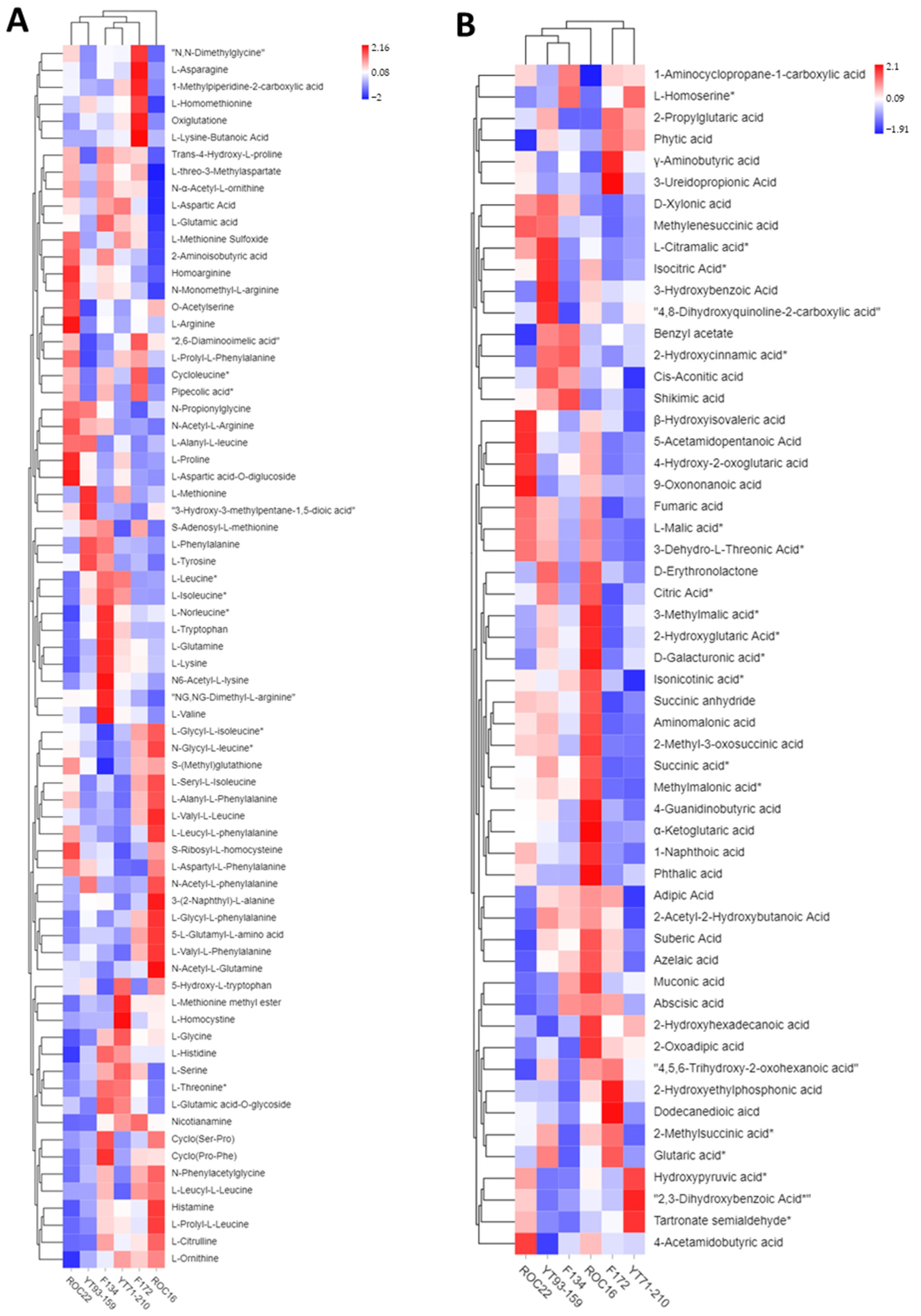
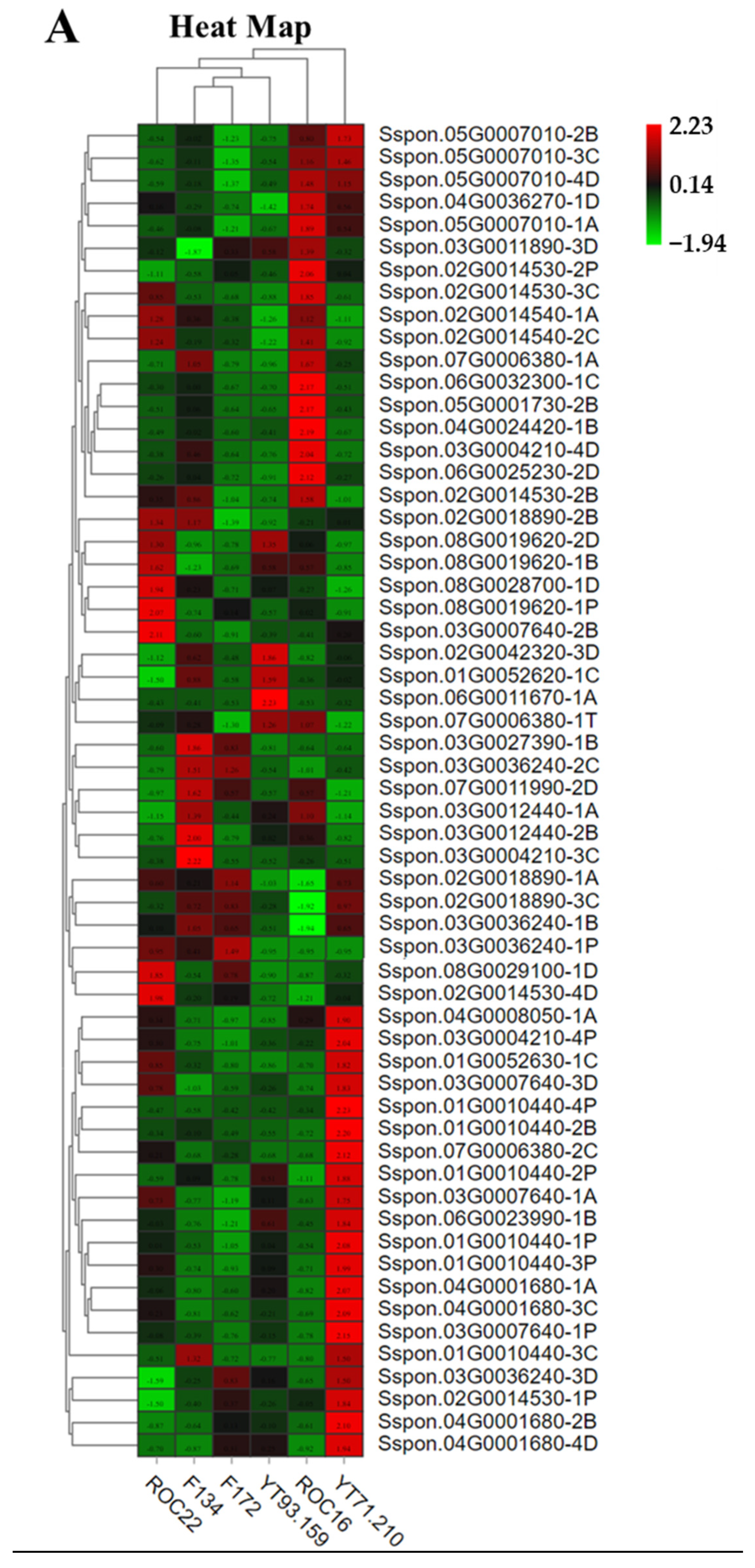
3.2. Hierarchical Cluster Analysis of Metabolites from the Six Sugarcane Varieties
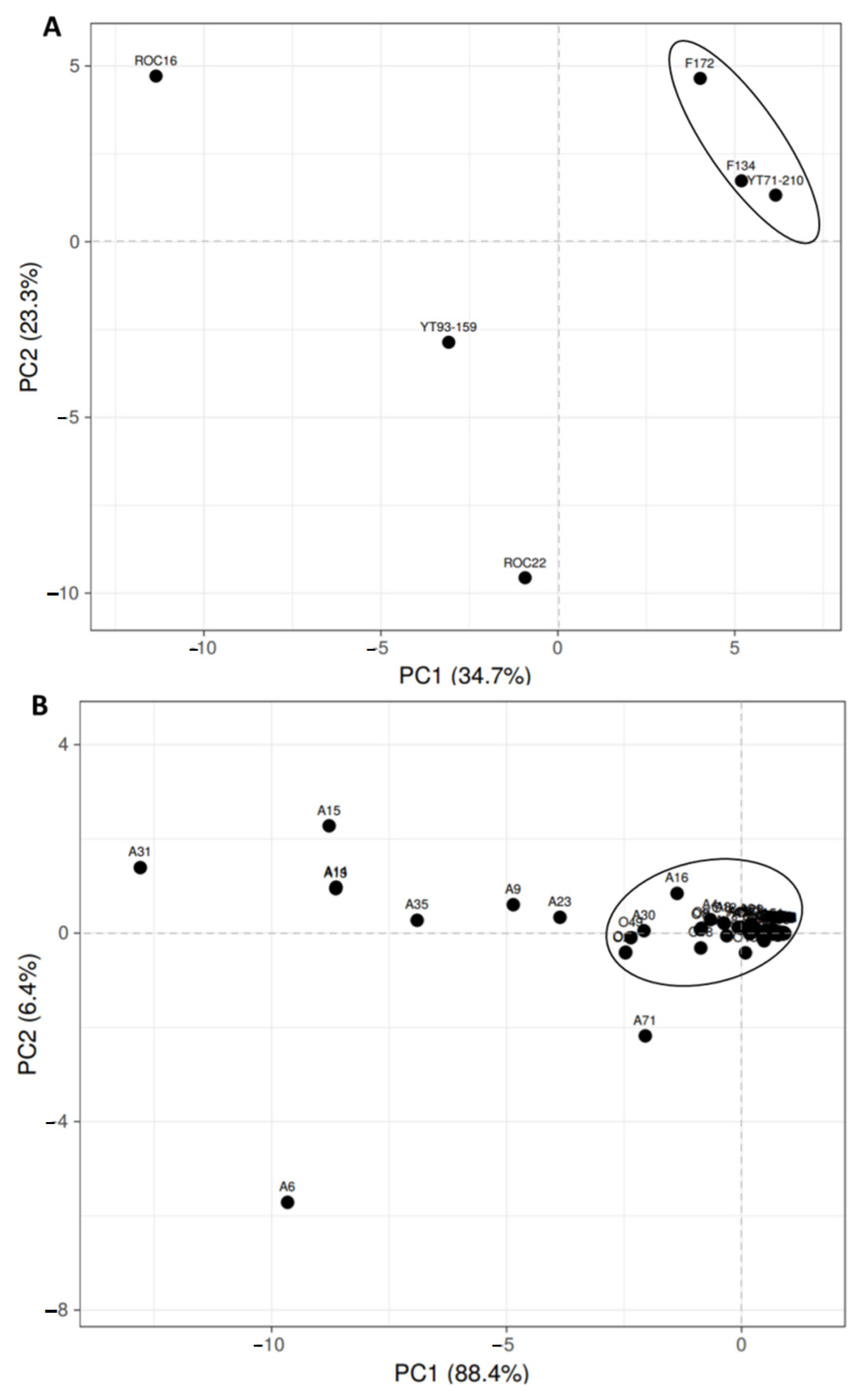
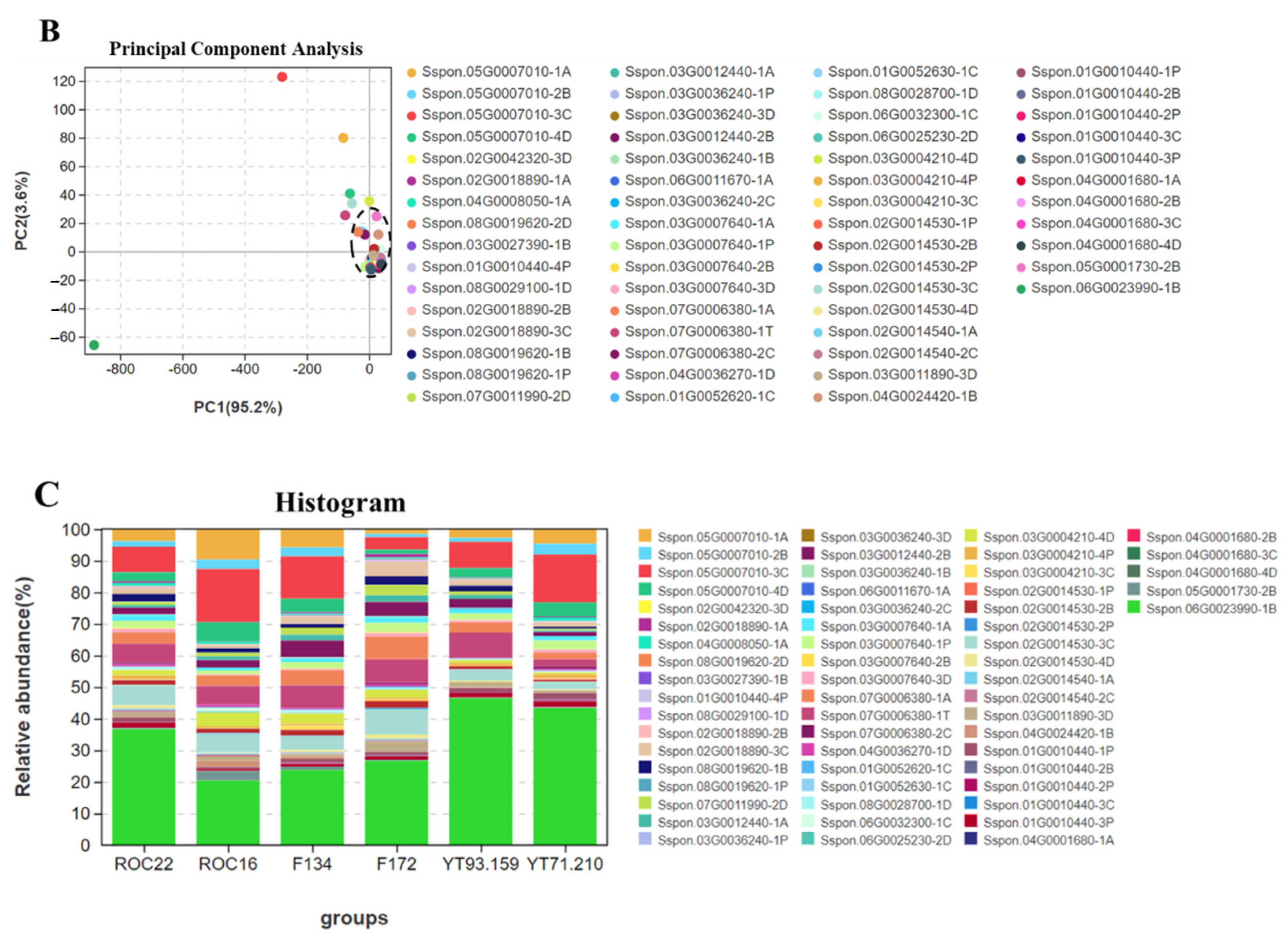
3.3. Principal Component Analysis (PCA) of Organic Acid and Amino Acid
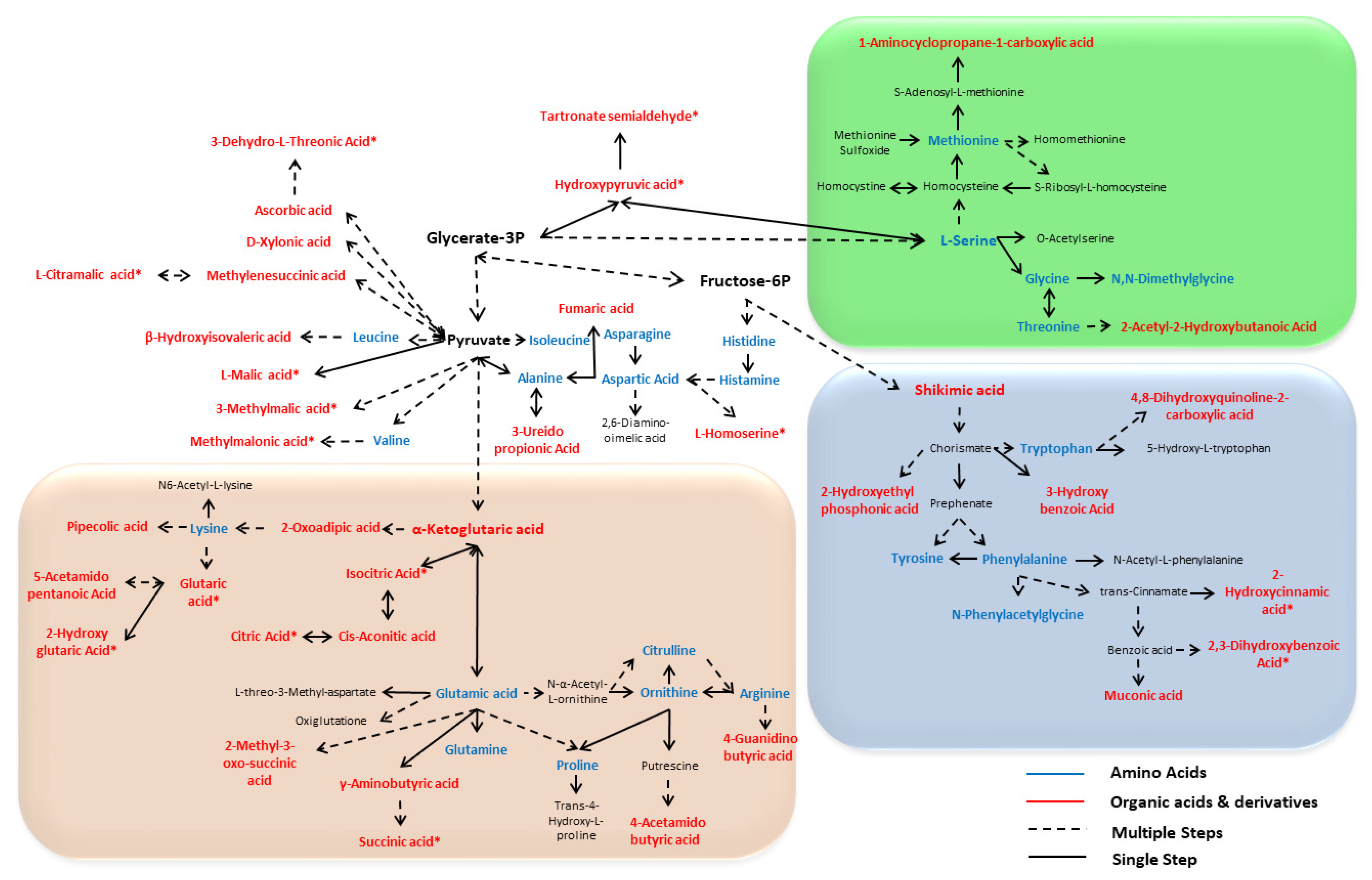
3.4. Comprehensive Amino and Organic Acid Biosynthesis Pathways
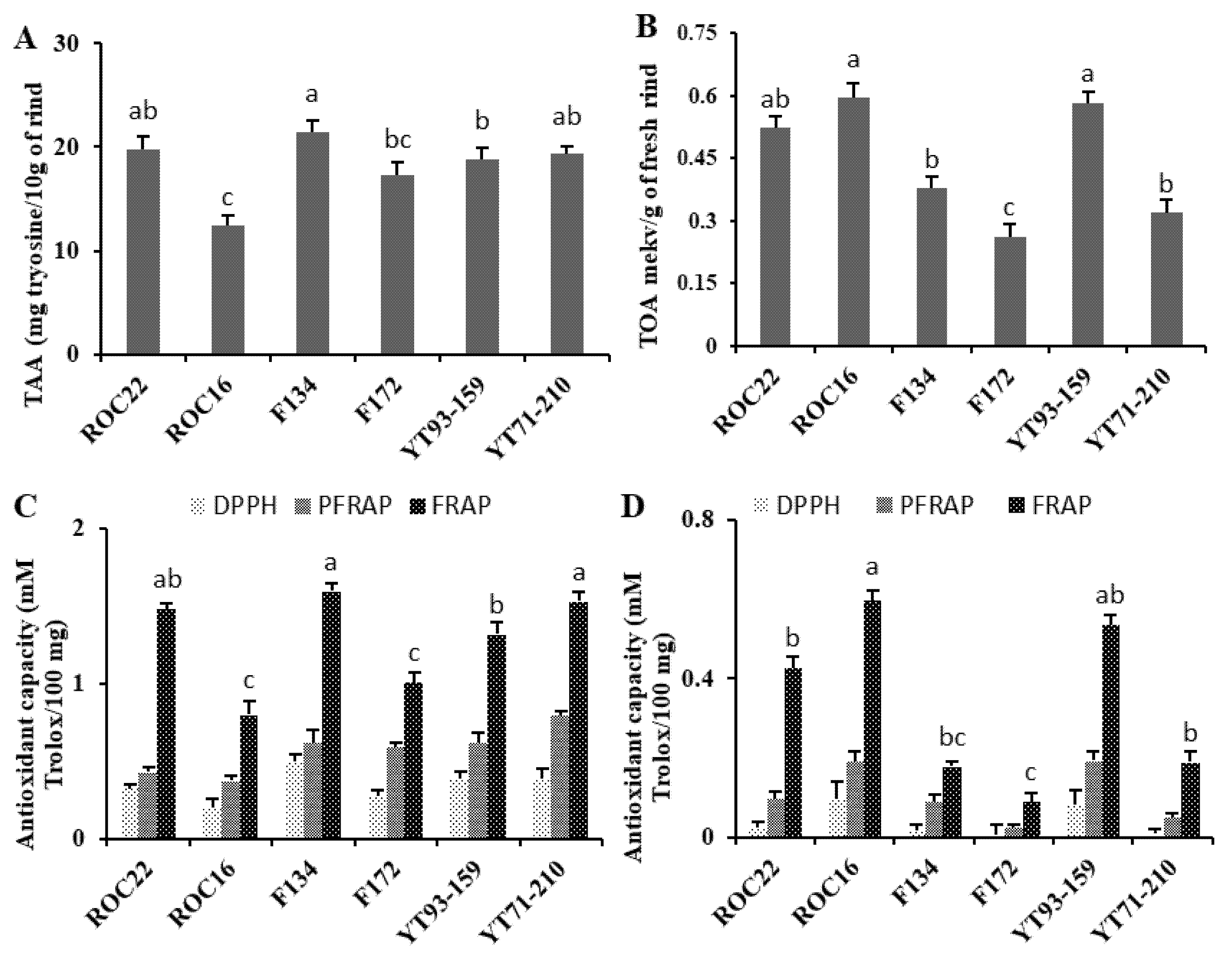
3.5. Bioactivity of Amino and Organic Acid in The Rind of Sugarcane
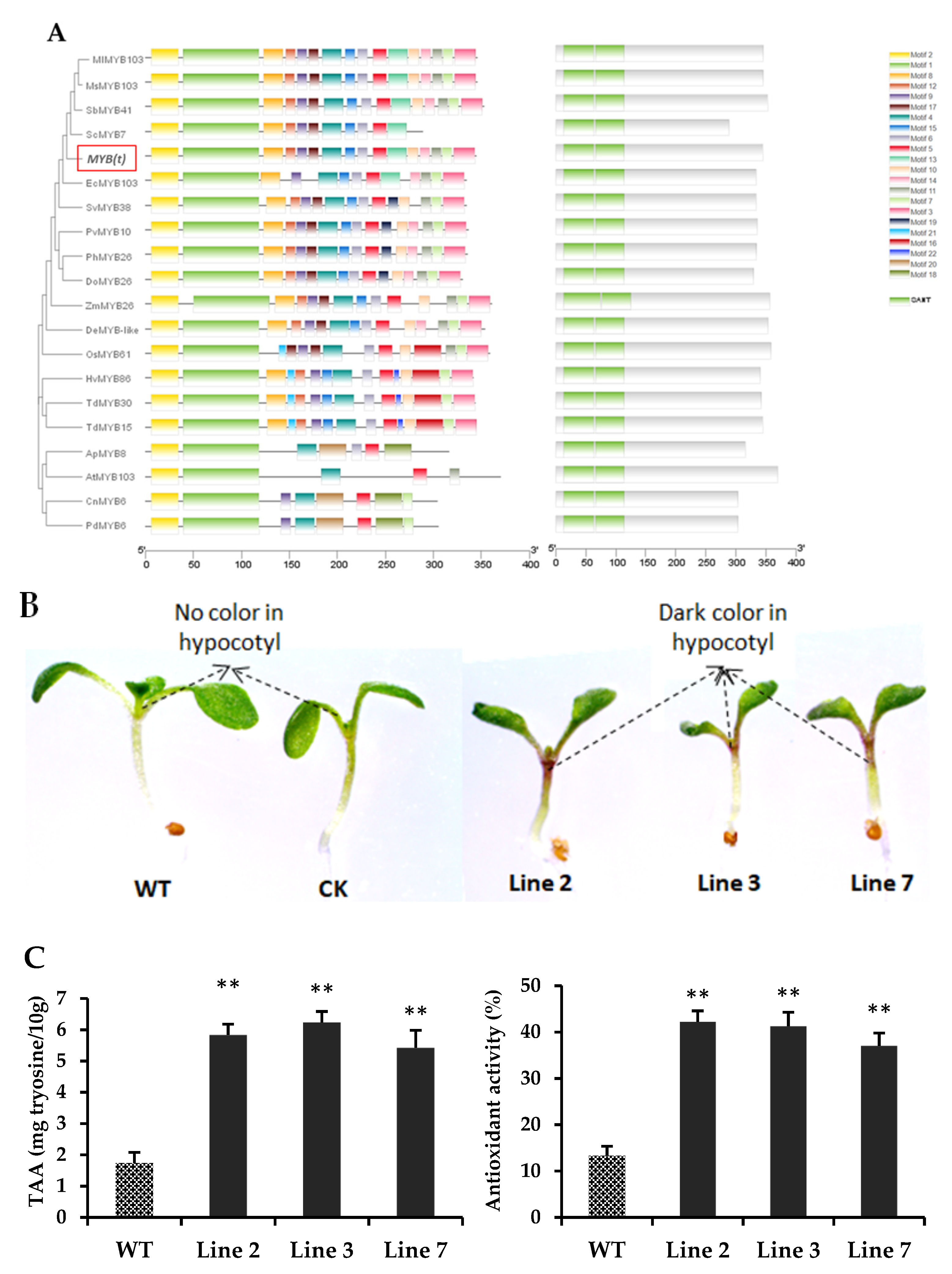
3.6. Transcriptomic Analysis of Genes in Regulating Amino Acid Biosynthesis
3.7. Overexpression of the Candidate MYB(t) Gene Showed a High Accumulation of Amino Acids in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Oliveira Bordonal, R.; Carvalho, J.L.N.; Lal, R.; de Figueiredo, E.B.; de Oliveira, B.G.; La Scala, N. Sustainability of sugarcane production in Brazil. A review. Agron. Sustain. Dev. 2018, 38, 13. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.J.; Duan, M.; Yang, M.; Fan, H.; Shen, S.; Hu, L.; Wang, L. Novel insights into anthocyanin metabolism and molecular characterization of associated genes in sugarcane rinds using the metabolome and transcriptome. Int. J. Mol. Sci. 2022, 23, 338. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Ahmed, U.; Ahmed, M.H.; Duan, M.; Wang, J.; Wang, Y.; Wang, L. Comparison and quantification of metabolites and their antioxidant activities in young and mature leaves of sugarcane. ACS Food Sci. Technol. 2021, 1, 362–373. [Google Scholar] [CrossRef]
- Geng, P.; Fang, Y.; Xie, R.; Hu, W.; Xi, X.; Chu, Q.; Dong, G.; Shaheen, N.; Wei, Y. Separation of phenolic acids from sugarcane rind by online solid-phase extraction with high-speed counter-current chromatography. J. Sep. Sci. 2017, 40, 991–998. [Google Scholar] [CrossRef]
- Rao, M.J.; Duan, M.; Wei, X.; Zuo, H.; Ma, L.; Tahir ul Qamar, M.; Li, M.; Han, S.; Hu, L.; Wang, L. LC-MS/MS-based metabolomics approach revealed novel phytocompounds from sugarcane rind having promising pharmacological value. J. Sci. Food Agric. 2022. [Google Scholar] [CrossRef]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Qiang, X. CsCYT75B1, a Citrus CYTOCHROME P450 gene, is involved in accumulation of antioxidant flavonoids and induces drought tolerance in transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef] [Green Version]
- Duarte-Almeida, J.M.; Salatino, A.; Genovese, M.I.; Lajolo, F.M. Phenolic composition and antioxidant activity of culms and sugarcane (Saccharum officinarum L.) products. Food Chem. 2011, 125, 660–664. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant. Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef]
- Sita, K.; Kumar, V. Role of Gamma Amino Butyric Acid (GABA) against abiotic stress tolerance in legumes: A review. Plant Physiol. Rep. 2020, 25, 654–663. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zheng, X.; Zhu, K.; Xu, Q.; Deng, X. Molecular characterization, critical amino acid identification, and promoter analysis of a lycopene β-cyclase gene from citrus. Tree Genet. Genomes 2016, 12, 106. [Google Scholar] [CrossRef]
- Zhang, B.; Xia, T.; Duan, W.; Zhang, Z.; Li, Y.; Fang, B.; Xia, M.; Wang, M. Effects of organic acids, amino acids and phenolic compounds on antioxidant characteristic of Zhenjiang aromatic vinegar. Molecules 2019, 24, 3799. [Google Scholar] [CrossRef] [Green Version]
- Brosnan, J.T.; Brosnan, M.E. The sulfur-containing amino acids: An overview. J. Nutr. 2006, 136, 1636S–1640S. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Gui, Q.; Shi, J.J.; Zhang, X.Y.; Chen, F.S. Analysis of variation of main components during aging process of Shanxi Aged Vinegar. Acetic Acid Bact. 2013, 2, e6. [Google Scholar] [CrossRef]
- Moosmann, B.; Behl, C. Cytoprotective antioxidant function of tyrosine and tryptophan residues in transmembrane proteins. Eur. J. Biochem. 2000, 267, 5687–5692. [Google Scholar] [CrossRef] [Green Version]
- Croteau, R.; Kutchan, T.M.; Lewis, N.G. Natural products (secondary metabolites). Biochem. Mol. Biol. Plants 2000, 24, 1250–1319. [Google Scholar]
- Barros, J.; Serrani-Yarce, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Razal, R.A.; Ellis, S.; Singh, S.; Lewis, N.G.; Towers, G.H.N. Nitrogen recycling in phenylpropanoid metabolism. Phytochemistry 1996, 41, 31–35. [Google Scholar] [CrossRef]
- Lopez-Bucio, J.; Nieto-Jacobo, M.F.; Ramırez-Rodrıguez, V.; Herrera-Estrella, L. Organic acid metabolism in plants: From adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 2000, 160, 1–13. [Google Scholar] [CrossRef]
- Albertini, M.V.; Carcouet, E.; Pailly, O.; Gambotti, C.; Luro, F.; Berti, L. Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. J. Agric. Food Chem. 2006, 54, 8335–8339. [Google Scholar] [CrossRef]
- Zheng, B.; Zhao, L.; Jiang, X.; Cherono, S.; Liu, J.; Ogutu, C.; Ntini, C.; Zhang, X.; Han, Y. Assessment of organic acid accumulation and its related genes in peach. Food Chem. 2021, 334, 127567. [Google Scholar] [CrossRef]
- Velazquez-martinez, V.; Valles-rosales, D.; Rodriguez-uribe, L.; Holguin, O.; Quintero-quiroz, J.; Reyes-jaquez, D.; Rodriguez-borbon, M.I.; Yazmin, L.; Delgado, E. Antimicrobial, Shelf-Life Stability, and Effect of Maltodextrin and Gum Arabic on the Encapsulation Efficiency of Sugarcane Bagasse Bioactive Compounds. Foods 2021, 10, 116. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelb. Ger. Eur. Mol. Biol. Lab. 2012, 10, f1000research. [Google Scholar]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Chanana, S.; Thomas, C.S.; Zhang, F.; Rajski, S.R.; Bugni, T.S. HCAPCA: Automated hierarchical clustering and principal component analysis of large metabolomic datasets in R. Metabolites 2020, 10, 297. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yemm, E.W.; Cocking, E.C.; Ricketts, R.E. The determination of amino-acids with ninhydrin. Analyst 1955, 80, 209–214. [Google Scholar] [CrossRef]
- Damiano, S.; Forino, M.; De, A.; Vitali, L.A.; Lupidi, G.; Taglialatela-Scafati, O. Antioxidant and antibiofilm activities of secondary metabolites from Ziziphus jujuba leaves used for infusion preparation. Food Chem. 2017, 230, 24–29. [Google Scholar] [CrossRef]
- Ozgen, M.; Scheerens, J.C.; Reese, R.N.; Miller, R.A. Total phenolic, anthocyanin contents and antioxidant capacity of selected elderberry (Sambucus canadensis L.) accessions. Pharmacogn. Mag. 2010, 6, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Oyaizu, M. Studies on products of browning reaction: Antioxidative activity of products of browning reaction. Jpn. J. Nutr 1986, 44, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Nešić, M.; Marković, M.; Trajković, R.; Pavlović, D.; Ilić, M.; Mitić, V.; Stankov-Jovanović, V. Total content of organic acids in plants from fire affected forest. Biol. Nyssana 2010, 1, 65–69. [Google Scholar]
- Al Mahmud, J.; Hasanuzzaman, M.; Nahar, K.; Bhuyan, M.H.M.B.; Fujita, M. Insights into citric acid-induced cadmium tolerance and phytoremediation in Brassica juncea L.: Coordinated functions of metal chelation, antioxidant defense and glyoxalase systems. Ecotoxicol. Environ. Saf. 2018, 147, 990–1001. [Google Scholar] [CrossRef]
- Rao, M.J.; Wu, S.; Duan, M.; Wang, L. Antioxidant metabolites in primitive, wild, and cultivated citrus and their role in stress tolerance. Molecules 2021, 26, 5801. [Google Scholar] [CrossRef]
- Rao, M.J.; Xu, Y.; Huang, Y.; Tang, X.; Deng, X.; Xu, Q. Ectopic expression of citrus UDP-GLUCOSYL TRANSFERASE gene enhances anthocyanin and proanthocyanidins contents and confers high light tolerance in Arabidopsis. BMC Plant Biol. 2019, 19, 603. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Öhman, D.; Demedts, B.; Kumar, M.; Gerber, L.; Gorzsás, A.; Goeminne, G.; Hedenström, M.; Ellis, B.; Boerjan, W.; Sundberg, B. MYB 103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in A rabidopsis stems. Plant J. 2013, 73, 63–76. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Lynch, J.H.; Dudareva, N. Aromatic amino acids: A complex network ripe for future exploration. Trends Plant Sci. 2020, 25, 670–681. [Google Scholar] [CrossRef]
- Galili, G.; Amir, R.; Fernie, A.R. The regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 2016, 67, 153–178. [Google Scholar] [CrossRef]
- Killiny, N.; Hijaz, F. Amino acids implicated in plant defense are higher in Candidatus Liberibacter asiaticus-tolerant citrus varieties. Plant Signal. Behav. 2016, 11, e1171449. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.J.; Ding, F.; Wang, N.; Deng, X.; Xu, Q. Metabolic mechanisms of host species against citrus huanglongbing (Greening Disease). Crit. Rev. Plant Sci. 2018, 37, 496–511. [Google Scholar] [CrossRef]
- Touraine, B.; Muller, B.; Grignon, C. Effect of phloem-translocated malate on NO3−uptake by roots of intact soybean plants. Plant Physiol. 1992, 99, 1118–1123. [Google Scholar] [CrossRef] [Green Version]
- El-Kereamy, A.; Bi, Y.-M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.J.; Duan, M.; Yang, M.; Li, M.; Wang, L. Sugarcane rind secondary metabolites and their antioxidant activities in eleven cultivated sugarcane varieties. Sugar Tech. 2022. [Google Scholar] [CrossRef]
- Cin, V.D.; Tieman, D.M.; Tohge, T.; McQuinn, R.; de Vos, R.C.H.; Osorio, S.; Schmelz, E.A.; Taylor, M.G.; Smits-Kroon, M.T.; Schuurink, R.C.; et al. Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 2011, 23, 2738–2753. [Google Scholar]
- Bovy, A.; De Vos, R.; Kemper, M.; Schijlen, E.; Almenar Pertejo, M.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [Green Version]
- Rao, M.J.; Wang, L. CRISPR/Cas9 technology for improving agronomic traits and future prospective in agriculture. Planta 2021, 254, 68. [Google Scholar] [CrossRef]
- Rao, M.J.; Zuo, H.; Xu, Q. Genomic insights into citrus domestication and its important agronomic traits. Plant Commun. 2021, 2, 100138. [Google Scholar] [CrossRef]
- Abbas, M.; Hernández-García, J.; Blanco-Touriñán, N.; Aliaga, N.; Minguet, E.G.; Alabadí, D.; Blázquez, M.A. Reduction of indole-3-acetic acid methyltransferase activity compensates for high-temperature male sterility in Arabidopsis. Plant Biotechnol. J. 2018, 16, 272–279. [Google Scholar] [CrossRef] [Green Version]

| Variables | Total Amino Acid | Total Organic Acid | DPPH | PFRAP | FRAP |
|---|---|---|---|---|---|
| Total amino acid | 1.00 | ||||
| Total organic acid | −0.37 | 1.00 | |||
| DPPH | 0.90 | −0.28 | 1.00 | ||
| PFRAP | 0.57 | −0.64 | 0.63 | 1.00 | |
| FRAP | 0.94 | −0.24 | 0.88 | 0.56 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, M.J.; Duan, M.; Wang, J.; Han, S.; Ma, L.; Mo, X.; Li, M.; Hu, L.; Wang, L. Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties. Antioxidants 2022, 11, 1319. https://doi.org/10.3390/antiox11071319
Rao MJ, Duan M, Wang J, Han S, Ma L, Mo X, Li M, Hu L, Wang L. Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties. Antioxidants. 2022; 11(7):1319. https://doi.org/10.3390/antiox11071319
Chicago/Turabian StyleRao, Muhammad Junaid, Mingzheng Duan, Jihong Wang, Shijian Han, Li Ma, Xinyi Mo, Min Li, Lihua Hu, and Lingqiang Wang. 2022. "Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties" Antioxidants 11, no. 7: 1319. https://doi.org/10.3390/antiox11071319
APA StyleRao, M. J., Duan, M., Wang, J., Han, S., Ma, L., Mo, X., Li, M., Hu, L., & Wang, L. (2022). Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties. Antioxidants, 11(7), 1319. https://doi.org/10.3390/antiox11071319








