Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Supplies
2.2. Preparation of Buffer Solutions and Reagents
2.3. Standard and Solutions
2.4. Patients and Serum Samples
2.5. Derivatization Procedure
2.6. Detection of H2S Species in Serum Samples
2.7. Instrumentation and Analytical Methods
2.8. Method Validation and Statistical Analysis
3. Results
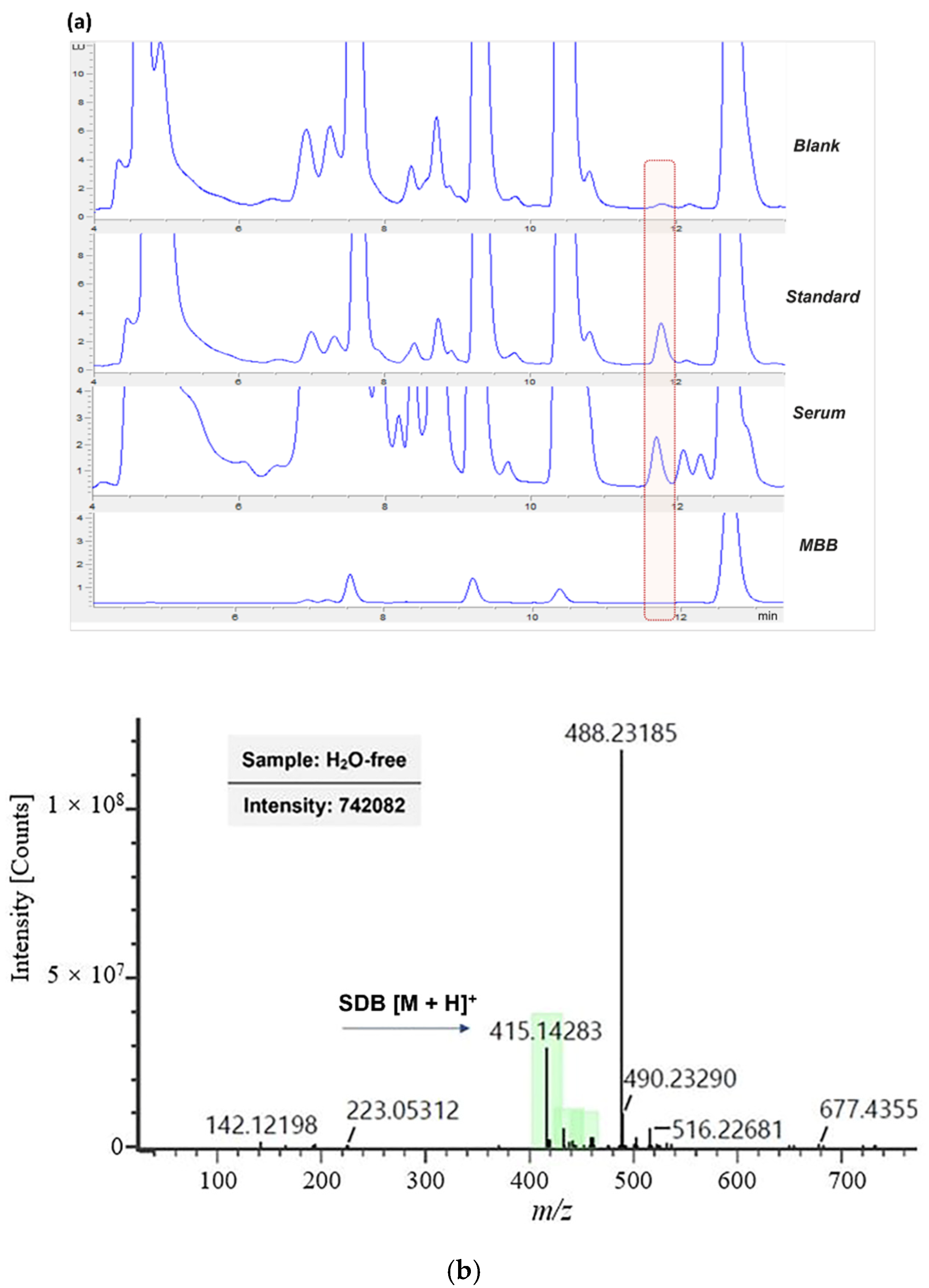
3.1. RP-HPLC-FLD Separation of Derivatization Product
3.2. Sulfide Levels Quantification, Method Development
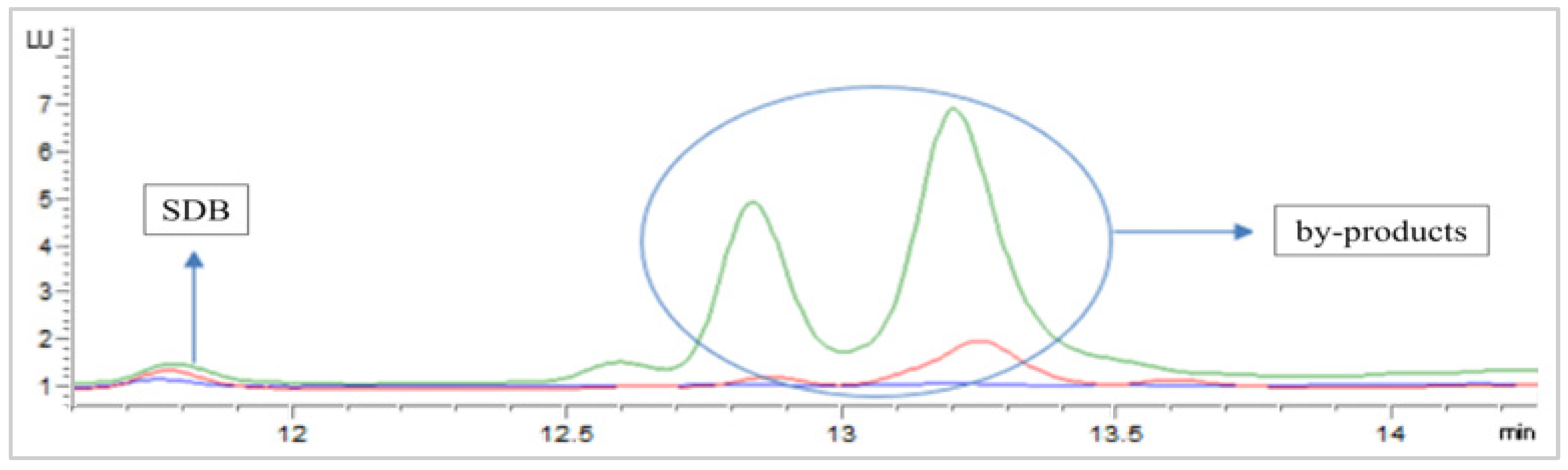
3.2.1. Derivatization Method Optimization
MBB Concentration
Temperature
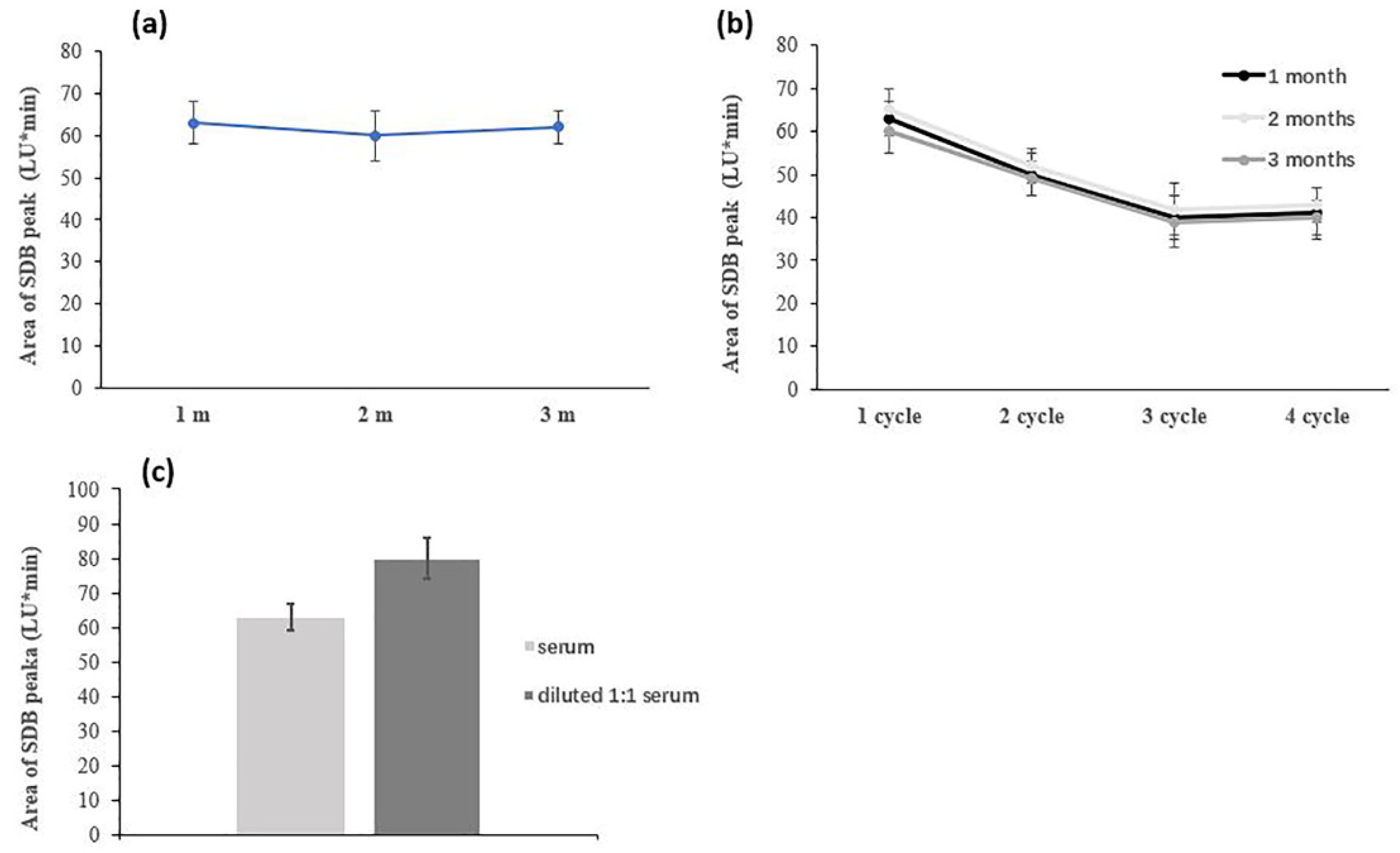
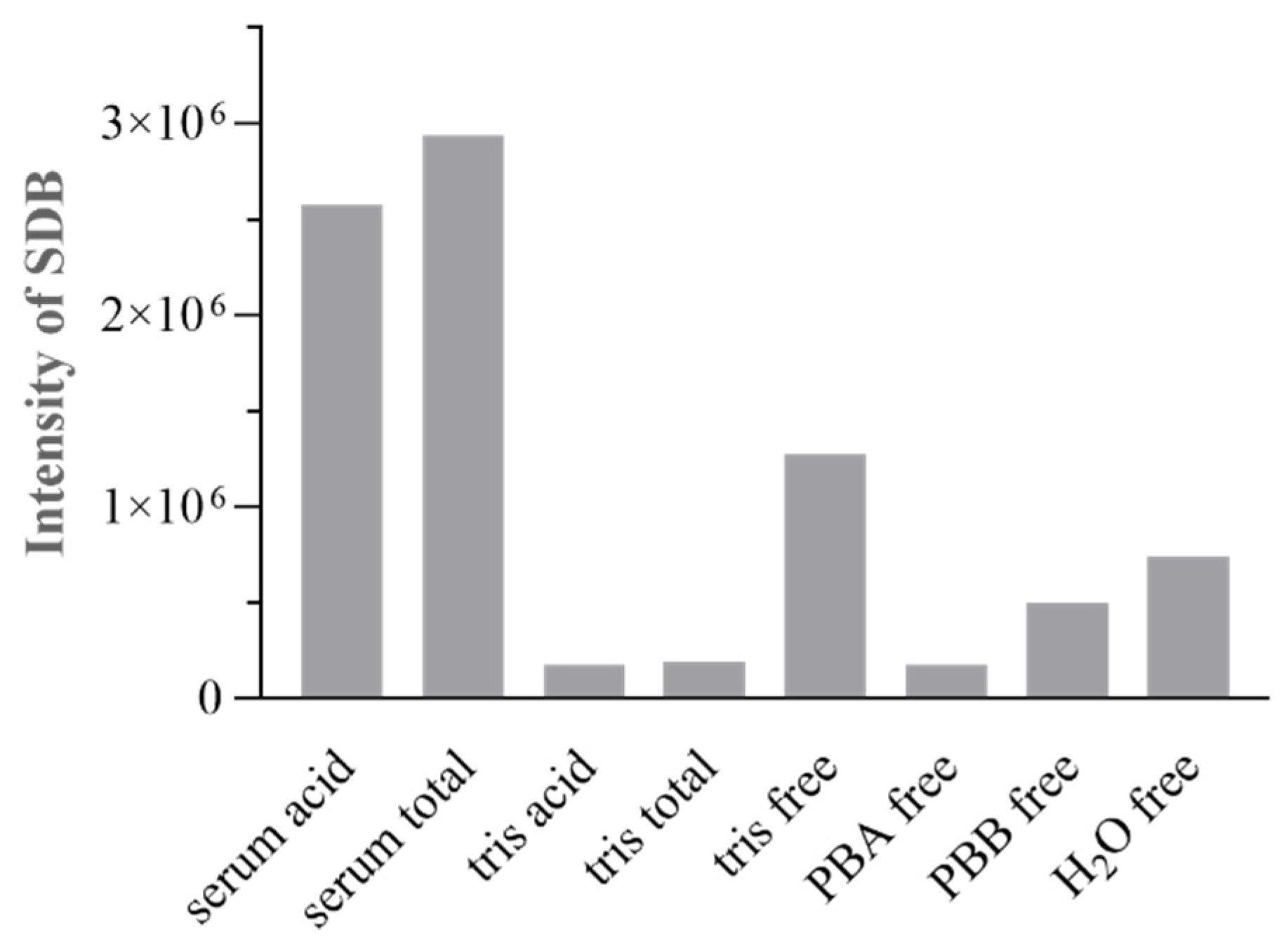
Serum Handling (Storage Conditions, Aging, Dilution)
Speciation Protocol Tubes
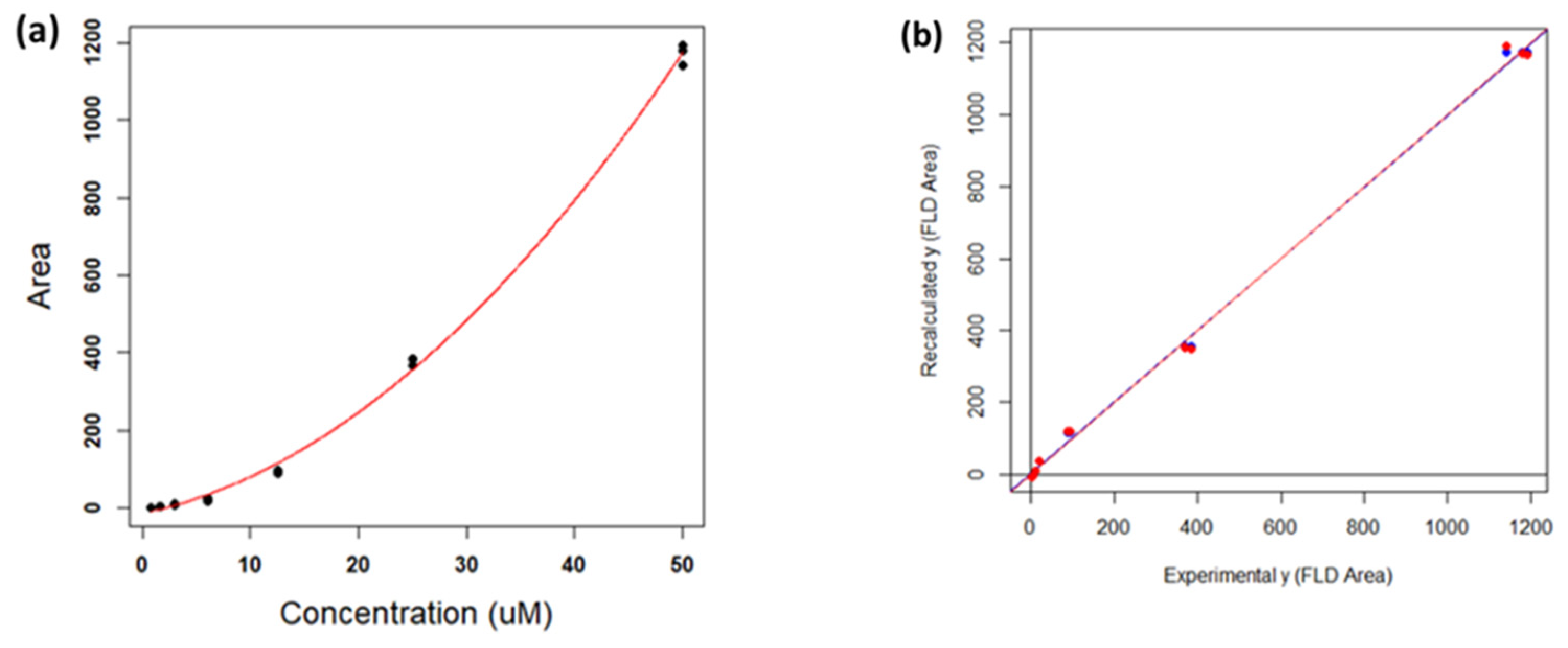
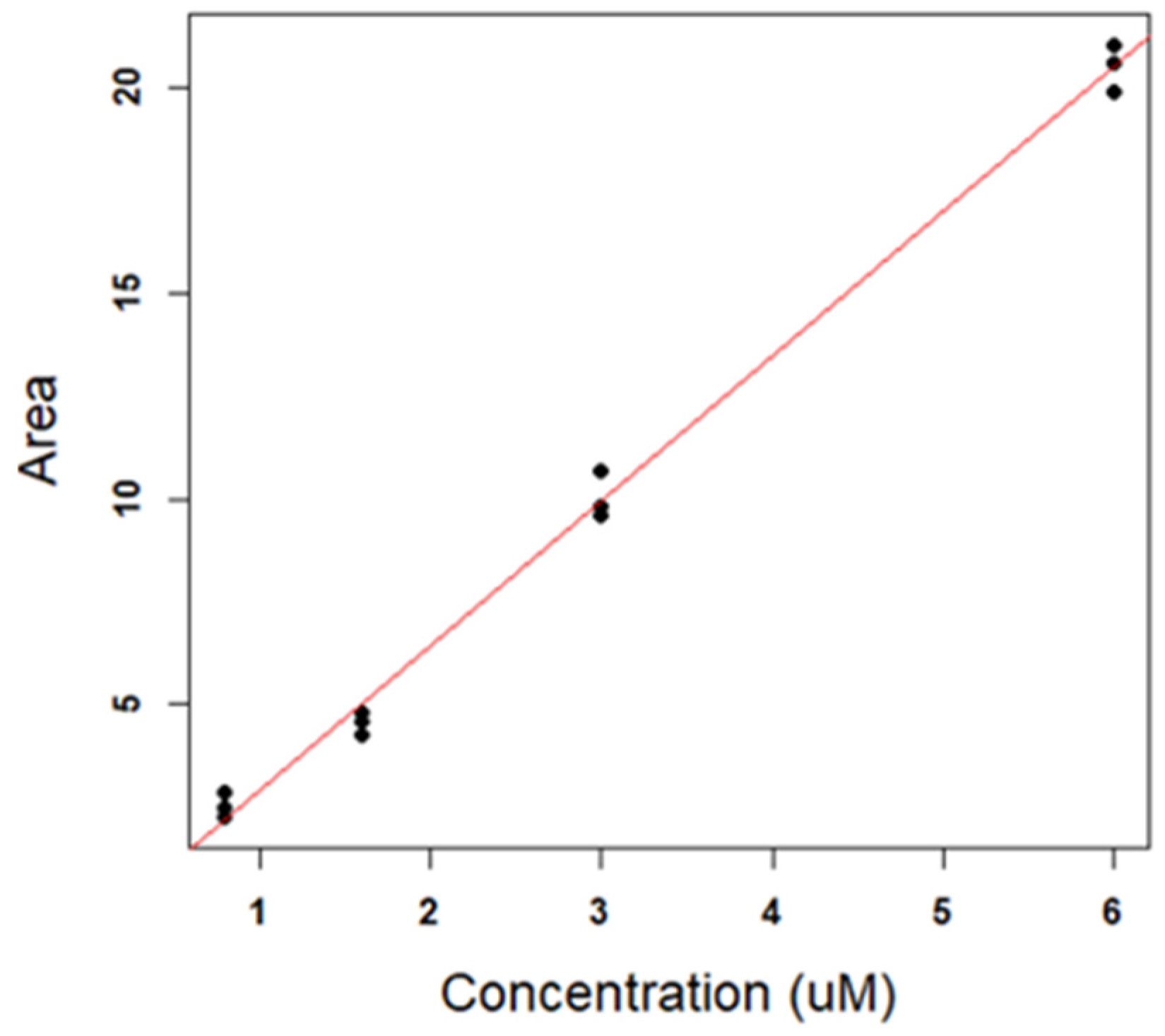
3.2.2. Calibration Curve: Optimization and Validation
3.3. Efficacy of the Method on Human Serum Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, G.; Wu, L.; Jiang, B.; Yang, W.; Qi, J.; Cao, K.; Meng, Q.; Mustafa, A.K.; Mu, W.; Zhang, S.; et al. H2S as a Physiologic Vasorelaxant: Hypertension in Mice with Deletion of Cystathionine γ-Lyase. Science 2008, 322, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.; Sandhir, R. Hydrogen Sulfide in Physiological and Pathological Mechanisms in Brain. CNS Neurol. Disord. Drug Targets 2018, 17, 654–670. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H. Hydrogen Sulfide as a Neuromodulator. Mol. Neurobiol. 2002, 26, 13–19. [Google Scholar] [CrossRef]
- Lobb, I.; Sonke, E.; Aboalsamh, G.; Sener, A. Hydrogen sulphide and the kidney: Important roles in renal physiology and pathogenesis and treatment of kidney injury and disease. Nitric Oxide 2015, 46, 55–65. [Google Scholar] [CrossRef]
- Chen, H.J.; Ngowi, E.E.; Qian, L.; Li, T.; Qin, Y.Z.; Zhou, J.J.; Li, K.; Ji, X.Y.; Wu, D.D. Role of Hydrogen Sulfide in the Endocrine System. Front. Endocrinol. 2021, 12, 704620. [Google Scholar] [CrossRef]
- Hine, C.; Zhu, Y.; Hollenberg, A.N.; Mitchell, J.R. Dietary and Endocrine Regulation of Endogenous Hydrogen Sulfide Production: Implications for Longevity. Antioxid. Redox Signal. 2018, 28, 1483–1502. [Google Scholar] [CrossRef]
- Jimenez, M.; Gil, V.; Martinez-Cutillas, M.; Mane, N.; Gallego, D. Hydrogen sulphide as a signalling molecule regulating physiopathological processes in gastrointestinal motility. Br. J. Pharmacol. 2017, 174, 2805–2817. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R. A Case for Hydrogen Sulfide Metabolism as an Oxygen Sensing Mechanism. Antioxidants 2021, 10, 1650. [Google Scholar] [CrossRef]
- Dong, Q.; Yang, B.; Han, J.G.; Zhang, M.M.; Liu, W.; Zhang, X.; Yu, H.L.; Liu, Z.G.; Zhang, S.H.; Li, T.; et al. A novel hydrogen sulfide-releasing donor, HA-ADT, suppresses the growth of human breast cancer cells through inhibiting the PI3K/AKT/mTOR and Ras/Raf/MEK/ERK signaling pathways. Cancer Lett. 2019, 455, 60–72. [Google Scholar] [CrossRef]
- Vitvitsky, V.; Kabil, O.; Banerjee, R. High turnover rates for hydrogen sulfide allow for rapid regulation of its tissue concentrations. Antioxid. Redox Signal. 2012, 17, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R. A practical look at the chemistry and biology of hydrogen sulfide. Antioxid. Redox Signal. 2012, 17, 32–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabil, O.; Banerjee, R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 2014, 20, 770–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Minkler, P.; Grove, D.; Wang, R.; Willard, B.; Dweik, R.; Hine, C. Non-enzymatic hydrogen sulfide production from cysteine in blood is catalyzed by iron and vitamin B6. Commun. Biol. 2019, 2, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolluru, G.K.; Shen, X.; Bir, S.C.; Kevil, C.G. Hydrogen sulfide chemical biology: Pathophysiological roles and detection. Nitric Oxide 2013, 35, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Greiner, R.; Palinkas, Z.; Basell, K.; Becher, D.; Antelmann, H.; Nagy, P.; Dick, T.P. Polysulfides link H2S to protein thiol oxidation. Antioxid. Redox Signal. 2013, 19, 1749–1765. [Google Scholar] [CrossRef] [Green Version]
- Kolluru, G.K.; Shen, X.; Kevil, C.G. Reactive Sulfur Species: A New Redox Player in Cardiovascular Pathophysiology. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 874–884. [Google Scholar] [CrossRef]
- Nagy, P.; Palinkas, Z.; Nagy, A.; Budai, B.; Toth, I.; Vasas, A. Chemical aspects of hydrogen sulfide measurements in physiological samples. Biochim. Biophys. Acta 2014, 1840, 876–891. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H. H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol 2012, 13, 499–507. [Google Scholar] [CrossRef]
- Ono, K.; Akaike, T.; Sawa, T.; Kumagai, Y.; Wink, D.A.; Tantillo, D.J.; Hobbs, A.J.; Nagy, P.; Xian, M.; Lin, J.; et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: Implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014, 77, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P. Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol. 2015, 554, 3–29. [Google Scholar] [CrossRef]
- Kabil, O.; Banerjee, R. Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 2010, 285, 21903–21907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Pattillo, C.B.; Pardue, S.; Bir, S.C.; Wang, R.; Kevil, C.G. Measurement of plasma hydrogen sulfide in vivo and in vitro. Free Radic. Biol. Med. 2011, 50, 1021–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, K.R. Is hydrogen sulfide a circulating “gasotransmitter” in vertebrate blood? Biochim. Biophys. Acta 2009, 1787, 856–863. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R.; DeLeon, E.R.; Liu, F. Controversies and conundrums in hydrogen sulfide biology. Nitric Oxide 2014, 41, 11–26. [Google Scholar] [CrossRef]
- Vitvitsky, V.; Banerjee, R. H2S analysis in biological samples using gas chromatography with sulfur chemiluminescence detection. Methods Enzymol. 2015, 554, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, C.K.; Lambert, J.P.; Molkentin, J.D.; Sadoshima, J.; Calvert, J.W. Thioredoxin 1 is essential for sodium sulfide-mediated cardioprotection in the setting of heart failure. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 744–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Kolluru, G.K.; Yuan, S.; Kevil, C.G. Measurement of H2S in vivo and in vitro by the monobromobimane method. Methods Enzymol. 2015, 554, 31–45. [Google Scholar] [CrossRef] [Green Version]
- Ditroi, T.; Nagy, A.; Martinelli, D.; Rosta, A.; Kozich, V.; Nagy, P. Comprehensive analysis of how experimental parameters affect H2S measurements by the monobromobimane method. Free Radic. Biol. Med. 2019, 136, 146–158. [Google Scholar] [CrossRef]
- Chen, B.; Li, W.; Lv, C.; Zhao, M.; Jin, H.; Jin, H.; Du, J.; Zhang, L.; Tang, X. Fluorescent probe for highly selective and sensitive detection of hydrogen sulfide in living cells and cardiac tissues. Analyst 2013, 138, 946–951. [Google Scholar] [CrossRef]
- Qian, Y.; Karpus, J.; Kabil, O.; Zhang, S.Y.; Zhu, H.L.; Banerjee, R.; Zhao, J.; He, C. Selective fluorescent probes for live-cell monitoring of sulphide. Nat. Commun. 2011, 2, 495. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.X.; Lian, K.Q.; Li, X.; Li, N.; Wang, W.; Kang, W.J.; Shi, H.M. Development of a derivatization method for the quantification of hydrogen sulfide and its application in vascular calcification rats. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1055–1056, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, H.; Zhang, X.; Chen, Z.; Zhao, R.; Hou, N.; Liu, J.; Xun, L.; Liu, H. Developing Polysulfide-Sensitive GFPs for Real-Time Analysis of Polysulfides in Live Cells and Subcellular Organelles. Anal. Chem. 2019, 91, 3893–3901. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Chen, Z.; Zhao, R.; Wang, Q.; Ran, M.; Xia, Y.; Hu, X.; Liu, J.; Xian, M.; et al. Using resonance synchronous spectroscopy to characterize the reactivity and electrophilicity of biologically relevant sulfane sulfur. Redox Biol. 2019, 24, 101179. [Google Scholar] [CrossRef] [PubMed]
- Newton, G.L.; Dorian, R.; Fahey, R. Analysis of biological thiols: Derivatization with monobrobimane and separation by Reverse-phase high-performance liquid chromatography. Anal. Biochem. 1981, 114, 383–387. [Google Scholar] [CrossRef]
- Shen, X.; Peter, E.A.; Bir, S.; Wang, R.; Kevil, C.G. Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic. Biol. Med. 2012, 52, 2276–2283. [Google Scholar] [CrossRef] [Green Version]
- Wintner, E.A.; Deckwerth, T.L.; Langston, W.; Bengtsson, A.; Leviten, D.; Hill, P.; Insko, M.A.; Dumpit, R.; VandenEkart, E.; Toombs, C.F.; et al. A monobromobimane-based assay to measure the pharmacokinetic profile of reactive sulphide species in blood. Br. J. Pharmacol. 2010, 160, 941–957. [Google Scholar] [CrossRef] [Green Version]
- Bogdandi, V.; Ida, T.; Sutton, T.R.; Bianco, C.; Ditroi, T.; Koster, G.; Henthorn, H.A.; Minnion, M.; Toscano, J.P.; van der Vliet, A.; et al. Speciation of reactive sulfur species and their reactions with alkylating agents: Do we have any clue about what is present inside the cell? Br. J. Pharmacol. 2019, 176, 646–670. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Chakraborty, S.; Dugas, T.R.; Kevil, C.G. Hydrogen sulfide measurement using sulfide dibimane: Critical evaluation with electrospray ion trap mass spectrometry. Nitric Oxide 2014, 41, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Brereton, R.G. Chemometrics for Pattern Recognition; Wiley: Chichester, UK, 2010. [Google Scholar]
- Mani, S.; Li, H.; Untereiner, A.; Wu, L.; Yang, G.; Austin, R.C.; Dickhout, J.G.; Lhotak, S.; Meng, Q.H.; Wang, R. Decreased endogenous production of hydrogen sulfide accelerates atherosclerosis. Circulation 2013, 127, 2523–2534. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Sagara, M.; Aoki, C.; Tanaka, S.; Aso, Y. Clinical Implication of Plasma Hydrogen Sulfide Levels in Japanese Patients with Type 2 Diabetes. Intern. Med. 2017, 56, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Longchamp, A.; MacArthur, M.R.; Trocha, K.; Ganahl, J.; Mann, C.G.; Kip, P.; King, W.W.; Sharma, G.; Tao, M.; Mitchell, S.J.; et al. Plasma Hydrogen Sulfide Is Positively Associated with Post-operative Survival in Patients Undergoing Surgical Revascularization. Front. Cardiovasc. Med. 2021, 8, 750926. [Google Scholar] [CrossRef] [PubMed]
- Rajpal, S.; Katikaneni, P.; Deshotels, M.; Pardue, S.; Glawe, J.; Shen, X.; Akkus, N.; Modi, K.; Bhandari, R.; Dominic, P.; et al. Total sulfane sulfur bioavailability reflects ethnic and gender disparities in cardiovascular disease. Redox Biol. 2018, 15, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Gambari, L.; Lisignoli, G.; Cattini, L.; Manferdini, C.; Facchini, A.; Grassi, F. Sodium hydrosulfide inhibits the differentiation of osteoclast progenitor cells via NRF2-dependent mechanism. Pharmacol. Res. 2014, 87, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, R.; Liu, X.; Zhou, Y.; Qu, C.; Kikuiri, T.; Wang, S.; Zandi, E.; Du, J.; Ambudkar, I.S.; et al. Hydrogen sulfide maintains mesenchymal stem cell function and bone homeostasis via regulation of Ca2+ channel sulfhydration. Cell Stem Cell 2014, 15, 66–78. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Shi, C.; Liu, Z.; Han, B.; Guo, L.; Zhu, L.; Ye, T. Hydrogen sulfide is a novel regulator implicated in glucocorticoidsinhibited bone formation. Aging 2019, 11, 7537–7552. [Google Scholar] [CrossRef]
- Grassi, F.; Tyagi, A.M.; Calvert, J.W.; Gambari, L.; Walker, L.D.; Yu, M.; Robinson, J.; Li, J.Y.; Lisignoli, G.; Vaccaro, C.; et al. Hydrogen Sulfide Is a Novel Regulator of Bone Formation Implicated in the Bone Loss Induced by Estrogen Deficiency. J. Bone Miner. Res. 2016, 31, 949–963. [Google Scholar] [CrossRef]
- Burguera, E.F.; Vela-Anero, A.; Gato-Calvo, L.; Vaamonde-Garcia, C.; Meijide-Failde, R.; Blanco, F.J. Hydrogen sulfide biosynthesis is impaired in the osteoarthritic joint. Int. J. Biometeorol. 2020, 64, 997–1010. [Google Scholar] [CrossRef]
- Fogo, J.K.; Popowsky, A.M. Spectrophotometric Determination of Hydrogen Sulfide. Anal. Chem. 1949, 21, 732–734. [Google Scholar] [CrossRef]
- Whitfield, N.L.; Kreimier, E.L.; Francys, C.; Verdial, F.C.; Skovgaard, N.; Olson, K.R. Rapid Estimation a Sulfide-Specific of Sulfide in Rumen and Blood with ion Electrode. Microchem. J. 1980, 25, 388–395. [Google Scholar]
- Xu, T.; Scafa, N.; Xu, L.P.; Zhou, S.; Abdullah Al-Ghanem, K.; Mahboob, S.; Fugetsu, B.; Zhang, X. Electrochemical hydrogen sulfide biosensors. Analyst 2016, 141, 1185–1195. [Google Scholar] [CrossRef] [Green Version]
- Levitt, M.D.; Abdel-Rehim, M.S.; Furne, J. Free and Acid-Labile Hydrogen Sulfide Concentrations in Mouse Tissues: Anomalously High Free Hydrogen Sulfide in Aortic Tissue. Forum Orig. Res. Commun. 2011, 15, 373–378. [Google Scholar] [CrossRef]
- Ibrahim, H.; Serag, A.; Farag, M.A. Emerging analytical tools for the detection of the third gasotransmitter H2S, a comprehensive review. J. Adv. Res. 2021, 27, 137–153. [Google Scholar] [CrossRef] [PubMed]

| SDB Peak Area (LU * min) | RT | 50 °C |
|---|---|---|
| Na2S 12.5 μM | 270 | 800 |
| Serum | 63 | 207 |
| SBF | 2 | 230 |
| Na2S 5 μM/ Serum+ Na2S 5 μM | Na2S 12.5 μM/ Serum+ Na2S 12.5 μM | Na2S 25 μM/ Serum+ Na2S 25 μM | |
|---|---|---|---|
| SDB peak area (LU * min) | 25/22 | 180/100 | 400/200 |
| Parameter | Value |
|---|---|
| a (AU) | −13.2 |
| sa (AU) | 6.61 |
| b (AU µM−1) | 5.87 |
| sb (AU µM−1) | 0.930 |
| c (AU µM−2) | 0.358 |
| sc (AU µM−2) | 0.0181 |
| R2 | 0.998 |
| RMSE (AU) | 18.16 |
| model p-value | <2.20 × 10−16 |
| CV | Calibration | |
|---|---|---|
| Intercept | − 0.108 | 0.429 |
| sIntercept | 0.685 | 0.520 |
| Slope | 0.999 | 0.998 |
| sSlope | 0.0122 | 0.00969 |
| R2 | 0.997 | 0.998 |
| RMSE | 22.3 | 17.8 |
| model p-value | <2.20 × 10−16 | <2.20 × 10−16 |
| Parameter | Value |
|---|---|
| Intercept (AU) | − 0.581 |
| sIntercept (AU) | 0.260 |
| Slope (AU µM−1) | 3.51 |
| sSlope (AU µM−1) | 0.0749 |
| R2 | 0.995 |
| RMSE (AU) | 0.514 |
| model p-value | 4.77 × 10−13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roda, B.; Zhang, N.; Gambari, L.; Grigolo, B.; Eller-Vainicher, C.; Gennari, L.; Zappi, A.; Giordani, S.; Marassi, V.; Zattoni, A.; et al. Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment. Antioxidants 2022, 11, 939. https://doi.org/10.3390/antiox11050939
Roda B, Zhang N, Gambari L, Grigolo B, Eller-Vainicher C, Gennari L, Zappi A, Giordani S, Marassi V, Zattoni A, et al. Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment. Antioxidants. 2022; 11(5):939. https://doi.org/10.3390/antiox11050939
Chicago/Turabian StyleRoda, Barbara, Nan Zhang, Laura Gambari, Brunella Grigolo, Cristina Eller-Vainicher, Luigi Gennari, Alessandro Zappi, Stefano Giordani, Valentina Marassi, Andrea Zattoni, and et al. 2022. "Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment" Antioxidants 11, no. 5: 939. https://doi.org/10.3390/antiox11050939
APA StyleRoda, B., Zhang, N., Gambari, L., Grigolo, B., Eller-Vainicher, C., Gennari, L., Zappi, A., Giordani, S., Marassi, V., Zattoni, A., Reschiglian, P., & Grassi, F. (2022). Optimization of a Monobromobimane (MBB) Derivatization and RP-HPLC-FLD Detection Method for Sulfur Species Measurement in Human Serum after Sulfur Inhalation Treatment. Antioxidants, 11(5), 939. https://doi.org/10.3390/antiox11050939











