Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots
Abstract
1. Introduction
2. Materials and Methods
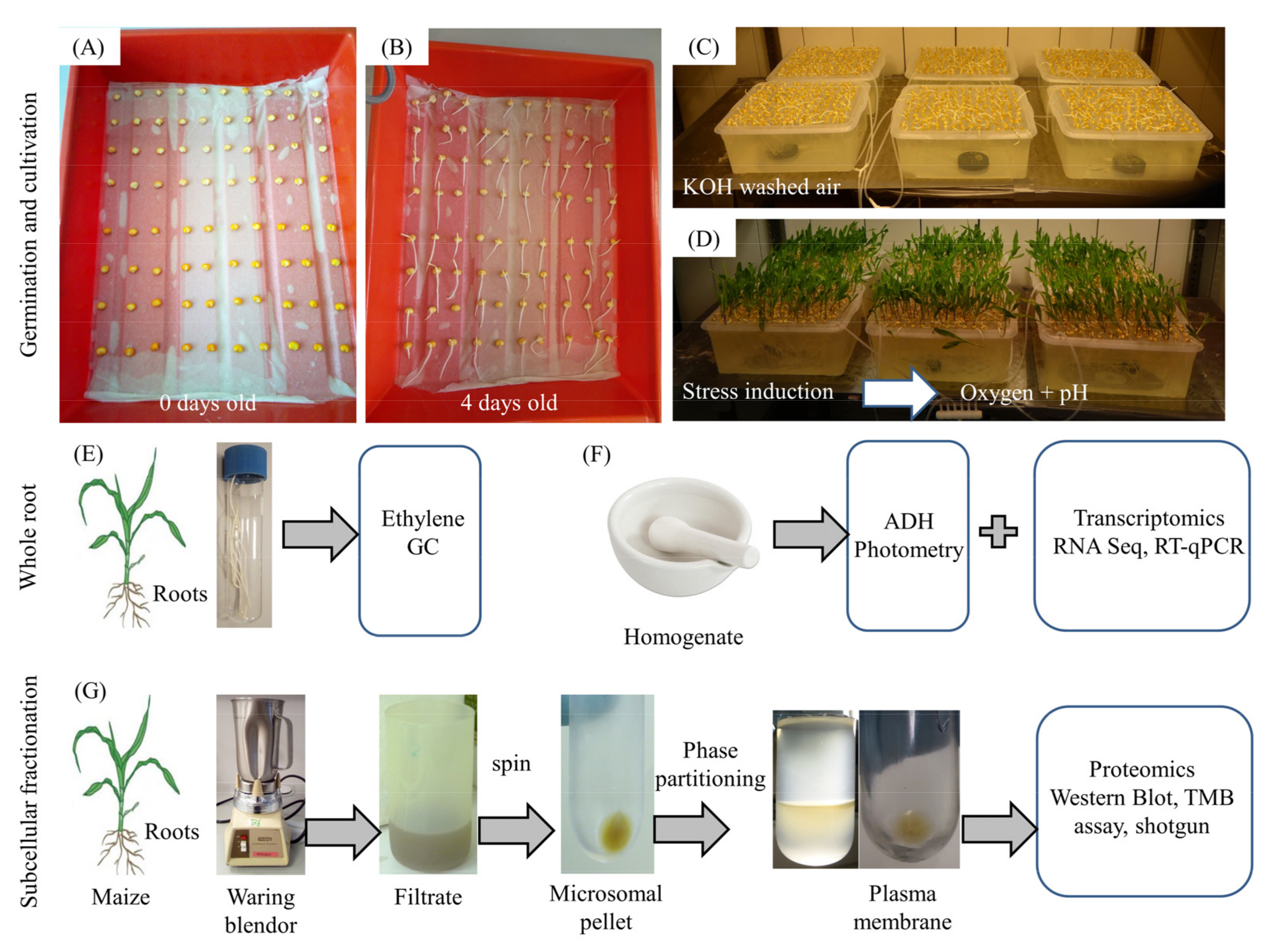
2.1. Plant Material and Growth Conditions
2.2. Ethylene and CO2
2.3. Alcohol Dehydrogenase (ADH) Activity
2.4. RNA Analysis
2.4.1. RNA Sequencing
2.4.2. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
2.5. Preparation of Subcellular Fractions
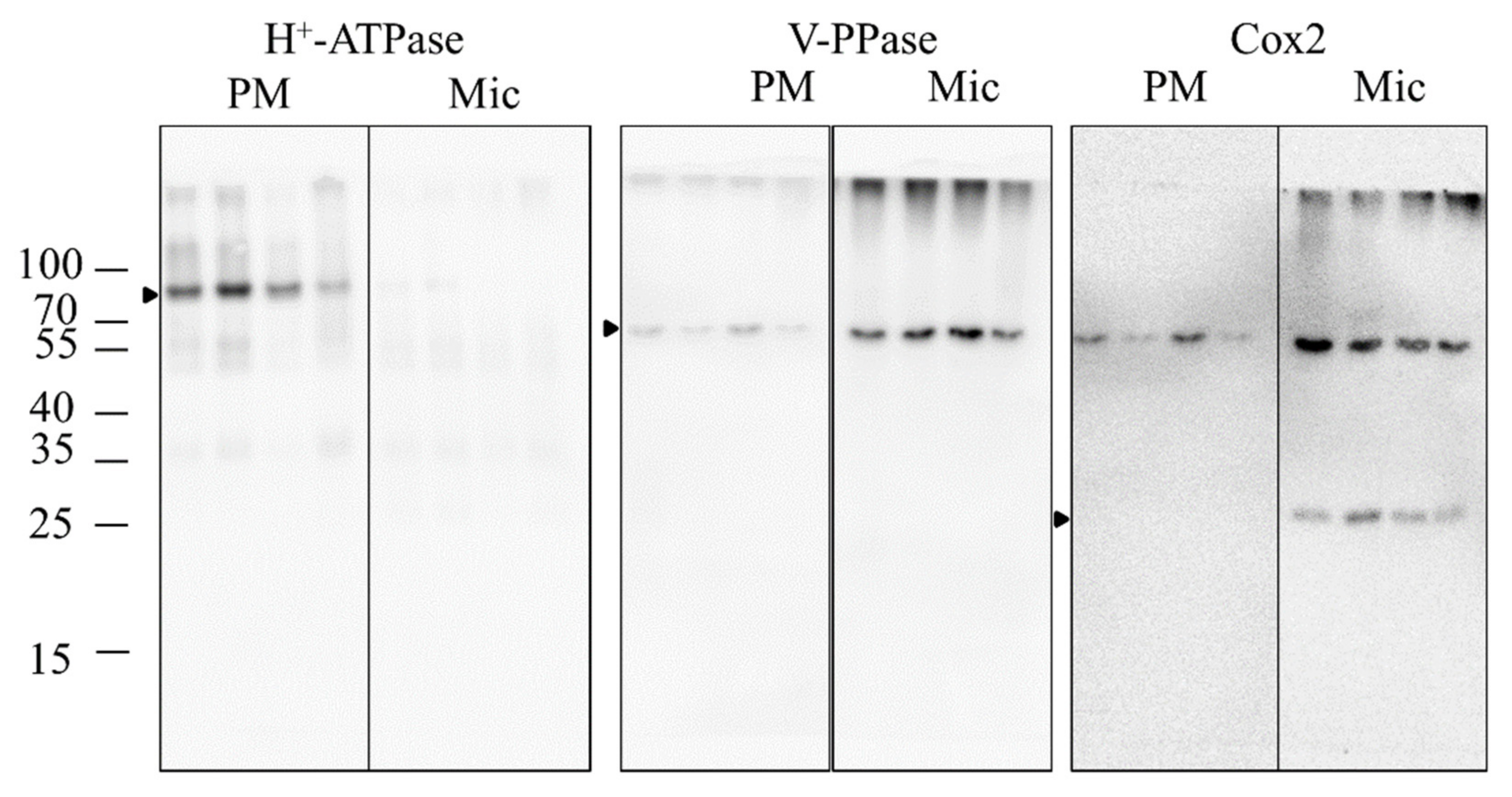
2.6. Purity Verification of the PM Fractions
2.7. Tetramethylbenzidine Oxidase Activity
2.8. Shotgun Proteomics of the Plasma Membrane and Soluble Fraction
2.9. Analyses of Shotgun Proteomics of the PMs and RNA Sequencing Data
3. Results
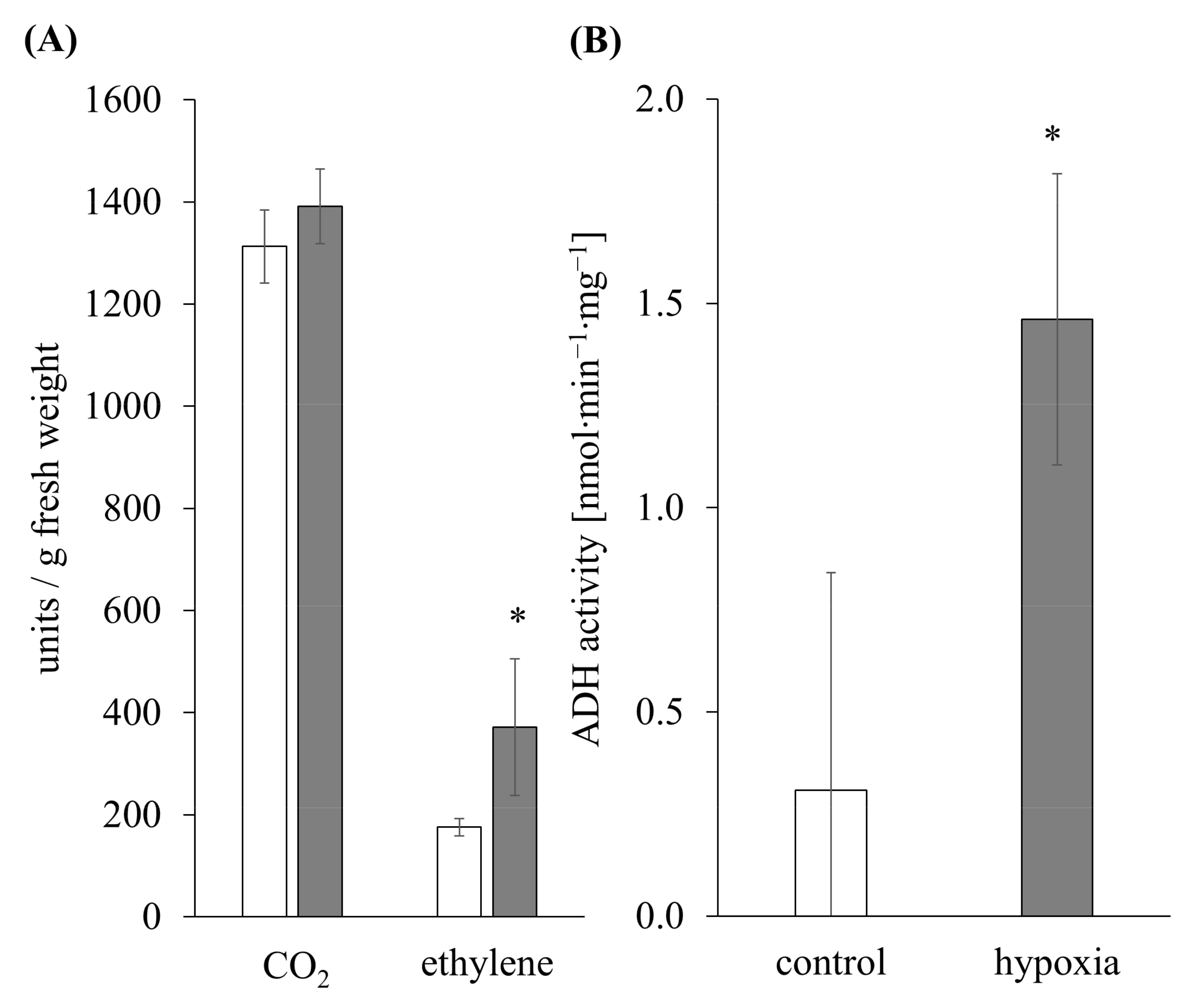
3.1. Ethylene Induction and Metabolic Switch as an Indicator for Hypoxia Stress
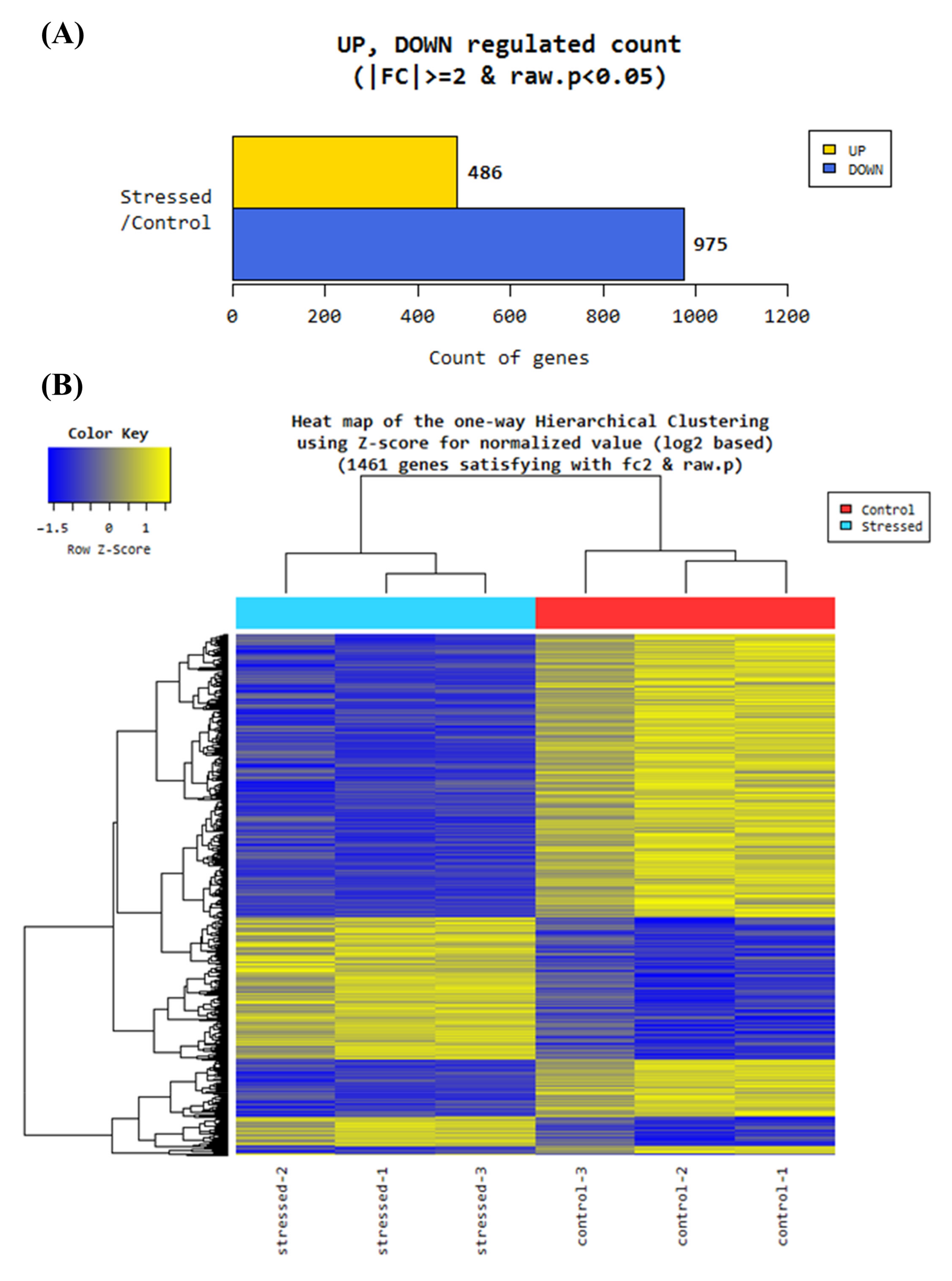
3.2. RNA Sequence Analysis
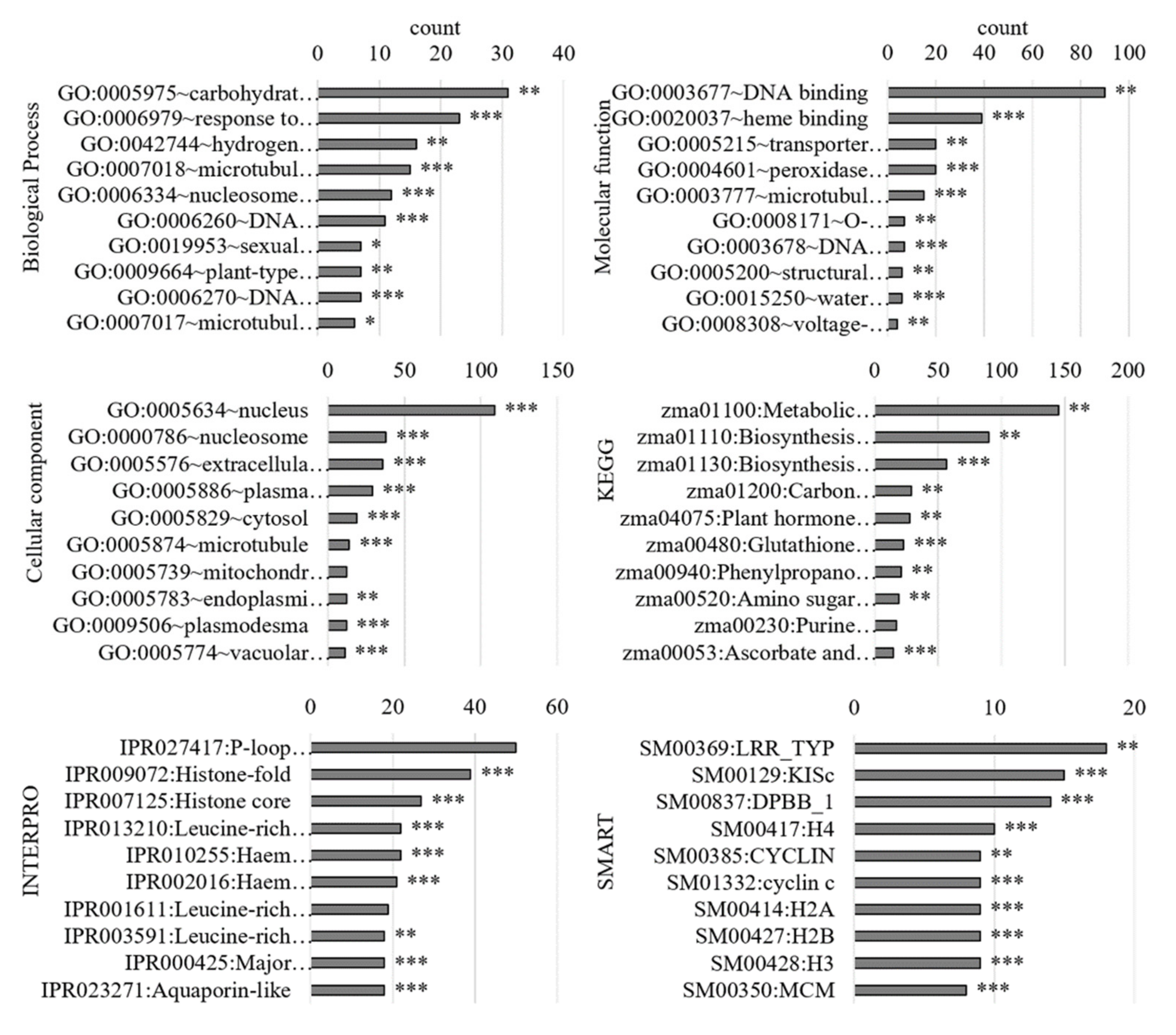
3.2.1. Gene Ontology Analysis of Differentially Regulated Genes
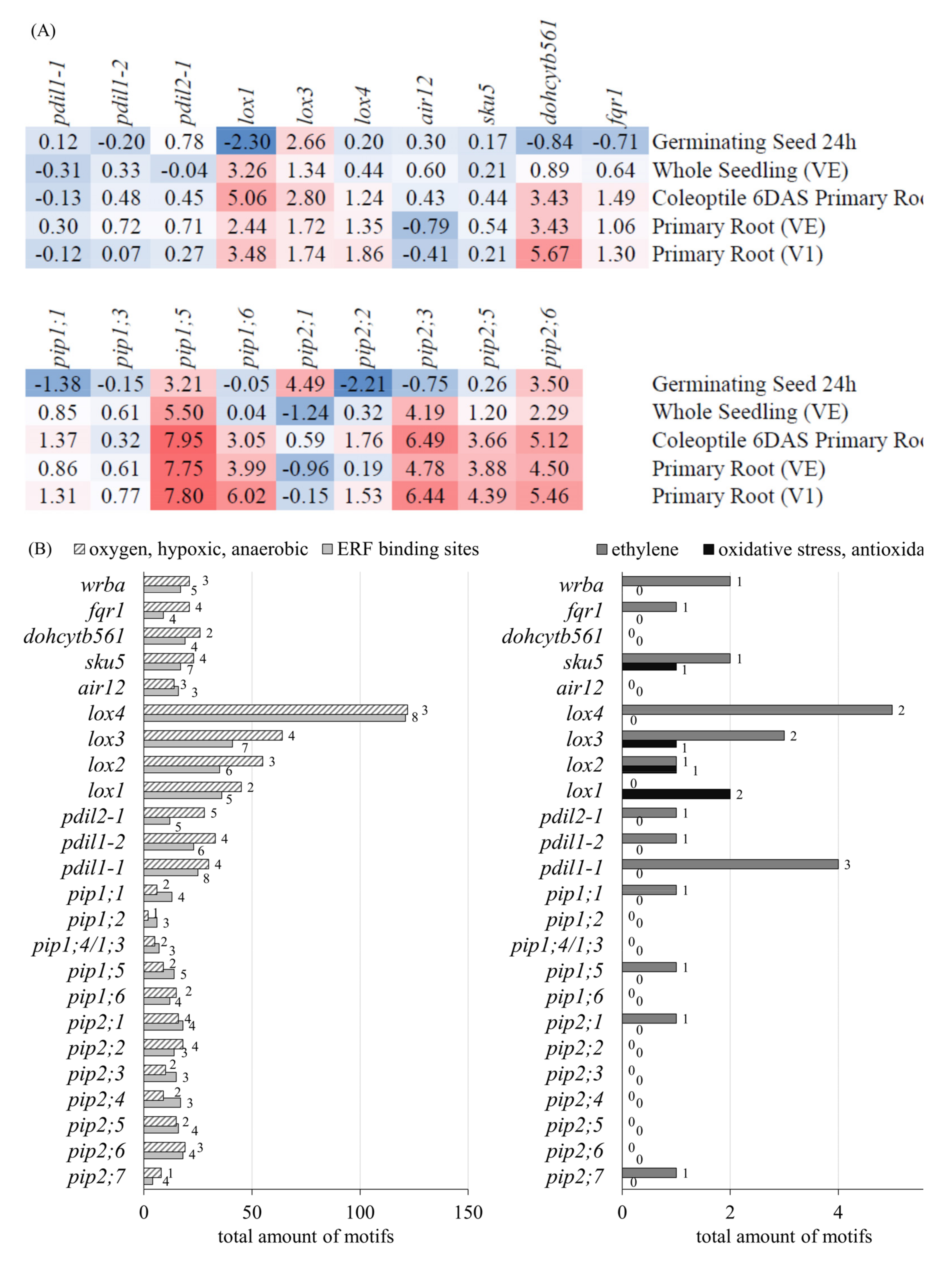
3.2.2. RNA Sequence Analysis of Genes Related to Redox System, Antioxidant Biosynthesis, and Transporters
3.2.3. Gene Expression and Promotor Analysis of PM-Bound Redox Systems and PIPs
3.2.4. RT-qPCR
3.3. Shotgun Analysis
3.3.1. Gene Ontology Analysis of the PM-Bound Proteins
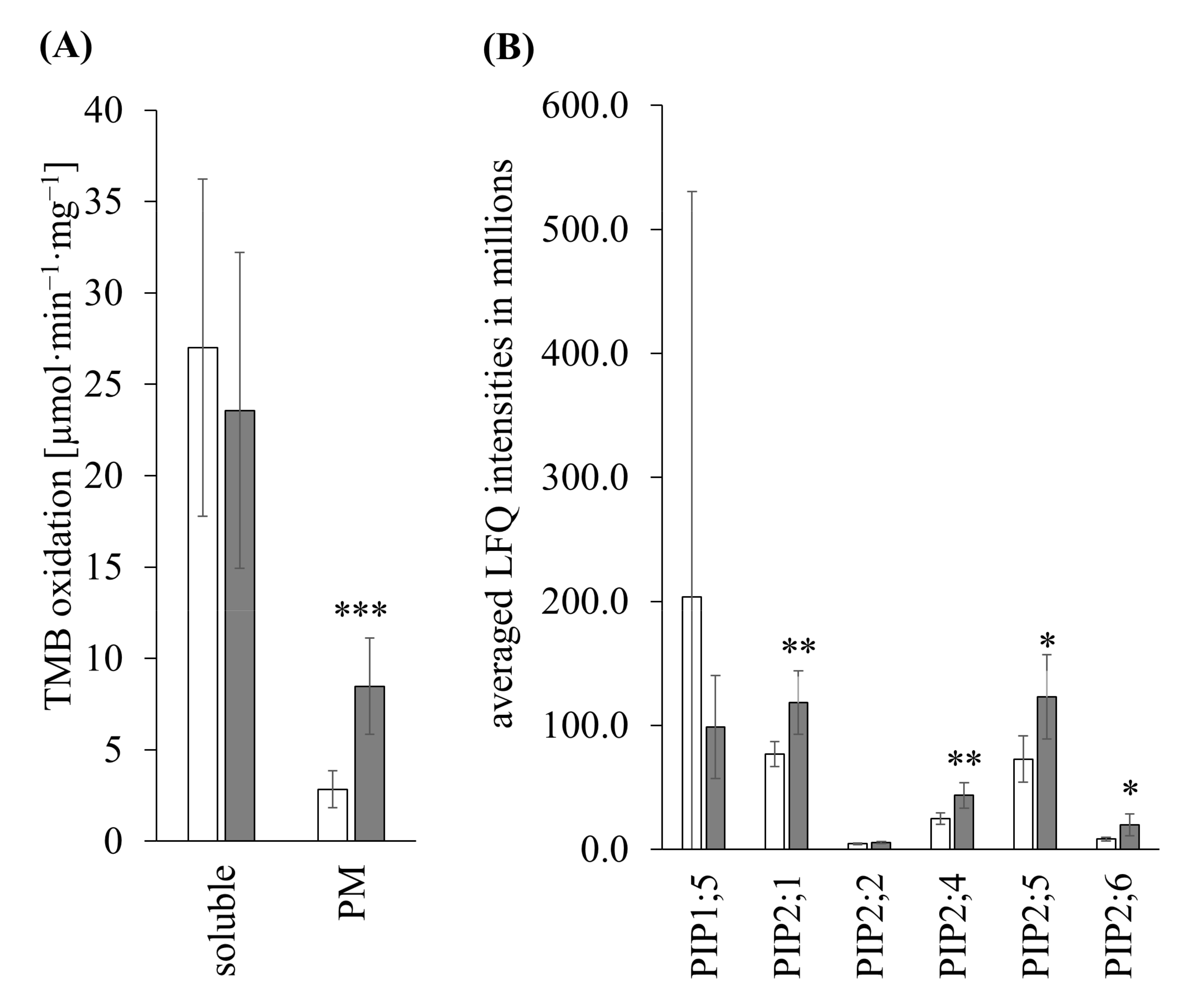
3.3.2. TMB Oxidation
3.3.3. Shotgun of PM
3.3.4. Shotgun of Soluble Fraction
4. Discussion
4.1. The Oil-Based Setup Induced Typical Hypoxia Stress Symptoms
4.2. Hypoxia-Induced Redox Systems Effected Root Architecture
4.3. Hypoxia-Induced PM Redox Systems Are Involved in ROS Production
4.4. Hypoxia-Induced PM Redox Systems Are Involved in Membrane Protection and Redox Homeostasis
4.5. Redox Regulation of H2O2 Transporting Aquaporins
4.6. Transcriptome to Proteome Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Visser, E.J.W.; Bögemann, G.M.; Blom, C.W.P.M.; Voesenek, L.A. Ethylene accumulation in waterlogged Rumex plants promotes formation of adventitious roots. J. Exp. Bot. 1996, 47, 403–410. [Google Scholar] [CrossRef]
- Pedersen, O.; Perata, P.; Voesenek, L. Flooding and low oxygen responses in plants. Funct. Plant Biol. 2017, 44, iii–vi. [Google Scholar] [CrossRef] [PubMed]
- Colmer, T.D.; Winkel, A.; Kotula, L.; Armstrong, W.; Revsbech, N.P.; Pedersen, O. Root O2 consumption, CO2 production and tissue concentration profiles in chickpea, as influenced by environmental hypoxia. New Phytol. 2020, 226, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Geisler-Lee, J.; Caldwell, C.; Gallie, D.R. Expression of the ethylene biosynthetic machinery in maize roots is regulated in response to hypoxia. J. Exp. Bot. 2009, 61, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Drew, M.; Jackson, M.B.; Giffard, S. Ethylene-promoted Adventitious Rooting and Development of Cortical Air Spaces (Aerenchyma) in Roots May be Adaptive Response to Flooding in Zea mays L. Planta 1979, 147, 83–88. [Google Scholar] [CrossRef]
- Sagee, O.; Goren, R.; Riov, J. Abscission of Citrus Leaf Explants. Plant Physiol. 1980, 66, 750–753. [Google Scholar] [CrossRef]
- Jackson, M.B.; Armstrong, W. Formation of Aerenchyma and the Processes of Plant Ventilation in Relation to Soil Flooding and Submergence. Plant Biol. 1999, 1, 274–287. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene-promoted elongation: An adaptation to submergence stress. Ann. Bot. 2008, 101, 229–248. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Life in the balance: A signaling network controlling survival of flooding. Curr. Opin. Plant Biol. 2010, 13, 489–494. [Google Scholar] [CrossRef]
- Sachs, M.M.; Freeling, M.; Okimoto, R. The Anaerobic Proteins of Maize. Cell 1980, 20, 761–767. [Google Scholar] [CrossRef]
- Andrews, D.L.; Cobb, G.; Johnson, J.R.; Drew, M.C. Hypoxic and Anoxic Induction of Alcohol Dehydrogenase in Roots and Shoots of Seedlings of Zea mays. Plant Physiol. 1993, 101, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Schippers, J.H.M.; Schmidt, R.R. Redox and low-oxygen stress: Signal integration and interplay. Plant Physiol. 2021, 186, 66–78. [Google Scholar] [CrossRef] [PubMed]
- De Gara, L. Class III peroxidases and ascorbate metabolism in plants. Phytochem. Rev. 2004, 3, 185–205. [Google Scholar] [CrossRef]
- Schaffer, W.M.; Bronnikova, T.V. Peroxidase-ROS interactions. Nonlinear Dyn. 2012, 68, 413–430. [Google Scholar] [CrossRef]
- Görlach, A.; Dimova, E.Y.; Petry, A.; Martinez-Ruiz, A.; Hernansanz-Agustin, P.; Rolo, A.P.; Palmeira, C.M.; Kietzmann, T. Reactive oxygen species, nutrition, hypoxia and diseases: Problems solved? Redox Biol. 2015, 6, 372–385. [Google Scholar] [CrossRef]
- Meisrimler, C.N.; Buck, F.; Lüthje, S. Alterations in soluble class III peroxidases of maize shoots by flooding stress. Proteomes 2014, 2, 303–322. [Google Scholar] [CrossRef]
- Hofmann, A.; Wienkoop, S.; Harder, S.; Bartlog, F.; Luthje, S. Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes. Int. J. Mol. Sci. 2020, 21, 8872. [Google Scholar] [CrossRef]
- May, M.J.; Vernoux, T.; Leaver, C.; van Montagu, M.; Inzé, D. Glutathione homeostasis in plants: Implications for environmental sensing and plant development. J. Exp. Bot. 1998, 49, 649–667. [Google Scholar] [CrossRef]
- Linster, C.L.; Clarke, S.G. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008, 13, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Zwiazek, J.J.; Xu, H.; Tan, X.; Navarro-Rodenas, A.; Morte, A. Significance of oxygen transport through aquaporins. Sci. Rep. 2017, 7, 40411. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, F.; Barrieu, F.; Wojcik, E.; Chrispeels, M.J.; Jung, R. Aquaporins Constitute a Large and Highly Divergent Protein Family in Maize. Plant Physiol. 2001, 125, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018, 51, 4. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in Plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Zelazny, E.; Borst, J.W.; Muylaert, M.; Batoko, H.; Hemminga, M.A.; Chaumon, F. FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 2007, 104, 12359–12364. [Google Scholar] [CrossRef]
- Hukin, D.; Doering-Saad, C.; Thomas, C.R.; Pritchard, J. Sensitivity of cell hydraulic conductivity to mercury is coincident with symplasmic isolation and expression of plasmalemma aquaporin genes in growing maize roots. Planta 2002, 215, 1047–1056. [Google Scholar] [CrossRef]
- Maurel, C. Plant aquaporins: Novel functions and regulation properties. FEBS Lett. 2007, 581, 2227–2236. [Google Scholar] [CrossRef]
- Chaumont, F.; Barrieu, F.; Jung, R.; Chrispeels, M.J. Plasma Membrane Intrinsic Proteins from Maize Cluster in Two Sequence Subgroups with Differential Aquaporin Activity. Plant Physiol. 2000, 122, 1025–1034. [Google Scholar] [CrossRef]
- Schuurmans, J.A.M.J.; van Dongen, J.T.; Rutjens, B.P.W.; Boonman, A.; Pieterse, C.M.J.; Borstlap, A.C. Members of the aquaporin family in the developing pea seed coat include representatives of the PIP, TIP, and NIP subfamilies. Plant Mol. Biol. 2003, 53, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Fetter, K.; van Wilder, V.; Moshelion, M.; Chaumont, F. Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 2004, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Bramley, H.; Turner, D.W.; Tyerman, S.D.; Turner, N.C. Water Flow in the Roots of Crop Species: The Influence of Root Structure, Aquaporin Activity, and Waterlogging. Adv. Agron. 2007, 96, 133–196. [Google Scholar] [CrossRef]
- Gomes, D.; Agasse, A.; Thiebaud, P.; Delrot, S.; Geros, H.; Chaumont, F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim. Biophys. Acta 2009, 1788, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Törnroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.-T.; Bligny, R.; Maurel, C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef]
- Henzler, T.; Ye, Q.; Steudle, E. Oxidative gating of water channels (aquaporins in Chara by hydroxyl radicals). Plant Cell Environ. 2004, 27, 1184–1195. [Google Scholar] [CrossRef]
- Ye, Q.; Steudle, E. Oxidative gating of water channels (aquaporins) in corn roots. Plant Cell Environ. 2006, 29, 459–470. [Google Scholar] [CrossRef]
- Bestetti, S.; Medrano-Fernandez, I.; Galli, M.; Ghitti, M.; Bienert, G.P.; Musco, G.; Orsi, A.; Rubartelli, A.; Sitia, R. A persulfidation-based mechanism controls aquaporin-8 conductance. Sci. Adv. 2018, 4, eaar5770. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; van Veen, H.; Yeung, E.; Voesenek, L. Signal Dynamics and Interactions during Flooding Stress. Plant Physiol. 2018, 176, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Bienfait, F.; Lüttge, U. On the function of two systems that can transfer electrons across the plasma membrane. Plant Physiol. Biochem. 1988, 26, 665–671. [Google Scholar]
- Lüthje, S.; Döring, O.; Heuer, S.; Lüthen, H.; Böttger, M. Oxidoreductases in plant plasma membranes. Biochim. Biophys. Acta 1997, 1331, 81–102. [Google Scholar] [CrossRef]
- Lüthje, S.; van Gestelen, P.; Córdoba-Pedregosa, M.C.; González-Reyes, J.A.; Asard, H.; Villalba, J.M.; Böttger, M. Quinones in plant plasma membranes—A missing link? Protoplasma 1998, 205, 43–51. [Google Scholar] [CrossRef]
- Asard, H.; Barbaro, R.; Trost, P.; Berczi, A. Cytochromes b561: Ascorbate-mediated trans-membrane electron transport. Antioxid. Redox Signal. 2013, 19, 1026–1035. [Google Scholar] [CrossRef]
- Menckhoff, M.; Lüthje, S. Transmembrane electron transport in sealed and NAD(P)H-loaded right-side-out plasma membrane vesicles isolated from maize (Zea mays L.) roots. J. Exp. Bot. 2004, 55, 1343–1349. [Google Scholar] [CrossRef][Green Version]
- Biniek, C.; Heyno, E.; Kruk, J.; Sparla, F.; Trost, P.; Krieger-Liszkay, A. Role of the NAD(P)H quinone oxidoreductase NQR and the cytochrome b AIR12 in controlling superoxide generation at the plasma membrane. Planta 2017, 245, 807–817. [Google Scholar] [CrossRef]
- Laszlo, A.; St. Lawrence, P. Parallel Induction and Synthesis of PDC and ADH in Anoxic Maize Roots. Mol. Gen. Genet. 1983, 192, 110–117. [Google Scholar] [CrossRef]
- Lüthje, S.; Meisrimler, C.N.; Hopff, D.; Schütze, T.; Koppe, J.; Heino, K. Class III peroxidases. Methods Mol. Biol. 2014, 1072, 687–706. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sun, X.; Weckwerth, W. COVAIN: A toolbox for uni- and multivariate statistics, time-series and correlation network analysis and inverse estimation of the differential Jacobian from metabolomics covariance data. Metabolomics 2012, 8, 81–93. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–178. [Google Scholar] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous Hypoxia in Lateral Root Primordia Controls Root Architecture by Antagonizing Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 538–551. [Google Scholar] [CrossRef]
- Gibson, S.W.; Todd, C.D. Arabidopsis AIR12 influences root development. Physiol. Mol. Biol. Plants 2015, 21, 479–489. [Google Scholar] [CrossRef]
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Masson, P.H.; Somerville, C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648. [Google Scholar] [CrossRef]
- Zhou, K. GPI-anchored SKS proteins regulated root development through controlling cell polar expansion and cell wall synthesis. Biochem. Biophys. Res. Commun. 2019, 509, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Walper, E.; Weiste, C.; Mueller, M.J.; Hamberg, M.; Droge-Laser, W. Screen Identifying Arabidopsis Transcription Factors Involved in the Response to 9-Lipoxygenase-Derived Oxylipins. PLoS ONE 2016, 11, e0153216. [Google Scholar] [CrossRef] [PubMed]
- Vellosillo, T.; Martinez, M.; Lopez, M.A.; Vicente, J.; Cascon, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef]
- Gao, X.; Starr, J.; Göbel, C.; Engelberth, J.; Feussner, I.; Tumlinson, J.; Kolomiets, M. Maize 9-Lipoxygenase ZmLOX3 Controls Development, Root-Specific Expression of Defense Genes, and Resistance to Root-Knot Nematodes. MPMI 2008, 21, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Abe, F.; Tsutsumi, N.; Nakazono, M. Root cortex provides a venue for gas-space formation and is essential for plant adaptation to waterlogging. Front. Plant Sci. 2019, 10, 259. [Google Scholar] [CrossRef]
- Ding, L.; Milhiet, T.; Couvreur, V.; Nelissen, H.; Meziane, A.; Parent, B.; Aesaert, S.; van Lijsebettens, M.; Inze, D.; Tardieu, F.; et al. Modification of the Expression of the Aquaporin ZmPIP2;5 Affects Water Relations and Plant Growth. Plant Physiol. 2020, 182, 2154–2165. [Google Scholar] [CrossRef] [PubMed]
- Kaldenhoff, R.; Grote, K.; Zhu, J.-J.; Zimmermann, U. Significane of plasmalemma aquaporins for water-transport in Arabidopsis thaliana. Plant J. 1998, 14, 121–128. [Google Scholar] [CrossRef]
- Moshelion, M.; Hachez, C.; Ye, Q.; Cavez, D.; Bajji, M.; Jung, R.; Chaumont, F. Membrane water permeability and aquaporin expression increase during growth of maize suspension cultured cells. Plant Cell Environ. 2009, 32, 1334–1345. [Google Scholar] [CrossRef]
- Birner, T.P.; Steudle, E. Effects of anaerobic conditions on water and solute relations, and on active transport inroots of maize (Zea mays L.). Planta 1992, 190, 474–483. [Google Scholar]
- Ehlert, C.; Maurel, C.; Tardieu, F.; Simonneau, T. Aquaporin-mediated reduction in maize root hydraulic conductivity impacts cell turgor and leaf elongation even without changing transpiration. Plant Physiol. 2009, 150, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, M.; Du, N.; Zwiazek, J.J. Ethylene enhances root water transport and aquaporin expression in trembling aspen (Populus tremuloides) exposed to root hypoxia. BMC Plant Biol. 2021, 21, 227. [Google Scholar] [CrossRef] [PubMed]
- Felle, H.H. pH regulation in anoxic plants. Ann. Bot. 2005, 96, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, F.; Bing, J.; Sun, W.; Feng, X.; Ma, X.; Zhou, Y.; Zhang, G. Overexpression of the Jojoba Aquaporin Gene, ScPIP1, Enhances Drought and Salt Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 153. [Google Scholar] [CrossRef]
- Xu, F.; Wang, K.; Yuan, W.; Xu, W.; Shuang, L.; Kronzucker, H.J.; Chen, G.; Miao, R.; Zhang, M.; Ding, M.; et al. Overexpression of rice aquaporin OsPIP1;2 improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J. Exp. Bot. 2019, 70, 671–681. [Google Scholar] [CrossRef]
- Lin, W.D.; Liao, Y.Y.; Yang, T.J.; Pan, C.Y.; Buckhout, T.J.; Schmidt, W. Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 2011, 155, 1383–1402. [Google Scholar] [CrossRef]
- Hachez, C.; Moshelion, M.; Zelazny, E.; Cavez, D.; Chaumont, F. Localization and quantification of plasma membrane aquaporin expression in maize primary root: A clue to understanding their role as cellular plumbers. Plant Mol. Biol. 2006, 62, 305–323. [Google Scholar] [CrossRef]
- Hachez, C.; Veselov, D.; Ye, Q.; Reinhardt, H.; Knipfer, T.; Fricke, W.; Chaumont, F. Short-term control of maize cell and root water permeability through plasma membrane aquaporin isoforms. Plant Cell Environ. 2012, 35, 185–198. [Google Scholar] [CrossRef]
- Bienert, G.P.; Heinen, R.B.; Berny, M.C.; Chaumont, F. Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 2014, 1838, 216–222. [Google Scholar] [CrossRef]
- Safavi-Rizi, V.; Herde, M.; Stohr, C. RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 2020, 10, 1692. [Google Scholar] [CrossRef]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 2006, 141, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Iacopino, S.; Licausi, F. The Contribution of Plant Dioxygenases to Hypoxia Signaling. Front. Plant Sci. 2020, 11, 1008. [Google Scholar] [CrossRef]
- Pucciariello, C.; Perata, P. The Oxidative Paradox in Low Oxygen Stress in Plants. Antioxidants 2021, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.E.; Ryan, D.; Levin, W. An Improved Staining Procedure for the Detection of the Peroxidase Activity of cytochrome P-450 on Sodium Dodecyl Sulfate Polyacrylamide Gels. Anal. Biochem. 1976, 75, 168–176. [Google Scholar] [CrossRef]
- Miller, D.J.; Nicholas, D.J.D. 3,3′,5,5′-TetramethyIbenzidine/H202 Staining Is Not Specific for Heme Proteins Separated by Gel Electrophoresis. Anal. Biochem. 1984, 140, 577–580. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH Oxidase RBOH Functions in Rice Roots during Lysigenous Aerenchyma Formation under Oxygen-Deficient Conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Y.; Sun, Y.Y.; Zhang, Y.J.; Lu, C.H.; Miao, J.F. Detection of biological thiols based on a colorimetric method. J. Zhejiang Univ. Sci. B 2016, 17, 807–812. [Google Scholar] [CrossRef]
- Lim, C.W.; Han, S.W.; Hwang, I.S.; Kim, D.S.; Hwang, B.K.; Lee, S.C. The Pepper Lipoxygenase CaLOX1 Plays a Role in Osmotic, Drought and High Salinity Stress Response. Plant Cell Physiol. 2015, 56, 930–942. [Google Scholar] [CrossRef]
- Hu, J.; Ren, B.; Dong, S.; Liu, P.; Zhao, B.; Zhang, J. Comparative proteomic analysis reveals that exogenous 6-benzyladenine (6-BA) improves the defense system activity of waterlogged summer maize. BMC Plant Biol. 2020, 20, 44. [Google Scholar] [CrossRef]
- Lopez, M.A.; Vicente, J.; Kulasekaran, S.; Vellosillo, T.; Martinez, M.; Irigoyen, M.L.; Cascon, T.; Bannenberg, G.; Hamberg, M.; Castresana, C. Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 2011, 67, 447–458. [Google Scholar] [CrossRef]
- Roberts, J.K.M.; Callis, J.; Jardetzky, O.; Walbot, V.; Freeling, M. Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc. Natl. Acad. Sci. USA 1984, 81, 6029–6033. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.K.M.; Hooks, M.A.; Miaullis, A.P.; Edwards, S.; Webster, C. Contribution of Malat and Amino Acid Metabolism to Cytoplasmic pH Regulaton in Hypoxic Maize Root Tips Studied Using Nuclear Magnetiv Resonance Spectroscopy. Plant Physiol. 1992, 88, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, K. Intracellular pH Regulation of Plant Cells Under Anaerobic Conditions. In Low-Oxygen Stress in Plants; Plant Cell Monographs; van Dongen, J., Licausi, F., Eds.; Springer: Vienna, Austria, 2014; Volume 21. [Google Scholar] [CrossRef]
- Smith, F.A.; Raven, J.A. Intracellular pH and its Regulation. Ann. Rev. Plant Physiol. 1979, 30, 289–311. [Google Scholar] [CrossRef]
- Menckhoff, L.; Mielke-Ehret, N.; Buck, F.; Vuletic, M.; Luthje, S. Plasma membrane-associated malate dehydrogenase of maize (Zea mays L.) roots: Native versus recombinant protein. J. Proteom. 2013, 80, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Savchenko, G.; Wiese, C.; Neimanis, S.; Hedrich, R.; Heber, U. pH regulation in apoplastic and cytoplasmic cell compartments of leaves. Planta 1999, 211, 246–255. [Google Scholar] [CrossRef]
- Córdoba-Pedregosa, M.C.; González-Reyes, J.A.; Serrano, A.; Villalba, J.M.; Navas, P.; Córdoba, F. Pasmalemma-associated malate dehydrogenase activity in onion root cells. Protoplasma 1998, 205, 29–36. [Google Scholar] [CrossRef]
- Lüthje, S.; Möller, B.; Perrineau, F.C.; Woltje, K. Plasma membrane electron pathways and oxidative stress. Antioxid. Redox Signal. 2013, 18, 2163–2183. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, F.; Liu, B.; Xu, C.; He, Q.; Cheng, W.; Zhao, X.; Ding, Z.; Zhang, W.; Zhang, K.; et al. ZmSKS13, a cupredoxin domain-containing protein, is required for maize kernel development via modulation of redox homeostasis. New Phytol. 2021, 229, 2163–2178. [Google Scholar] [CrossRef]
- Canfield, L.M.; Davy, L.A.; Thomas, G.L. Anti-Oxidant/Pro-Oxidant Reactions of Vitamin K. Biochem. Biophys. Res. Commun. 1985, 128, 211–219. [Google Scholar] [CrossRef]
- Lüthje, S. Plasma Membrane Redox Systems: Lipid Rafts and Protein Assemblies. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2008; pp. 169–200. [Google Scholar]
- Gu, X.; Chen, I.G.; Harding, S.A.; Nyamdari, B.; Ortega, M.A.; Clermont, K.; Westwood, J.H.; Tsai, C.J. Plasma membrane phylloquinone biosynthesis in nonphotosynthetic parasitic plants. Plant Physiol. 2021, 185, 1443–1456. [Google Scholar] [CrossRef]
- Ohyashiki, T.; Yabunaka, Y.; Matsui, K. Antioxidant Effect of Vitamin K Homologues on Ascorbic Acid/Fe2+-Induced Lipid Peroxidation of Lecithin Liposomes. Chem. Pharm. Bull. 1991, 39, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zwiazek, J.J. Stable expression of aquaporins and hypoxia-responsive genes in adventitious roots are linked to maintaining hydraulic conductance in tobacco (Nicotiana tabacum) exposed to root hypoxia. PLoS ONE 2019, 14, e0212059. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Xue, J.; Li, Y.; Liu, X.; Dai, F.; Jia, W.; Luo, Y.; Gao, J. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef]
- Zhu, C.; Schraut, D.; Hartung, W.; Schaffner, A.R. Differential responses of maize MIP genes to salt stress and ABA. J. Exp. Bot. 2005, 56, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Kjell, J.; Rasmusson, A.G.; Larsson, H.; Widell, S. Protein complexes of the plant plasma membrane resolved by Blue Native PAGE. Physiol. Plant. 2004, 121, 546–555. [Google Scholar] [CrossRef]
- Sjöhamn, J.; Hedfalk, K. Unraveling aquaporin interaction partners. Biochim. Biophys. Acta 2014, 1840, 1614–1623. [Google Scholar] [CrossRef]
- Piotrovskii, M.S.; Lapshin, N.K.; Andreev, I.M.; Trofimova, M.S. Role of PIP-Aquaporin Phosphorylation in Redox-Dependent Modulation of Osmotic Water Permeability in Plasmalemma from Roots of Pea Seedlings. Russ. J. Plant Physiol. 2019, 66, 637–645. [Google Scholar] [CrossRef]
- Wudick, M.M.; Li, X.; Valentini, V.; Geldner, N.; Chory, J.; Lin, J.; Maurel, C.; Luu, D.T. Subcellular Redistribution of Root Aquaporins Induced by Hydrogen Peroxide. Mol. Plant 2015, 8, 1103–1114. [Google Scholar] [CrossRef]
- Martre, P.; Morillon, R.; Barrieu, F.; North, G.B.; Nobel, P.S.; Chrispeels, M.J. Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 2002, 130, 2101–2110. [Google Scholar] [CrossRef]
- Qing, D.; Yang, Z.; Li, M.; Wong, W.S.; Guo, G.; Liu, S.; Guo, H.; Li, N. Quantitative and Functional Phosphoproteomic Analysis Reveals that Ethylene Regulates Water Transport via the C-Terminal Phosphorylation of Aquaporin PIP2;1 in Arabidopsis. Mol. Plant 2016, 9, 158–174. [Google Scholar] [CrossRef]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Schoebel, S.; Schmitz, F.; Dong, H.; Hedfalk, K. Characterization of aquaporin-driven hydrogen peroxide transport. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183065. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Rhee, J.Y.; Chung, G.C.; Kang, H. Aquaporin as a membrane transporter of hydrogen peroxide in plant response to stresses. Plant Signal. Behav. 2012, 7, 1180–1181. [Google Scholar] [CrossRef]
- Hooijmaijers, C.; Rhee, J.Y.; Kwak, K.J.; Chung, G.C.; Horie, T.; Katsuhara, M.; Kang, H. Hydrogen peroxide permeability of plasma membrane aquaporins of Arabidopsis thaliana. J. Plant Res. 2012, 125, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shi, H.; Li, N.; Wei, N.; Tian, Y.; Peng, J.; Chen, X.; Zhang, L.; Zhang, M.; Dong, H. Aquaporin OsPIP2;2 Links the H2O2 Signal and a Membrane-anchored Transcription Factor to Promote Plant Defense. Plant Physiol. 2021, 188, 2325–2341. [Google Scholar] [CrossRef]
- Bellati, J.; Champeyroux, C.; Hem, S.; Rofidal, V.; Krouk, G.; Maurel, C.; Santoni, V. Novel Aquaporin Regulatory Mechanisms Revealed by Interactomics. Mol. Cell Proteom. 2016, 15, 3473–3487. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, B.; Furt, F.; Hartmann, M.A.; Michaelson, L.V.; Carde, J.P.; Sargueil-Boiron, F.; Rossignol, M.; Napier, J.A.; Cullimore, J.; Bessoule, J.J.; et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007, 144, 402–418. [Google Scholar] [CrossRef]
- Yepes-Molina, L.; Carvajal, M.; Martinez-Ballesta, M.C. Detergent Resistant Membrane Domains in Broccoli Plasma Membrane Associated to the Response to Salinity Stress. Int. J. Mol. Sci. 2020, 21, 7694. [Google Scholar] [CrossRef]
- Champeyroux, C.; Bellati, J.; Barberon, M.; Rofidal, V.; Maurel, C.; Santoni, V. Regulation of a plant aquaporin by a Casparian strip membrane domain protein-like. Plant Cell Environ. 2019, 42, 1788–1801. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Johnson, E.A.; Aggarwal, M.; Gates, L.; Wu, H.M.; Cheung, A.Y. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 2014, 5, 3129. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Frick, A.; Jarva, M.; Ekvall, M.; Uzdavinys, P.; Nyblom, M.; Tornroth-Horsefield, S. Mercury increases water permeability of a plant aquaporin through a non-cysteine-related mechanism. Biochem. J. 2013, 454, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Laohavisit, A.; Wakatake, T.; Ishihama, N.; Mulvey, H.; Takizawa, K.; Suzuki, T.; Shirasu, K. Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature 2020, 587, 92–97. [Google Scholar] [CrossRef]
- Furt, F.; Oostende, C.; Widhalm, J.R.; Dale, M.A.; Wertz, J.; Basset, G.J. A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K(1)) in chloroplasts. Plant J. 2010, 64, 38–46. [Google Scholar] [CrossRef]
- Karamoko, M.; Cline, S.; Redding, K.; Ruiz, N.; Hamel, P.P. Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 2011, 23, 4462–4475. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, H.R.; Li, H.; Cui, H.R.; Feng, Y.G.; Wang, X.Y. A chloroplast membrane protein LTO1/AtVKOR involving in redox regulation and ROS homeostasis. Plant Cell Rep. 2013, 32, 1427–1440. [Google Scholar] [CrossRef]
- Ivanova, D.; Zhelev, Z.; Getsov, P.; Nikolova, B.; Aoki, I.; Higashi, T.; Bakalova, R. Vitamin K: Redox-modulation, prevention of mitochondrial dysfunction and anticancer effect. Redox Biol. 2018, 16, 352–358. [Google Scholar] [CrossRef]
- Ghezzi, P.; Bonetto, V.; Fratelli, M. Thiol–Disulfide Balance: From the Concept of Oxidative Stress to that of Redox Regulation. Antioxid. Redox Signal. 2005, 7, 964–972. [Google Scholar] [CrossRef]
- Bresnick, E.H.; Sanchez, E.R.; Harrison, R.W.; Pratt, W.B. Hydrogen Peroxide Stabilizes the Steroid-Binding State of Rat Liver Glucocorticoid Receptors by Promoting Disulfide Bond Formation. Am. Chem. Soc. 1988, 27, 2866–2872. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Cavez, D.; Besserer, A.; Berny, M.C.; Gilis, D.; Rooman, M.; Chaumont, F. A conserved cysteine residue is involved in disulfide bond formation between plant plasma membrane aquaporin monomers. Biochem. J. 2012, 445, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Xu, S.; Zou, X.; Zheng, Y.; Qiu, F. Changes of Antioxidative Enzymes and Lipid Peroxidation in Leaves and Roots of Waterlogging-Tolerant and Waterlogging-Sensitive Maize Genotypes at Seedling Stage. Agric. Sci. China 2010, 9, 651–661. [Google Scholar] [CrossRef]
- Aroca, R.; Amodeo, G.; Fernández-Illescas, S.; Herman, E.M.; Chaumont, F.O.; Chrispeels, M.J. The Role of Aquaporins and Membrane Damage in Chilling and Hydrogen Peroxide Induced Changes in the Hydraulic Conductance of Maize Roots. Plant Physiol. 2005, 137, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Marulanda, A.; Azcon, R.; Chaumont, F.; Ruiz-Lozano, J.M.; Aroca, R. Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 2010, 232, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Chee, N.T.; Lohse, I.; Brothers, S.P. mRNA-to-protein translation in hypoxia. Mol. Cancer 2019, 18, 49. [Google Scholar] [CrossRef]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Tian, Q.; Stepaniants, S.B.; Mao, M.; Weng, L.; Feetham, M.C.; Doyle, M.J.; Yi, E.C.; Dai, H.; Thorsson, V.; Eng, J.; et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol. Cell Proteomics 2004, 3, 960–969. [Google Scholar] [CrossRef]
- Nouri, M.Z.; Komatsu, S. Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 2010, 10, 1930–1945. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed]
- Everaert, J.; Podina, I.R.; Koster, E.H.W. A comprehensive meta-analysis of interpretation biases in depression. Clin. Psychol. Rev. 2017, 58, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef]
- Zhu, G.; Li, W.; Zhang, F.; Guo, W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom. 2018, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Kabran, P.; Rossignol, T.; Gaillardin, C.; Nicaud, J.M.; Neuveglise, C. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res. 2012, 19, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Comellas, A.P.; Dada, L.A.; Lecuona, E.; Pesce, L.M.; Chandel, N.S.; Quesada, N.; Budinger, G.R.; Strous, G.J.; Ciechanover, A.; Sznajder, J.I. Hypoxia-mediated degradation of Na,K-ATPase via mitochondrial reactive oxygen species and the ubiquitin-conjugating system. Circ. Res. 2006, 98, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Berger, N.; Demolombe, V.; Hem, S.; Rofidal, V.; Steinmann, L.; Krouk, G.; Crabos, A.; Nacry, P.; Verdoucq, L.; Santoni, V. Root membrane ubiquitinome under short-term osmotic stress. Int. J. Mol. Sci. 2021, 23, 1956. [Google Scholar] [CrossRef] [PubMed]
- Uniacke, J.; Holterman, C.; Lachance, G.; Franovic, A.; Jacob, M.D.; Fabian, M.R.; Payette, J.; Holcik, M.; Pause, A.; Lee, S. An oxygen-regulated switch in the protein synthesis machinery. Nature 2012, 486, 126–129. [Google Scholar] [CrossRef]
- Liu, L.; Cash, T.P.; Jones, R.G.; Keith, B.; Thompson, C.B.; Simon, M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 2006, 21, 521–531. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]


| Gene ID | Name | Sequence forward Primer 5′-3′ | Sequence Reverse Primer 5′-3′ |
|---|---|---|---|
| 542619 | PIP2-5 | ACTGGATCTTCTGGGTGGGT | CGATCTAGCGGCTGAAGGAG |
| 541888 | PIP2-1 | CACTGGATCTTCTGGGTGGG | GATGGCATTCTCCTCGCTCAC |
| 542644 | PIP2-2 | TCGATCTAGCGTGGGGAGAG | AACAAAAGCGACCGACGAGA |
| 100285512 | DoHCytb561 | GCCGTTGTTCAGAGAGACAT | AGGAGTACAGACTACAGAGGC |
| 100383470 | SKU5 | TCTACTTCCCACCCCTTGGT | AGGTGCGTGTGGTTCATCTT |
| 100285365 | FQR1 | GGGTTCAGCCTGATCTACACAT | GAGCCACACAACATCCAGAC |
| 541856 | LOX1 | TACACGCTGCTCTACCCCAA | CCACACTTCACGGAACGGAA |
| 100037802 | LOX2 | GCCAGCGTTTCACCCAAAAA | CCAGCACCAGTACACCAAGG |
| 542495 | LOX3 | CAGCCTCACACAGACACCAA | GATGATCCCGCTCAGCATCT |
| 100273405 | EF-TuM | CGCAGTTGATGAGTACATCC | AACACGCCCAGTAACAACAG |
| Protein Name | Gene ID | Gene fc | UniProt Acc. | Protein fc | MW (1) | pI (1) | TMH (2) |
|---|---|---|---|---|---|---|---|
| Auxin induced in root cell cultures, AIR12 | 100280845 | −1.95 * | B6SN55 | −1.30 | 25.1 | 9.39 | 0/GPI |
| Auxin induced in root cell cultures, AIR12 | 100285927 | −4.05 * | B6UBU0 | n.d. | 26.7 | 9.34 | 0/GPI |
| Auxin induced in root cell cultures, AIR12 | 103653191 | −1.33 | A0A3L6EZL2 | n.d. | 24.0 | 9.39 | 0/GPI |
| Auxin induced in root cell cultures, AIR12 | 100286361 | −1.88 * | K7VGM8 | n.d. | 22.2 | 6.90 | 0/GPI |
| DoHcytb561 | 100285512 | −1.17 | B6U5U8 | 2.78 | 41.0 | 9.57 | 5 |
| H+-ATPase | 542052 | −1.03 | K7TX67 | 1.41 | 104.9 | 6.23 | 8 |
| H+-ATPase | 100502231 | −2.14 | A0A1D6MV33 | 1.03 | 105.4 | 6.20 | 8 |
| H+-ATPase | 542048 | −1.75 * | A0A1D6DVJ7 | 1.78 | 81.0 | 5.68 | 5 |
| Lipoxygenase 2, LOX1 | 541856 | −1.82 | Q9LKL4 | 2.17 | 98.2 | 6.25 | 0 |
| Linoleate 9S-lipoxygenase 2, LOX2 | 100037802 | −4.09 | A1XCH8 | 1.99 | 98.4 | 6.17 | 0 |
| Lipoxygenase, LOX3 | 542495 | 1.33 | Q8W0V2 | 2.03 | 96.5 | 5.72 | 0 |
| Lipoxygenase, LOX4 | 100037803 | 3.38 * | C0P840 | −1.06 | 100.4 | 6.18 | 0 |
| Malate dehydrogenase, MDH | 542598 | −1.18 | Q08062 | 2.33 * | 35.6 | 5.75 | 0 |
| Malate dehydrogenase, cMDH | 100280767 | −1.42 * | A0A1D6GPH0 | 1.73 | 33.3 | 7.00 | 0 |
| MDHAR | 100501585 | −1.30 ** | C4J4E4 | n.d. | 46.7 | 5.45 | 0 |
| Monocopper oxidase-like protein, SKU5 | 100383470 | −2.08 ** | C0PG78 | 1.49 | 65.8 | 6.01 | 1 |
| NAD(P)H dehydrogenase (quinone), FQR1 | 100285365 | −1.55 * | B6U474 | 2.36 | 21.5 | 6.06 | 0 |
| NAD(P)H dehydrogenase (quinone), WrbA | 100280914 | 1.17 | B6SPB2 | 1.44 | 25.7 | 6.98 | 0 |
| Peroxidase 3 | 542505 | −2.27 | A0A1D6LYW3 | 3.18 | 38.7 | 6.52 | 0 |
| Peroxidase 24 | 542464 | −1.17 | B4FHG3 | 4.72 | 37.9 | 5.51 | 0 |
| Peroxidase 81 | 100193733 | −2.89 | B4FG39 | 12.57 * | 36.6 | 8.08 | 1 |
| Peroxidase 85 | 100279351 | 1.53 * | A0A1D6E530 | 5.74 | 35.5 | 5.35 | 1 |
| Respiratory burst oxidase 10, Rboh10 | 100381459 | 5.91 | C0HG64 | n.d. | 45.2 | 9.22 | 2 |
| Nucleobase-ascorbate transporter 6 | 100272944 | −1.21 | B4F8S3 | n.d. | 57.7 | 9.37 | 11 |
| Nucleobase-ascorbate transporter 6 | 100282103 | 1.48 | C0PH14 | n.d. | 58.1 | 9.31 | 11 |
| Permease I | 100283406 | −1.39 | B4FIZ3 | n.d. | 57.4 | 9.54 | 11 |
| Nucleobase-ascorbate transporter 2 | 103652969 | −1.14 | K7U0A9 | n.d. | 57.3 | 8.65 | 9 |
| Nucleobase-ascorbate transporter 3 | 100279038 | 1.30 | A0A096QFX2 | n.d. | 60.5 | 9.13 | 11 |
| Nucleobase-ascorbate transporter 2 | 100279209 | 1.14 | A0A1D6EP29 | n.d. | 76.6 | 9.60 | 8 |
| Nucleobase-ascorbate transporter 12 | 100279219 | −1.34 | A0A1D6G835 | n.d. | 77.8 | 9.33 | 10 |
| PIP1;1 active with PIP1;2 | 542434 | −3.21 * | Q41870 | n.d. | 30.9 | 9.47 | 6 |
| PIP1;2 active with PIP1;1, PIP2;1, PIP2;4, or PIP2;5 | 541779 | −4.68 *** | Q9XF59 | n.d. | 30.8 | 9.00 | 6 |
| PIP1;3/1;4 | 541886 | −2.22 * | Q9AQU5 | n.d. | 31.0 | 8.83 | 6 |
| PIP1;5 | 542014 | −2.65 | Q9AR14 | −2.06 | 30.7 | 8.30 | 6 |
| PIP1;6 | 113523644 | n.d. | Q9ATN0 | n.d. | 31.0 | 6.70 | 6 |
| PIP2;1 active with PIP1;2 | 541888 | −1.94 | Q84RL7 | 1.54 * | 30.2 | 7.69 | 6 |
| PIP2;2 | 542644 | −1.92 | Q9ATM8 | 1.23 | 30.3 | 8.29 | 6 |
| PIP2;3 | 541889 | −2.55 * | Q9ATM7 | n.d. | 30.4 | 6.95 | 6 |
| PIP2;4 active with PIP1;2 | 541890 | −3.65 * | Q9ATM6 | 1.75 * | 30.3 | 6.50 | 6 |
| PIP2;5 active with PIP1;2, impaired by Hg2+ | 542619 | −4.29 ** | Q9XF58 | 1.69 | 29.8 | 7.70 | 6 |
| PIP2;6 | 541891 | −2.12 ** | Q9ATM5 | 2.39 | 30.2 | 8.38 | 7 |
| PIP2;7 | 542645 | n.d. | Q9ATM4 | n.d. | 30.8 | 8.26 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofmann, A.; Wienkoop, S.; Lüthje, S. Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants 2022, 11, 836. https://doi.org/10.3390/antiox11050836
Hofmann A, Wienkoop S, Lüthje S. Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants. 2022; 11(5):836. https://doi.org/10.3390/antiox11050836
Chicago/Turabian StyleHofmann, Anne, Stefanie Wienkoop, and Sabine Lüthje. 2022. "Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots" Antioxidants 11, no. 5: 836. https://doi.org/10.3390/antiox11050836
APA StyleHofmann, A., Wienkoop, S., & Lüthje, S. (2022). Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants, 11(5), 836. https://doi.org/10.3390/antiox11050836