Abstract
Mucopolysaccharidosis, type IIIB (MPS IIIB) is a rare disease caused by mutations in the N-alpha-acetylglucosaminidase (NAGLU) gene resulting in decreased or absent enzyme activity. On the cellular level, the disorder is characterized by the massive lysosomal storage of heparan sulfate (HS)—one species of glycosaminoglycans. HS is a sulfur-rich macromolecule, and its accumulation should affect the turnover of total sulfur in cells; according to the studies presented here, it, indeed, does. The lysosomal degradation of HS in cells produces monosaccharides and inorganic sulfate (SO42−). Sulfate is a product of L-cysteine metabolism, and any disruption of its levels affects the entire L-cysteine catabolism pathway, which was first reported in 2019. It is known that L-cysteine level is elevated in cells with the Naglu−/− gene mutation and in selected tissues of individuals with MPS IIIB. The level of glutathione and the Naglu−/− cells’ antioxidant potential are significantly reduced, as well as the activity of 3-mercaptopyruvate sulfurtransferase (MPST, EC 2.8.1.2) and the level of sulfane sulfur-containing compounds. The direct reason is not yet known. This paper attempts to identify some of cause-and-effect correlations that may lead to this condition and identifies research directions that should be explored.
1. Introduction
Rare diseases are disorders with a very low probability of occurrence in the population (the European Union Registration on Orphan Medicinal Products has estimated the rare disease prevalence as less than 1 person per 2000 [1]). They are usually genetic, chronic and severe, appearing mostly in childhood. Only a small group of people (about 5%) suffering from a rare disease are treated pharmacologically, which means that for the vast majority of them there is no effective therapy. One of these diseases is mucopolysaccharidosis, type IIIB (MPS IIIB) [OMIM 252920], which has an estimated incidence of 1/200,000 newborns [National Organization of Rare Disease]. Currently, there is no cure for MPS IIIB because there are no effective therapies which can stop or reverse it [2,3,4,5,6,7,8,9]. The disease itself is the result of a disruption of an ongoing process in the body of a healthy person. In this process, new molecules of polysaccharides made up of repeating disaccharide units—glycosaminoglycans (GAGs)—are continually produced, while older molecules are continually broken down. MPS IIIB is caused by a mutation in the N-alpha-acetylglucosaminidase (NAGLU) gene, resulting in the absence or decreased activity of the enzyme that is involved in the catabolism of GAGs. On the cellular level, the disorder is characterized by the massive lysosomal storage of heparan sulfate (HS)—a sulfur-rich macromolecule. Accumulation of such molecules should affect the turnover of total sulfur in the cells, and, as Kaczor-Kamińska and others showed [10,11], indeed, it does.
2. Glycosaminoglycans (GAGs)
GAGs (previously called mucopolysaccharides) accompany us from the beginning of fetal life [12,13,14], and this is true of all living organisms. From the chemical point of view, these are long, linear, unbranched heteropolysaccharides mostly bearing SO3− groups [15]. This makes them biopolymers that are negatively charged and thus have good water solubility under physiological conditions. The standard for identifying individual GAGs is the presence of alternating repetitions of the corresponding disaccharide sequences in the macromolecules. At this point, liquid chromatography with MS detection is the only tool to effectively identify these compounds [16,17]. The GAGs known in the animal kingdom include [18,19] heparin, heparan sulfate (HS), chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS) and hyaluronic acid (HA). Analysis of the names of these compounds alone indicates that they are closely related to the tissues in which they occur in significant amounts (CS—from chondrocytes, DS—from skin/dermal tissue, KS from keratinocytes) or where they were first detected (heparin and HS from hepar—liver in Latin). An exception to the general rule in the case of the presence of sulfur in the GAGs molecules is HA, which does not have SO3− groups and the negative charge is due to the presence of carboxyl groups [20].
Biosynthesis of GAGs is a very complex process involving many enzymes, and is still not completely understood [21]. GAGs are synthesized in cells of many tissues in which they play physiologically important functions. Universally, they are an important component of the intercellular matrix. In multicellular organisms this matrix is classically composed of fibrillar proteins such as collagen or elastin and mixtures of GAGs covalently linked to protein components [22,23]. There are organs in the body in which levels of these macromolecules are significantly higher, such as the eye [24] or heart muscle [25].
GAGs are macromolecules of which, compared to nucleic acids or proteins, biological significance has been underestimated until recently [26]. Classically they were perceived only as a non-protein component of proteoglycans in which the protein part is responsible for the biological activity of the whole molecule. This perception has changed considerably over the last decade and the concept of activity is no longer associated solely with proteins [26]. The participation of GAGs in so many biological processes, and their presence in almost all tissues, makes it reasonable to consider the option of using them as active substances in medicine [27]. The great importance and potential of GAGs in medicine may be evidenced by the fact that attempts have been made to realize ex vivo synthesis within the tools of classical organic synthesis of these compounds. Working with sugars is difficult due to the presence of multiple OH groups in the repeating unit, which require blocking and usually numerous steps [28] in order for them not to react. Nevertheless, the potential value of these compounds is so great for life sciences that attempts have been made to synthesize GAG fragments such as pentasaccharidic analogs of heparin [29] and longer structures [30,31]. Even simplified analogues of GAG are being developed, i.e., polymers with SO3− containing groups as side chains, such as PAMPS [poly(2-acrylamido-2-methyl-1-propanesulfonic acid)]. These polymers exhibit some of the properties of GAGs, especially heparin, i.e., anticoagulant and anti-angiogenic properties [32].
Heparin is one of the most commonly used GAGs in medicine, among others, in the treatment of cardiovascular diseases and in some interventional cardiology procedures. The anticoagulant action of heparin results from its ability to bind with plasma protein antithrombin, which interacts with several serine proteases of the coagulation system, most importantly factors IIa (thrombin), Xa and IXa [33]. In terms of anticoagulant properties, the sequence of five sugar units is critical; therefore, similar properties to heparin are exhibited by its lower molecular mass analogues, low molecular weight heparin and synthetic pentasaccharide (Idraparinux). The pharmacokinetic properties of these systems are different, more desirable from the standpoint of chronic use, and their performance appears to be more predictable [34,35] to the point that they can be used by the patient without direct supervision by trained personnel. In addition to the commonly known anticoagulant properties of heparin, it (also low molecular weight heparin—LMWH) may also affect blood lipid balance [36].
Most of the physico-chemical and biological properties of GAGs are related to their structure or sequence (monosaccharide constituents and bonds between the disaccharide repeating units), conformation, chain flexibility, molecular weight and charge density [37,38]. Among these properties, the degree of GAGs sulfation (sulfation code), which is inherent in almost all GAGs, is crucial. Activities and functions of GAGs are mainly triggered by their interactions with a wide variety of biological molecules. Structural changes in GAGs that have no effect at the cellular level often have a huge impact at the organ/tissue or organism level, and changes in the GAG sulfation patterns are often associated with different disorders such as Alzheimer’s disease and cancer [38,39]. On the other hand, irregularities in normal physiological GAG degradation, leading to, e.g., endolysosomal HS accumulation, cause a group of less known rare diseases identified collectively as mucopolysaccharidoses (type III).
3. Heparan Sulfate (HS)
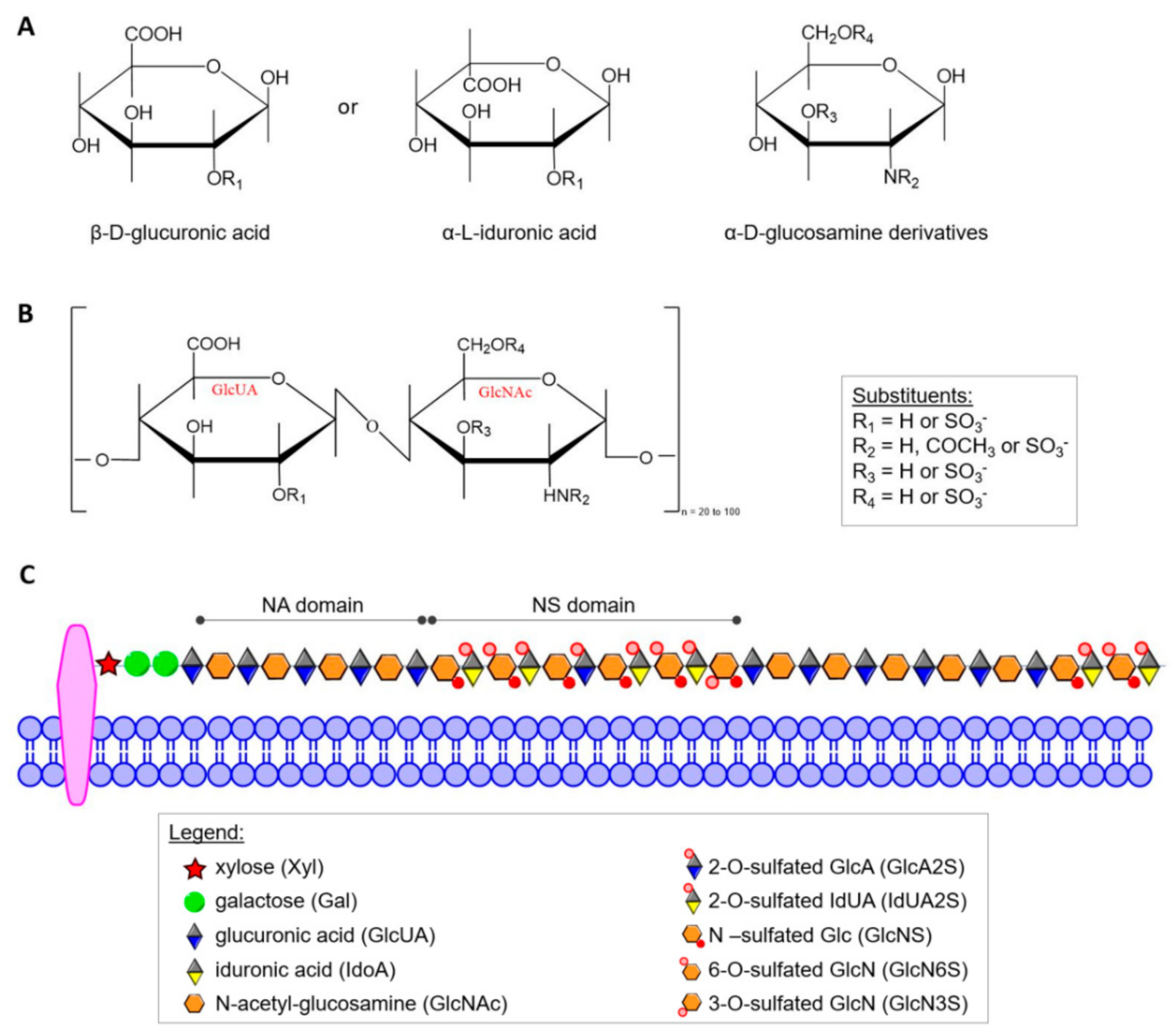
HS was defined as a distinct molecular entity by Jorpes and Gardell in 1948 [40] but elucidation of its structure has been a slow, painstaking process that is still underway. Today, it is known that HS is structurally closely related to heparin, and both differ from other GAGs in several important ways. α-d-Glucosamine (GlcN) and/or its derivatives and β-d-glucuronic acid (GlcUA) or its C5 epimer α-L-iduronic acid (IdUA) form the characteristic disaccharide repeat unit shown in Figure 1A,B. In contrast to most other GAGs, heparin and HS contain α-glycosidic linkages. In heparin almost all glucosamine residues (GlcN) contain sulfamide linkages, but a smaller number of GlcN are N-acetylated. Sulfate groups are linked by ester bonds to certain monosaccharides or by amide bonds to the amino group of GlcN. In addition to N-sulfate and O-sulfate on C6 of GlcN, heparin can also contain sulfate on C3 of the hexosamine and C2 of the uronic acid [41]. HS contains more N-acetyl groups, fewer N-sulfate groups, and a lower degree of O-sulfate groups. The sulfate content of heparin, although variable, approaches 2.7 sulfate per disaccharide unit in preparations with the highest biological activity [37], while HS contains only about one sulfate group per disaccharide. However, individual HS chains may have higher contents of this group and, additionally, often contain domains of extended sequences having low or high sulfation (Figure 1C) [42]. On account of structural heterogeneity, variability and polydisperse, HS cannot be considered a single compound but rather a family of related polymers [43]. Up to now, no rigorous formula of free HS has been established because within the carbohydrate chain of a given GAG there may be considerable variations of the constituent sugars, of the degree of sulfation and of the position of sulfate groups.
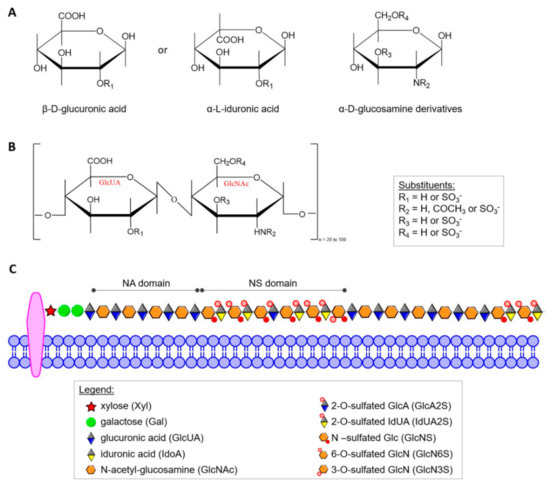
Figure 1.
(A) Haworth projection of the heparin/HS monosaccharide units; (B) repeating GlcUA-GlcNAc disaccharide units of HS; (C) scheme of structural organization of the residues in HS. In the mature form of HS, two main types of domains can be distinguished: NA domains build mostly by non-modified N-acetyl-glucosamine moiety linked mainly to glucuronic acid residues, and NS domains that are composed by highly sulfated disaccharides units).
The structural features of HS chains, including length, uronic acid epimerization degree and sulfation patterns, play crucial roles in various biological processes [44]. In a given HS polysaccharide, negatively charged sulfate and carboxyl groups are arranged in various types of domains (Figure 1C) [43], but the domain organization of substituents along the HS chain and the composition of HS produced are distinctly regulated by different cells. The large HS structural diversity results from a tightly controlled biosynthetic pathway that is differently regulated in various organs, stages of development and pathologies, including cancer [45,46]. The current distinction between heparin and HS (and various GAGs in general) is not only based on carbohydrate structure [42,44,47], but also includes proteoglycan type and cellular distribution. Unlike other GAGs, heparin is an intracellular component of mast cells [43,48,49] and functions predominantly as an anticoagulant [32] and lipid-clearing agent [37]. HS may be extracellular or an integral and ubiquitous component of the cell surface in many tissues [43,50] including endothelium of blood vessels [51], amyloid [49,52], and brain [53]. HS on the cell surface provides cells with a mechanism to capture a wide range of extracellular effectors without requiring multiple new binding proteins [54].
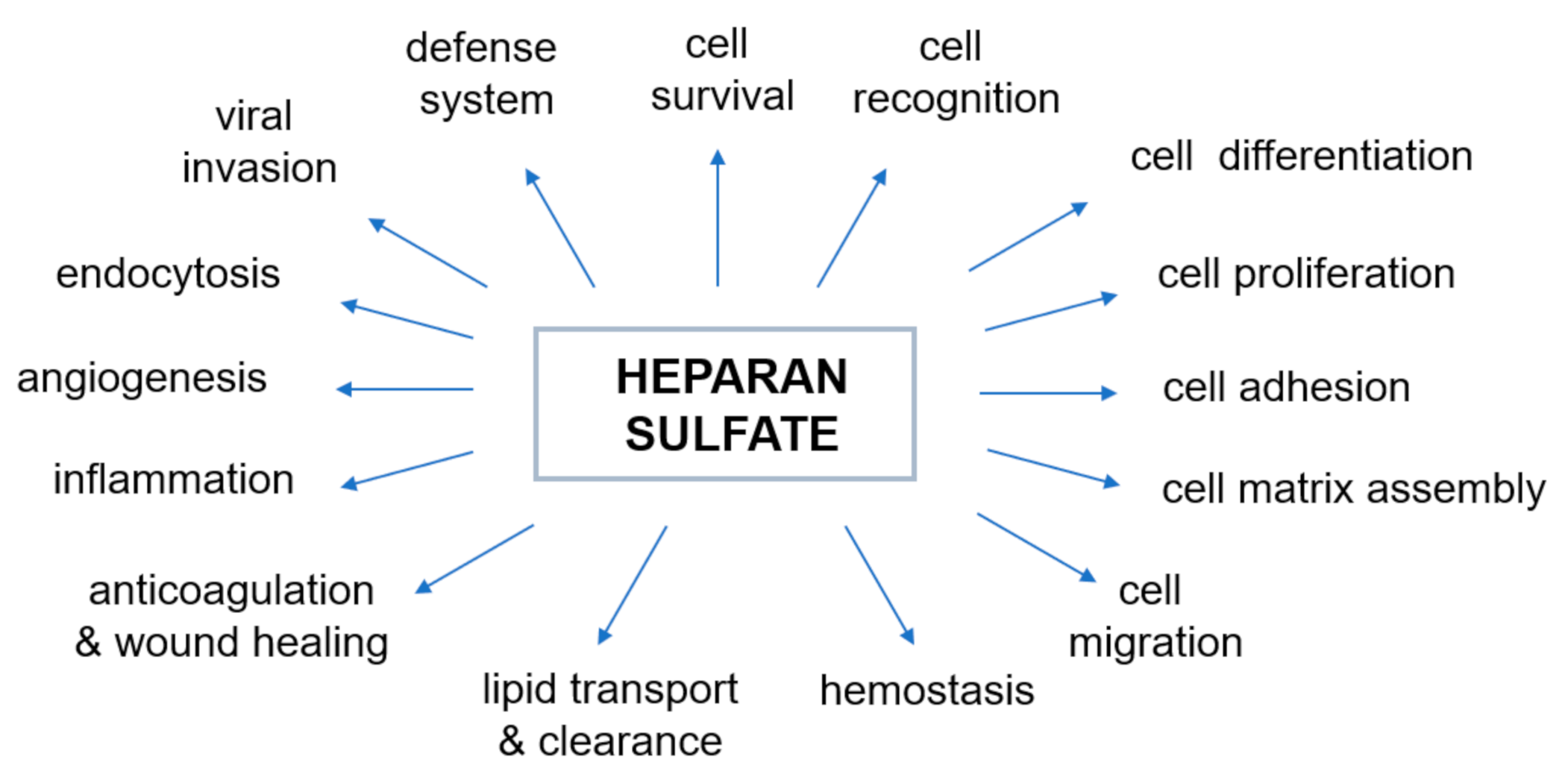
The negative charge of HS largely determines the nature of interactions between proteoglycans and other molecules and ions of the extracellular matrix. The majority of physiological, as well as pathophysiological [45], activities of HS are due to interactions with various proteins. The most prominent type of interaction between HS and a protein is electrostatic. However, in some cases there is a significant contribution to the binding by non-electrostatic interaction such as hydrogen bonding and hydrophobic interactions [37]. The mechanism by which HS changes the activity of proteins through such relations has proved elusive. Over the last few decades, heparin and HS have been shown to bind and regulate the activities of many biologically important proteins, including enzymes, growth factors, extracellular matrix proteins, and the cell surface proteins of pathogens [54,55], and therefore play an essential role in various physiological and pathophysiological processes (Figure 2, prepared based on the information included in [32,46,55,56,57,58,59,60,61,62]). Genetic studies have shown that the properties of heparin and HS reflect their ability to regulate various biological process (Figure 2) such as, but not limited to, cell signaling and morphogenesis in vivo, growth regulation and tumor suppression [59,60,61,62]. It has also recently been discovered that excess of HS (and/or its derivatives) in specific cells and tissues affects sulfur metabolism disruption [10,11]. Therefore, our understanding of the nature of the heparin/HS-protein interactions at the molecular level offers a large number of potential therapeutic applications for these compounds and is necessary to understand normal physiological and pathophysiological processes.
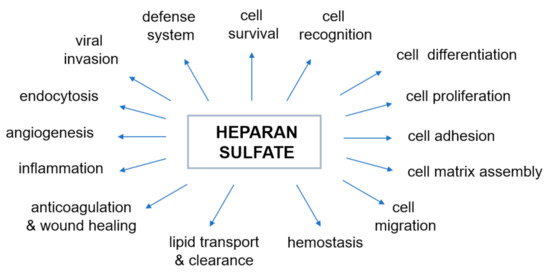
Figure 2.
Biological activities modulated by the interaction of heparan sulfate with proteins. The diagram was prepared based on the information included in the following papers: [32,46,55,56,57,58,59,60,61,62].
4. Decomposition of Heparan Sulfate
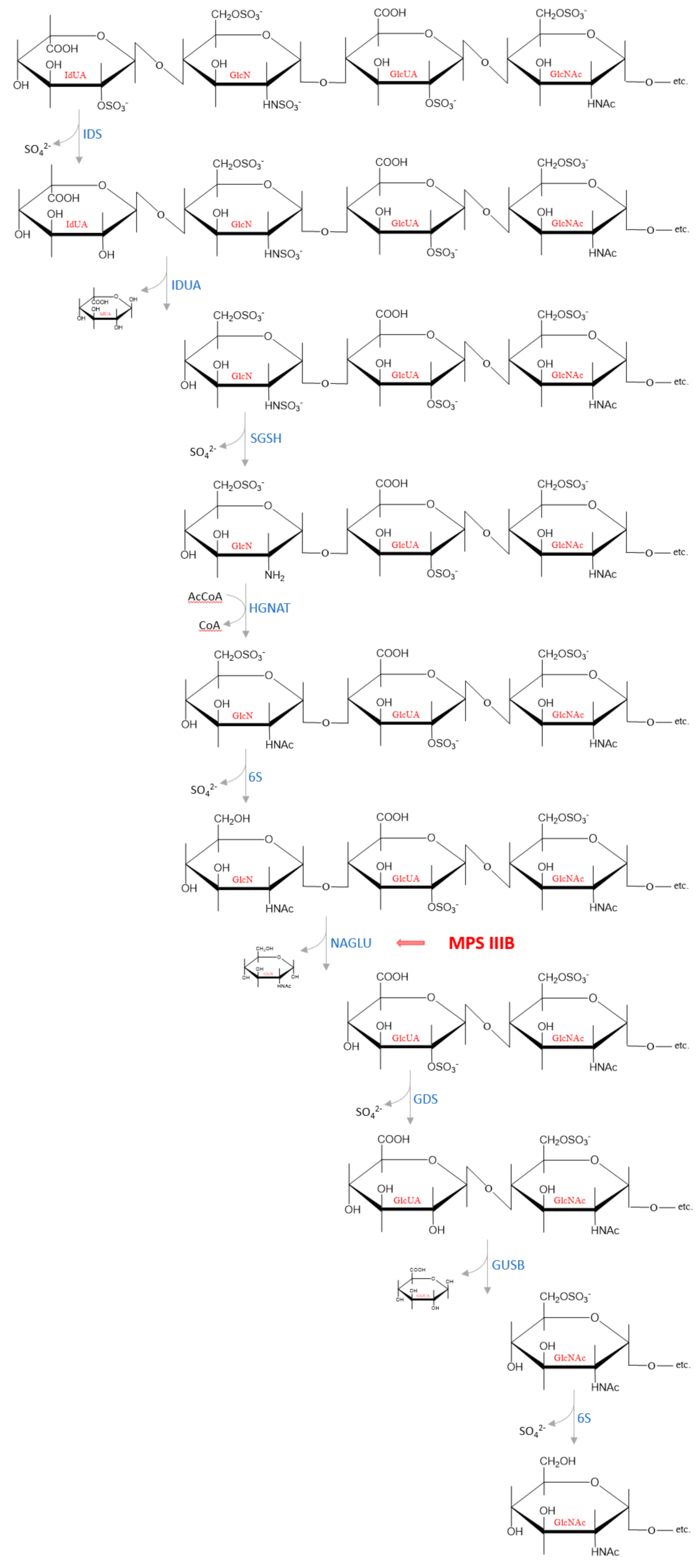
GAGs are present in cells in the form of proteoglycans, which are complex macromolecules composed of one or more GAG chains covalently attached to a protein core. Degradation of the protein moiety and the polysaccharide chains occurs independently of each other. This process begins with the proteolytic removal of the protein core of proteoglycans on the cell surface. Hydrolysis of some peptide bonds by proteases may take place extracellularly but the carbohydrate chain is degraded only within lysosomes [63,64]. The released GAG chains are then partially cleaved by enzymes such as endoglucuronidases or endohexosaminidases, which cut through the GAG macromolecules at several specific sites, resulting in formation of HS fragments and oligosaccharide fragments [65,66]. There are three lysosomal glycosidases that degrade HS oligosaccharides to monosaccharides: α-L-iduronidase (IDUA, EC 3.2.1.76) responsible for hydrolysis of unsulfated α-L-iduronosidic linkages; β-d-glucuronidase (GUSB, EC 3.2.1.31) that catalyzes hydrolysis of β-d-glucuronic acid from the non-reducing end of GAGs and α-N-acetylglucosaminidase (NAGLU, EC 3.2.1.50) that cleaves the N-acetyl-d-glucosamine α, 1 → 4 linkage between N-acetylglucosamine and the neighboring uronic acid from the non-reducing end in HS [64,66]. Complete degradation is achieved by the sequential action of endosulfatases, endodeacetylases and endoglycosidases at the non-reducing end of the chain [63,67]. An example of the subsequential cleavage of a HS chain by lysosomal enzymes is presented in Figure 3. The reaction products—monosaccharides and sulfate ions—are small enough to leave the lysosomal compartment. The degradation products are partially used in other metabolic processes, e.g., glucuronate and sulfate are used by the liver to conjugate with non-polar substances (e.g., some drugs such as diazepam, verapamil, and carbamazepine) or as final metabolites are excreted with the urine.
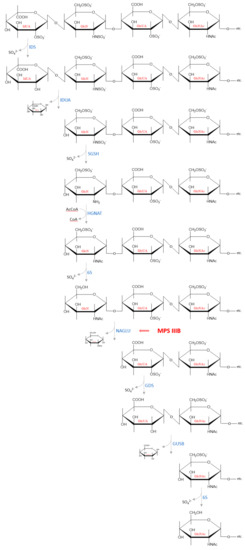
Figure 3.
An example of how the subsequential degradation from the non-reducing end of the tetrasaccharide IdUA2S (α 1−4)—GlcNS6S (α 1−4)—GlcA2S (β 1−4)—GlcNAc6S occurs in the lysosome by HS degrading enzymes. Abbreviations: IdUA = iduronic acid; GlcN = glucosamine; GlcUA = glucuronic acid; GlcNAc = N−acetylglucosamine; IDS—iduronate−2−sulfatase (EC 3.1.6.13); IDUA—α−L−iduronidase (EC 3.2.1.76); SGSH—N−sulfoglucosamine sulfohydrolase (EC 3.10.1.1); HGNAT heparan−α−glucosaminide N−acetyltransferase (EC 2.3.1.78); 6S—N−acetylglucosamine−6−sulfatase (EC 3.1.6.14); NAGLU—α−N−acetylglucosaminidase (EC 3.2.1.50); GDS—glucuronate−2−sulfatase (EC 3.1.6.18); GUSB—β−glucuronidase (EC 3.2.1.21).
All HS-degrading enzymes in the lysosomes are exoactive, meaning they must act sequentially to fully breakdown the oligosaccharide chains. The HS chains derived from the same biosynthetic pool (tissue-, cells- and function-dependent) show similar distribution of NA-, NS-, as well as NA/NS- domains ratio (Figure 1C), and each domain type retains a typical substitution pattern. However, even though such a pattern is characterized by a defined GlcUA/IdUA moieties ratio and levels of different sulfate groups (linked by N-, 2-O-, 3-O, 6-O-), there is no evidence for the generation of predominant sequences of variously modified/substituted monosaccharide units during degradation process [43]. Therefore, depending on the initial HS sequence, there are many various structures of the oligosaccharide fragments that can accumulate in people suffering from MPS IIIB. However, the basic mechanisms of chain breakdown are thought to be similar in different organisms, but little is known about the actual structure of the entire decomposition process. A complete understanding of this enzymatic machinery responsible for HS degradation seems crucial for designing therapies for related diseases such as mucopolysaccharidoses.
5. Mucopolysaccharidosis, Type IIIB
Mucopolysaccharidoses are disorders caused by faulty degradation of sulfated mucopolysaccharides (sulfated GAGs), because of absent or decreased activity of an enzyme involved in this process (there is one exception—MPS type IX—where non-sulfated compound—HA is accumulated). The degradation process continues until a linkage occurs at the terminal end for which the deficient enzyme is specific. The genetic defect of the enzyme clearly indicates the stage at which the process of breaking down GAGs is stopped, as well as the subtype of MPS. Any further breakdowns are then stopped except for those brought about by endoglycosidases [63], such as NAGLU glycosidase. A deficiency, or mutations, in NAGLU has been linked to MPS IIIB (also known as Sanfilippo B syndrome) and is characterized by the lysosomal accumulation and urinary excretion of HS. The incidence of the mucopolysaccharidosis III is reported to be between 0.28 and 4.1 cases per 100,000 births, but its subtype B affects approximately 1 in 200,000 neonates (National Organization of Rare Disease).
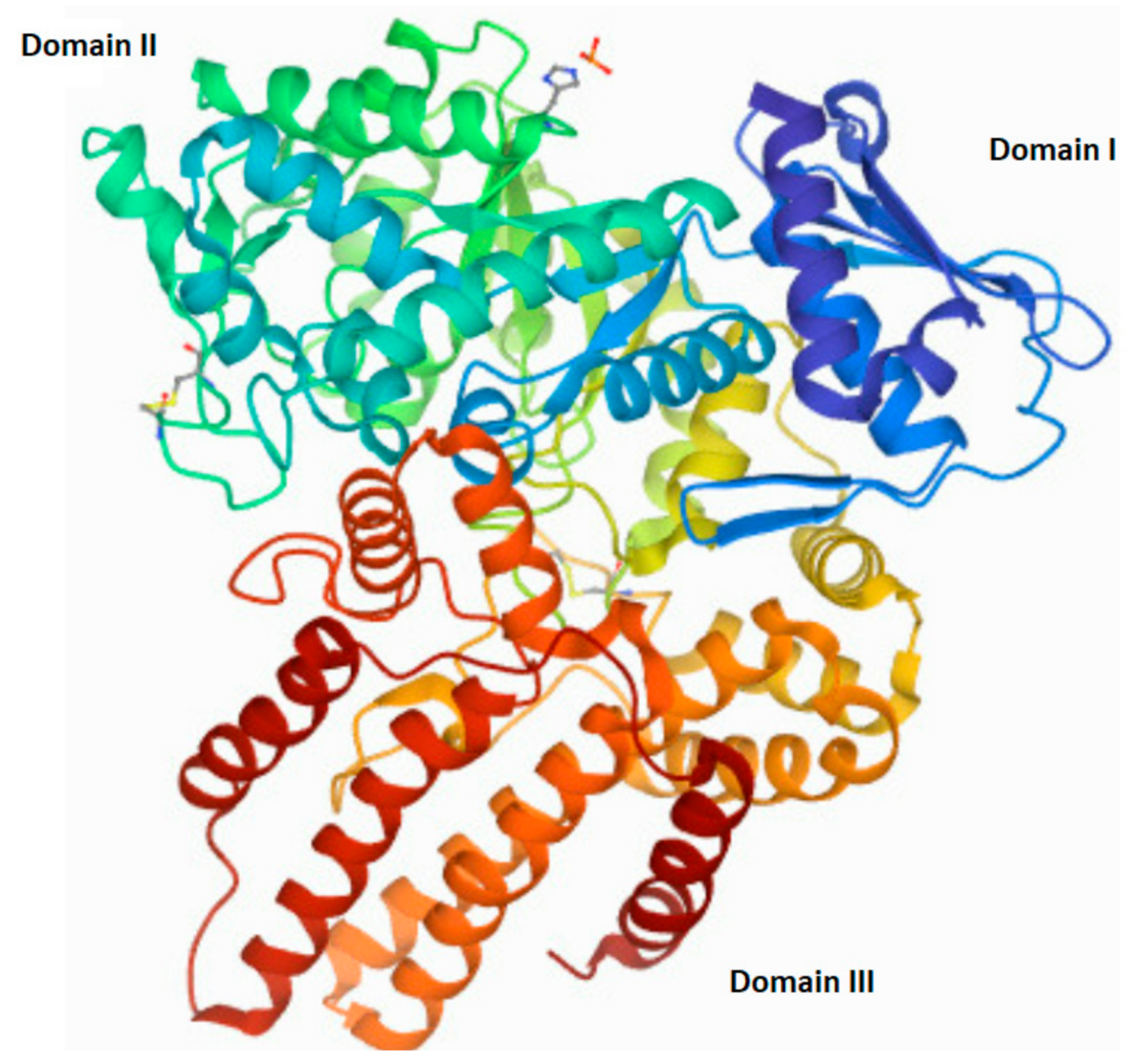
The human NAGLU gene is localized on chromosome 17q21.1 (gen length—10,212 bp) and consist of 7 exons (The National Center for Biotechnology Information, gene ID: 4669, accessed on 31 January 2022). In humans, alternative folding can result in five different transcript variants of NAGLU (four predicted and one established) that differ in the number of nucleotide pairs and lead to proteins differing in the number of amino acids. The one established NALGU cDNA (NM_000263.4) encodes a 743-amino-acid protein (NP_000254) that has six potential N-glycosylation sites. Using the NCBI database, as well as the National Institute of Environmental Health Sciences (NIH) website (https://manticore.niehs.nih.gov/snpinfo/snpinfo.html (accessed on 27 February 2022)), we checked whether there are changes (single nucleotide polymorphism—SNP) in the human NAGLU DNA that appear too often, to talk about the occurrence of a random mutation (differences in the population with a frequency above 1%). These analyses revealed, a significant number of single nucleotide polymorphisms present in the entire gene (3017 SNPs) as well as in its coding regions (cSNPs)—821 SNPs. Types of mutations found in the NAGLU coding region are as follows: 38 frame shift mutations, 285 synonymous mutations, 561 missense and 32 nonsense mutations, eight insertions and five deletions (according to the NIH database). Any non-synonymous mutation in the NAGLU gene may interfere with proper functioning, mostly by influence on expression and/or activity changes. Mutations located in the active site of the NAGLU enzymes, or those that result in the enzyme misfolding (causes their direction to proteasomal degradation instead of their cellular destination—lysosome), impair enzyme function. Mutations outside of the active site may alter the overall stability and/or function of the molecule [68]. Moreover, the number and/or location of the mutation correlates with the severity of the disease phenotype Clinical manifestations of MPS IIIB range from mild to severe and can be divided into three phases. In the first phase, there is a slight delay in the child’s development between 1 and 4 years of age. The second phase, which occurs around age 3–4, is characterized by hyperactivity, aggression, and a gradual loss of skills acquired during development, as well as a progressive intellectual decline. In the final stage, the child’s range of motion decreases significantly, even to the point where the patient loses mobility. Severe dementia and dysphagia occur [69,70,71,72,73,74]. Death usually occurs by the age of twenty, although there are some exceptions [75,76]. The structure of the native human NAGLU protein is shown in Figure 4.
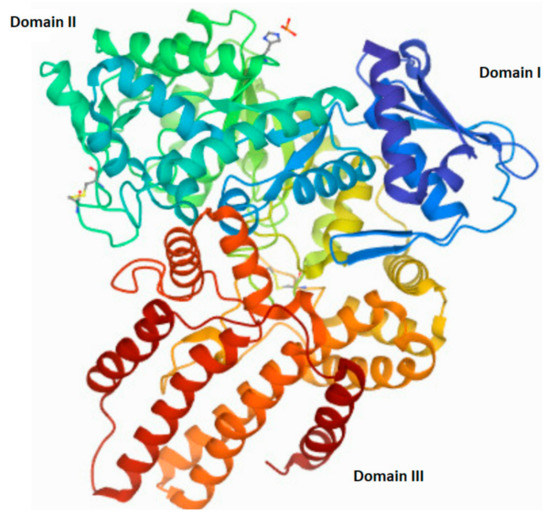
Figure 4.
The tertiary structure of human N-acetyl-alpha-glucosaminidase (EC 3.2.1.50). A modified image from the RCSB PDB (rcsb.org) of PDB ID 4XWH_1 [68].
The principal biochemical abnormality caused by the NAGLU gene or activity deficiency in MPS IIIB is the accumulation of lysosomal HS (the mechanism of HS storage has not yet been fully understood) and the elimination of this polysaccharide, or fragments derived from it, in body fluids [63]. Additionally, secondary storage products, such as: GM2 and GM3 gangliosides that play a role in central nervous systems pathology [63,77], inflammatory cytokines, reactive oxygen species [78] and globotriaosylsphingosine (LysoGb3) [79] have also been described. There are hypotheses concerning secondary storage product accumulation. Some have argued that this phenomenon may be due to the inhibition of relevant lysosomal enzymes by HS/GAGs, which when accumulating can selectively bind various hydrolases causing a decrease in their activity [79]. Others claim that they can cause critical disruption of the internal environment of the lysosome (for example, by changes in pH) and thus lead to reduced degradation of additional substrates [80].
However, many aspects of the MPS IIIB biochemistry are still unclear or even unknown. Therefore, understanding the roles of HS/GAGs role their effect on various biochemical processes in normal and pathogenic states seems to be very important, because no effective treatment for this disease is currently available [2], although a number of methods are being explored [3,4,5,6,7,8,9]. These methods include enzyme replacement therapies, substrate reduction therapies, gene therapy, hematopoietic stem cell transplantation and enzyme enhancement therapy [8,63]. Recently, Pearse and Iacovino [81] described clinical results of therapies for MPS IIIB such as genistein, BMN 250 and others [81]. Nevertheless, there is still a lack of approved MPS IIIB treatments, despite significant advances in research providing tools to understand their molecular basis, as well as legislation providing regulatory and economic incentives to accelerate the development of specific therapies.
6. GAGs as a Source of Sulfate
Lysosomal degradation of the carbohydrate portion of glycoproteins and GAGs produces monosaccharides and the greatest amount of inorganic sulfate (SO42−) in cells compared to other sources such as catabolism of sulfur amino acids released during proteins breakdown and dietary supply [82,83,84]. Lysosomal inorganic sulfate can be reused in other biosynthetic pathways. Sulfates exit the lysosome either by secretion, i.e., through fusion of the lysosome with the cell membrane, or by entering the cytoplasm via sulfate transporters. This requires lysosomal acidic pH for optimal activity [82,85], which is maintained by the action of the lysosomal H+-ATPase. Some evidence suggests that SO42− transport is modulated by ATP independently from the lysosomal H+-ATPase action [82,86]. There are no data in the literature to indicate that sulfate is accumulated in cells and tissues, even though there is chronic exposition/ingestion of ‘above-normal’ sulfate levels [83]. Therefore, it can be assumed that inorganic sulfates are incorporated into several types of biomolecules, such as glycoproteins, glycosaminoglycans and glycolipids. Sulfate excreted by the kidneys derived mostly from oxidation of sulfur-containing amino acids [83].
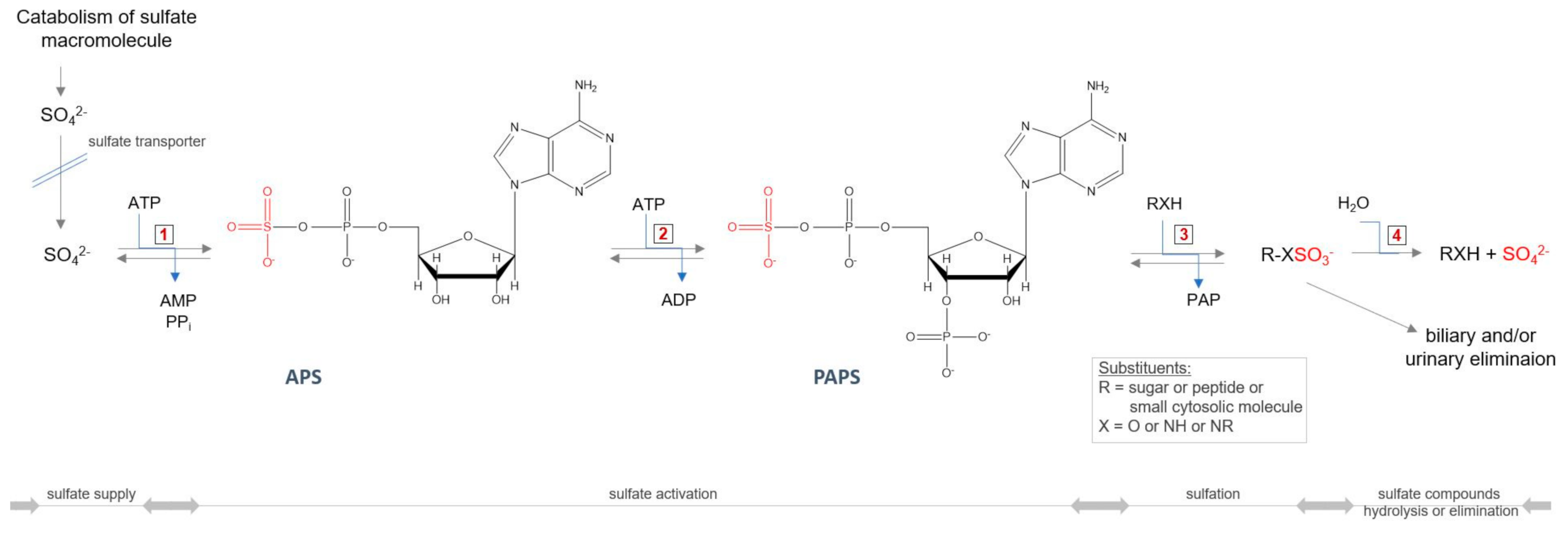
All GAGs (except HA), during the synthesis process, require an additional modification step—sulfation—taking place in the Golgi apparatus. The incorporation of this group into the molecule requires the active sulfate donor compound 3’-phosphoadenosine-5’-phosphosulfate (PAPS) (Figure 5). The level of PAPS is tissue-dependent, and in humans is in the range 3.6–22.6 nmol/g tissue [87,88]. The intracellular PAPS availability may itself be under at least three multiple levels of control. The first control point is in the biosynthetic pathway of PAPS. The availability of PAPS for sulfation of GAGs significantly affects the rate of production of sulfated GAGs [89]. Thus, a defective conversion of 3′-adenosine phosphosulfate to PAPS, resulting in reduced levels of intracellular PAPS, leads to a synthesis of undersulfated proteoglycans in brachymorphic mice [90]. Another possible control level may be on the PAPS transport pathway to the Golgi apparatus, where sulfation occurs. A further factor that may limit the synthesis of PAPS is intracellular level of SO42−—the third option. The Michaelis constant (Km) for SO42− at saturating MgATP in PAPS synthesis reaction is relatively high and equals 0.29 mM [91,92], and is on the order of magnitude of extracellular inorganic sulfate concentration, which is <100 µM [93] (the plasma concentration of inorganic sulfate is higher and maintained at levels 0.3–0.5 mM [94]). Thus, it is possible that PAPS synthesis is limited by the magnitude of the intracellular SO42− pool, and factors affecting the size of this pool may influence the PAPS levels and ultimately sulfation [90].
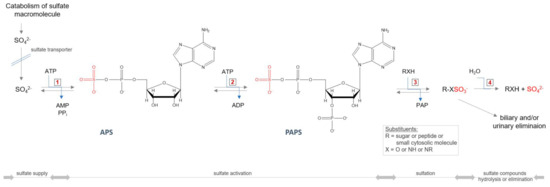
Figure 5.
Cellular sulfate reuse pathway. Abbreviations: ATP—adenosine triphosphate; ADP—adenosine diphosphate; AMP—adenosine monophosphate; PPi—pyrophosphate; APS—adenosine−5′−phosphosulfate; PAPS—3′−phosphoadenosine−5′−phosphosulfate, PAP—3′−phosphoadenosine−5′−monophosphate; 1—ATP−sulfurylase (EC 2.7.7.4); 2—APS kinase (EC 2.7.1.25); 3—sulfotransferase (EC 2.8.2.-); 4—sulfatase (3.1.6.-).
The intracellular sulfate pool size depends on extracellular uptake through membrane transporters, catabolism of sulfur-containing amino acids and other thiols with the formation of sulfides that are then oxidized to sulfates, and degradation of sulfated molecules/macromolecules by sulfatases [95]. Although sulfate transporters are widely distributed in organs, various tissues may utilize specific compounds/groups of compounds as a major source of sulfate (e.g., lung cells utilize cysteine, kidney cells utilize cysteine and glutathione) [96]. All of the sulfate sources listed above work together to maintain the intracellular sulfate pool, but with different prevalence depending on the cell types [88]. Any disruption of cellular sulfate levels leads to a shift in the equilibrium state position of the reactions responsible for maintaining physiological levels of SO42−, and indirectly, affects all reactions associated with these processes, as well as cellular sulfur metabolism in general (Figure 6). MPS IIIB is an example of a disorder in which there is an alteration in the amount of SO42− transferred from lysosomes to the cytoplasm after catabolism of HS macromolecules, as a result of some sulfate groups trapping in undegraded HS fragments deposited in lysosomes. It is known that undegraded GAGs are powerful inhibitors of GAGs sulfatases (Ki of HS is ≈50 nM) because the affinity of GAGs sulfatases for oligomers fragments of HS chain are usually higher than for the monomeric fragments [18].
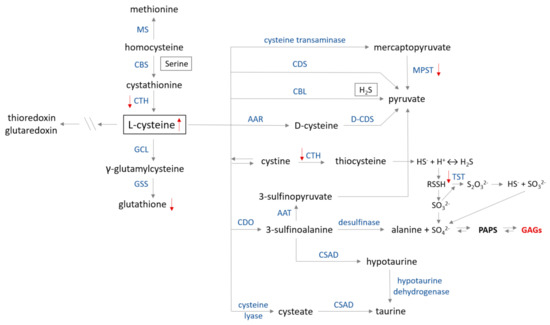
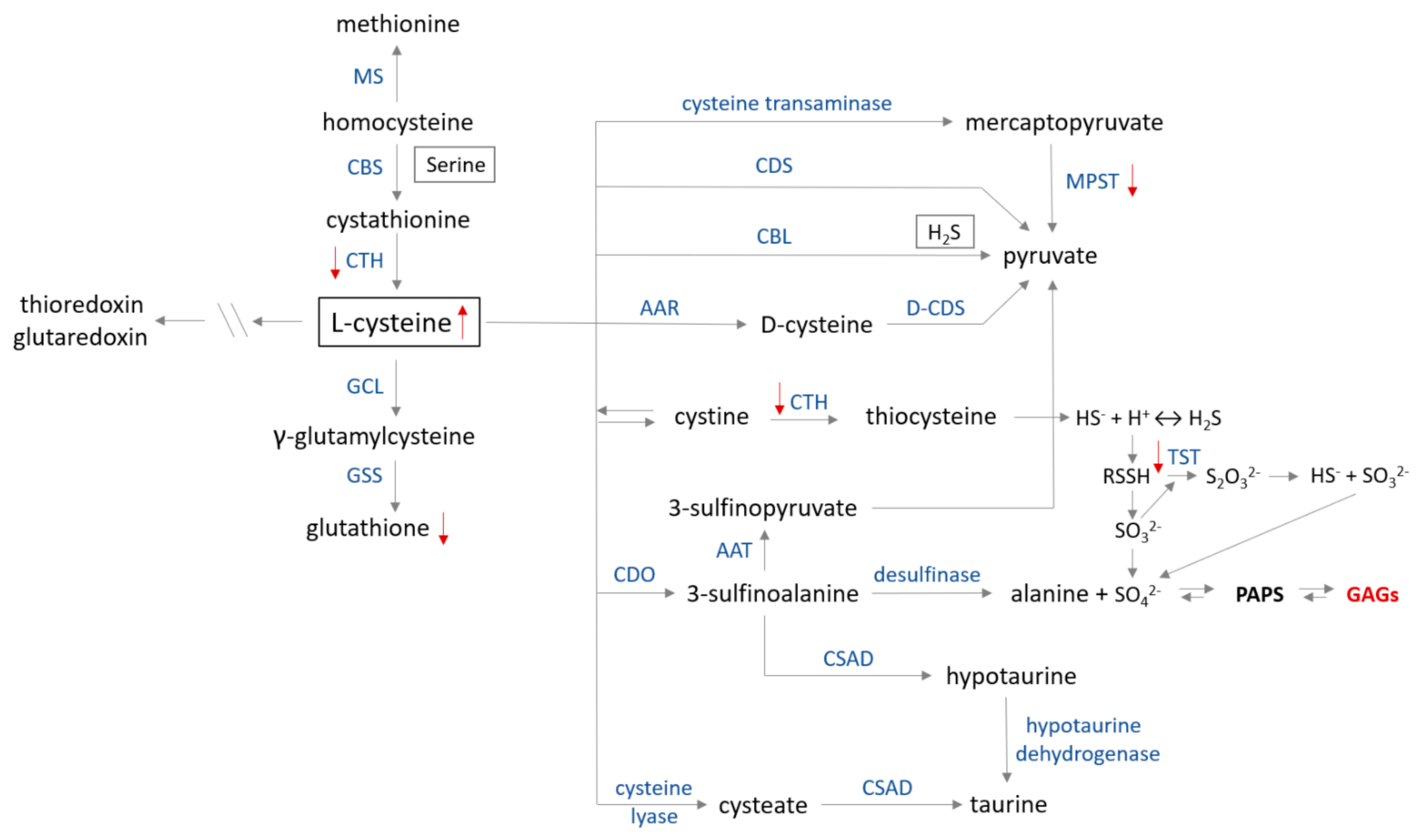
Figure 6.
Cysteine metabolism map. The catabolism of cysteine involves seven pathways producing pyruvate, hydrogen sulfide, alanine or taurine. (1) In the first step, cysteine is converted to mercaptopyruvate in deamination reaction catalyzed by cysteine transaminase (CGT, EC 2.6.1.3). Subsequently, mercaptopyruvate is changed to pyruvate on the trans-sulfuration way by 3−mercaptopyruvate sulfurtransferase (MPST, EC 2.8.1.2)—the ‘mercaptopyruvate pathway’. Cysteine in reactions catalyzed by (2) cysteine desulfhydrase (CDS, EC 4.4.1.15) and (3) cystathionine β−lyase (CBL, EC 4.4.1.8) is directly converted to pyruvate and H2S (only CBL). (4) D−cysteine is formed from L−cysteine by the action of amino acid racemase (AAR, EC 5.1.1.10). (5) Cysteine is in dynamic equilibrium with cystine, which can be converted to thiocysteine in a reaction catalyzed by cystathionine γ−lyase (CTH, EC 4.4.1.1) and, in the following step, into H2S or persulfide. Conversion of persulfide into thiosulfate is catalyzed by thiosulfate sulfurtransferase (TST, EC 2.8.1.1) in the ‘thiosulfate cycle’. (6) Cysteine is transformed to 3−sulfinoalanine by cysteine dioxygenase (CDO, EC 1.13.11.20) via oxidation of a sulfhydryl group. Then, 3−sulfinoalanine can be converted to: (a) 3−sulfinopyruvate in a reaction catalyzed by aspartate transaminase (AAT, EC 2.6.1.1), which is further transformed to pyruvate; (b) to alanine by desulfinase (EC 4.1.1.12) in the ‘alanine pathway’; and (c) to taurine by sulfinoalanine decarboxylase (CSAD, EC 4.1.1.29) and hypotaurine dehydrogenase (EC 1.8.1.3) in the ‘taurine pathway’. (7) Cysteine can also be transformed to cysteate in reaction catalyzed by cysteine lyase (EC 4.4.1.10) and then to taurine by action of CSAD. Glutamate cystine ligase (GCL, EC 6.3.2.2) and glutathione synthase (GSS, EC 6.3.2.3) catalyze glutathione synthesis from cysteine. Further metabolism between methionine and cysteine is regulated by CTH, cystathionine β−synthase (CBS, EC 4.2.1.22) and methionine synthase (MS, EC 2.1.1.13). Red arrows show changes in sulfur metabolism caused by the GAGs accumulation (modified according to [97]).
However, there are no scientific data on intracellular sulfate levels determined in individuals with MPS IIIB. Based on the information we have, it could be argued that the overall pool of sulfate derived from GAGs degradation in lysosomes of cells with MPS IIIB may be lower compared to the same cell type but without the genetic defect causing the disease, because partially undegraded HS fragments are trapped in lysosomes. On the other hand, the total amount of sulfate could remain unchanged, due to existence of various sources of inorganic sulfate in cells. Any perturbation related to the SO42− levels can be compensated for to some extent. The idea that sulfur metabolism may be one of the mechanisms used by cells to maintain/restore constant sulfate levels is supported by the fact that in the selected tissues (liver, kidney, heart, spleen) of MPS IIIB individuals, a positive correlation between increased cellular GAGs levels and changes of some parameters characteristic for sulfur metabolism was observed (Figure 6, modified according to [97]) [10]. The third option, where the sulfate levels increase in cells with MPS IIIB, seems the least likely. Nevertheless, it cannot be completely ruled out because in a healthy cell there is a certain pool of compounds that need to be sulfated for the cell to function properly. Therefore, three different classes of sulfotransferases can be distinguished in the eukaryotic cell: lysosomal sulfotransferases, which participate in catabolism of sulfated macromolecules; cytosolic sulfotransferases, that are responsible for sulfation of small molecules such as hormones, amines, drugs and xenobiotics; and Golgi-associated sulfotransferases that sulfate larger compounds involved in the secretory pathway [88,98]. It seems that the fastest way to raise the sulfate concentration in the cell, when sulfate synthesis is deficient, is to transport SO42− from the extracellular space. Furthermore, cellular uptake of exogenous labeled HS by cells with MPS IIIB has been documented in the literature [11]. This is an interesting observation, showing that despite the accumulation of HS resulting from a mutation in the NAGLU gene, the cell takes up HS (and/or its derivatives) from the environment and localizes it both in the cytoplasm and in lysosomes [11]. Directing the absorbed HS into lysosomes is intended to completely degrade the molecule, resulting in the release of the SO42− groups it contains and replenishing the sulfate pool present in the cytoplasm. However, little is yet known about how sulfate levels affect total sulfur turnover in cells with MPS IIIB.
7. Effects of GAGs/HS Accumulation on Sulfur Metabolism
It was only in 2019 that information regarding changes in sulfur metabolism in MPS IIIB sufferers appeared for the first time [10,11]. As was demonstrated in both MPS IIIB mouse tissues (liver, spleen, heart, kidney) [10] and in the cellular model of MPS IIIB (the Naglu−/− cells) [11], the cysteine level was elevated in the majority of studied cases (cysteine catabolism routes are presented in Figure 6). There may be many reasons for this condition because cysteine is a “metabolic hub” for biosynthesis of other S-containing small molecules (glutathione, coenzyme A and others) in cells; participates in many processes such as the synthesis of cysteinyl-tRNA required for protein synthesis, thiol-modified tRNA and is also a sulfur donor for the synthesis of Fe-S cluster cofactors or molybdenum cofactors [84,97]. However, it can be stated that, in general, the cysteine pool in the cells is increased under oxidizing conditions [99,100,101], but the research to-date does not demonstrate signs of oxidative stress in the Naglu−/− cells [11] or in the majority of examined tissues [10]. In the Naglu−/− cells, the ratio of reduced to oxidized glutathione concentration ([GSH]/[GSSG])—a major redox couple that determines the antioxidation capacity of the cells—is maintained, but the concentration values of both the reduced and oxidized forms of glutathione are significantly decreased (Figure 6) compared to the wild-type line [11]. Therefore, it may be assumed that cysteine (a substrate for the glutathione synthesis) is not used to synthesize glutathione molecules because the antioxidant potential of the Naglu−/− cells was significantly reduced [11]. This naturally raises the question of what happens to the excess cysteine in the cells and what is the direct cause.
Based on the scientific data [97,102] it is known that excess of cysteine inhibits its metabolism by the transamination pathway (Figure 6). In normal conditions, cysteine overabundance is oxidized by cysteine dioxygenase and further transformed into taurine and sulfate, as well as is metabolized by desulfhydration pathways, resulting in hydrogen sulfide (H2S) excess and thiosulfate (S2O32−) production (Figure 6) [97]. Unfortunately, no data are available for the parameters mentioned above measured in the population suffering from MPS. Therefore, based on the information we have [10,11], it can only be speculated that cysteine is diverted to the desulfuration pathway leading to the formation of compounds containing the so-called sulfane sulfur (a highly reactive, reduced sulfur atom at 0 or −1 oxidation state). However, preliminary research demonstrated [11] that the level of sulfane sulfur-containing compounds present in the Naglu−/− cells was reduced as compared to the wild type cells. A similar observation was made in the liver, spleen and kidney of animals with MPS IIIB [10]. At the moment, these results are inconclusive, and it is not known whether the sulfane sulfur-containing compound levels in the MPS IIIB cells are reduced or whether there were simply fewer of these compounds there (which could also partly explain the low antioxidant potential of the GAGs-overloaded cells).
The next significant observation reported in both the MPS IIIB models by Kaczor-Kamińska and others [10,11] was a consistent reduction of the 3-mercaptopyruvate sulfurtransferase (MPST, EC 2.8.1.2, Figure 6) activity. It is known [101], that MPST activity is regulated by redox change. The activity is inhibited by oxidation resulting in the suppression of cysteine degradation, which may explain the observed elevated level of cysteine in the Naglu−/− cells and tissues. MPST activity is regulated at the post-transcriptional level via redox-sensing molecular switches [101]. These switches can be classified into three subtypes: thioredoxin- glutathione-and glutaredoxin-specific, depending on reducing agents, demonstrating differences in their redox potential. If any data on these parameters were examined in patients with MPS IIIB, we might get closer to answering the question concerning possible causes of reduced MPST activity. Moreover, this could also provide information about redistribution of excess cysteine to other pathways, including the production of glutathione. Based on the available data, we know [11], that the level of glutathione in the Naglu−/− cells is reduced. However, at the moment, it is unknown whether the reason for this is the inhibition/slowdown of glutathione production or the intensification of processes in the cell leading to the consumption of this low molecular weight antioxidant. MPST contains cysteine residues at the catalytic site [103]. Every single oxidative modification of the enzyme of this type changes or inhibits its activity; therefore, efficiently functioning cellular antioxidant defense systems are important. The formation of disulfide protects them against irreversible oxidation, as S-S bonds can be reduced by the thioredoxin/thioredoxin reductase (EC 1.8.1.9) system or the glutathione/glutharedoxin/glutathione reductase (EC 1.8.1.7) systems. Thus, MPST, as well as other two sulfurtransferases—thiosulfate sulfurtransferase (TST, also known as rhodanese, EC 2.8.1.1, Figure 6) and cystathionine γ-lyase (CTH, EC 4.4.1.1, Figure 6), partly contribute to maintaining the redox homeostasis and serves as an antioxidant protein [101,103,104,105,106]. Their action helps to maintain homeostasis of the sulfane sulfur-containing compounds levels in cells. Thus, expanded research is needed to explain why, despite the increased level of cysteine and decreased level of glutathione, there is a depletion of sulfurtransferases activity in the Naglu−/− cells (MPST, TST, CTH) [11] and MPS IIIB tissues (MPST—kidney, spleen; TST—spleen and CTH—liver) [10].
It should also be mentioned that MPST and TST are regarded as sulfur-delivering enzymes for Fe-S cluster formation (the components of e.g., proteins mediating in electron transport processes). Therefore, their lower expression and/or activity (in the both tested MPS IIIB models, expression of MPST and TST remained at the control level, there was only one exception—the male Naglu−/− kidney, where the MPST expression was significantly higher as compared to the control group [10,11]) can result in an increase of cysteine level, a decrease of sulfur available for regeneration/formation of Fe-S clusters and a decrease of ATP production/level. As previously mentioned, ATP is required for proper activity of PAPS (Figure 5). Thus, a decrease in ATP availability may result in inhibition/slowdown of PAPS formation causing a consequent reduction in sulfated compound levels in which synthesis is indirectly dependent on ATP, and an increase in the cellular sulfate level itself. As demonstrated, little is known about the GAGs-induced changes affecting sulfur metabolism in individuals with MPS IIIB (with MPS in general).
8. Perspectives
This paper aims to draw the attention of the scientific community to a research problem that has not been sufficiently explored to date. To this purpose, we collected available data on documented changes occurring in sulfur metabolism in MPS IIIB patients and attempted to identify some causal correlations and indicate directions that should be explored. The observations presented in the paper are new and can significantly contribute to the expansion of knowledge related to MPS IIIB. They may point to new directions of research or therapeutic approaches, which, we hope, in the future will, if not cure, improve the comfort of patients affected by this disease.
Author Contributions
Conceptualization, M.K.-K.; writing—original draft preparation, M.K.-K. and K.K.; writing—review and editing, M.K.-K., K.K. and M.W.; visualization, M.K.-K.; supervision, M.K.-K., K.K. and M.W.; project administration, M.K.-K.; funding acquisition, M.K.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Polish National Science Center (grant number: UMO-2017/01/X/NZ3/00346).
Acknowledgments
M.K.-K. thanks to Kinga Kaszuba (Jagiellonian University, Collegium Medium, Cracow) for help in editing English version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Regulation (EC) No 141/2000 of the European Parliament and of the Council of 16 December 1999 on Orphan Medicinal Products; The Official Journal of the European Union; European Union: Maastricht, The Netherlands, 2000; pp. 1–5.
- Gaffke, L.; Pierzynowska, K.; Piotrowska, E.; Węgrzyn, G. How close are we to therapies for Sanfilippo disease? Metab. Brain Dis. 2018, 33, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, H.; Dirosario, J.; Killedar, S.; Zarapse, K.; McCarty, D.M. Correction of neurological disease of mucopolysaccharidosis IIIB in adult mice by rAAV9 trans-blood-brain barrier gene delivery. Mol. Ther. 2011, 19, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.H.; Aoyagai-Scharber, M.; Le, S.Q.; Vincelette, J.; Ohmi, K.; Bullens, S.; Wendt, D.J.; Christianson, T.M.; Tiger, P.M.N.; Brown, J.R.; et al. Delivery of an enzyme-IGFII fusion protein to the mouse brain is therapeutic for mucopolysaccharidosis type IIIB. Proc. Natl. Acad. Sci. USA 2014, 111, 14870–14875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kan, S.H.; Troitskaya, L.A.; Sinow, C.S.; Haitz, K.; Todd, A.K.; Di Stefano, A.; Le, S.Q.; Dickson, P.I.; Tippin, B.L. Insulin-like growth factor II peptide fusion enables uptake and lysosomal delivery of α-N-acetylglucosaminidase to mucopolysaccharidosis type IIIB fibroblasts. Biochem. J. 2014, 458, 281–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrey, D.A.; Naughton, B.J.; Duncan, F.J.; Meadows, A.S.; Ware, T.A.; Campbell, K.J.; Bremer, W.G.; Walker, C.M.; Goodchild, L.; Bolon, B.; et al. Feasibility and safety of systemic rAAV9-hNAGLU delivery for treating mucopolysaccharidosis IIIB: Toxicology, biodistribution, and immunological assessments in primates. Hum. Gene Ther. Clin. Dev. 2014, 25, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Marcó, S.; Haurigot, V.; Bosch, F. In vivo gene therapy for mucopolysaccharidosis type III (Sanfilippo syndrome): A new treatment horizon). Hum. Gene Ther. 2019, 30, 1211–1221. [Google Scholar] [CrossRef]
- Kong, W.; Yao, Y.; Zhang, J.; Lu, C.; Ding, Y.; Meng, Y. Update of treatment for mucopolysaccharidosis type III (Sanfilippo syndrome). Eur. J. Pharmacol. 2020, 888, 173562. [Google Scholar] [CrossRef]
- Tambuyzer, E.; Vandendriessche, B.; Austin, C.A.; Brooks, P.J.; Larsson, K.; Miller Needleman, K.I.; Valentine, J.; Davies, K.; Groft, S.C.; Preti, R.; et al. Therapies for rare diseases: Therapieutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 2020, 19, 93–111. [Google Scholar] [CrossRef]
- Kaczor-Kamińska, M.; Kamiński, K.; Staińska, K.; Wróbel, M.; Feldman, A. Effect of glycosaminoglycans accumulation on the non-oxidative sulfur metabolism in mouse model of Sanfilippo syndrome, type B. Acta Biochim. Pol. 2019, 66, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Kaczor-Kamińska, M.; Stalińska, K.; Kamiński, K.; Pisarek, A.; Maziarz, U.; Feldman, A.; Wróbel, M. Murine cellular model of mucopolysaccharidosis, type IIIb (MPS (IIIB)—A preliminary study with particular emphasis on the non-oxidative L-cysteine metabolism. Biochimie 2020, 174, 84–94. [Google Scholar] [CrossRef]
- Breen, M.; Weindtein, H.G.; Johnson, R.L.; Veis, A.; Marshall, T.R. Acidic glycosaminoglycans in human skin during fatal development and adult life. Biochim. Biophys. Acta 1970, 201, 54–60. [Google Scholar] [CrossRef]
- Lee, T.Y.; Jamieson, A.; Schafer, I. Changes in the composition and structure of glycosaminoglycans in the human placenta during development. Pediatr. Res. 1973, 7, 965–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Praus, R.; Brettschneider, I. Glycosaminoglycans in embryonic and postnatal human cornea. Ophthalmic Res. 1975, 7, 452–458. [Google Scholar] [CrossRef]
- Pepi, L.E.; Sasiene, Z.J.; Mendis, P.M.; Jackson, G.P.; Amster, J.I. Structural characterization of sulfated glycosaminoglycans using charge-transfer dissociation. J. Am. Soc. Mass. Spectrom. 2020, 31, 10. [Google Scholar] [CrossRef]
- Zaia, J.; Costello, C.E. Compositional Analysis of glycosaminoglycans by electrospray mass spectrometry. Anal. Chem. 2001, 73, 233–239. [Google Scholar] [CrossRef]
- Staples, G.O.; Zaia, J. Analysis of glycosaminoglycans using mass spectrometry. Curr. Proteomics 2011, 8, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Huxtable, R.J. Biochemistry of Sulfur; Frieden, E., Ed.; Plenum Press: New York, NY, USA, 1986. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, U.; Couchman, J.; Kimata, K.; Esko, J.D. Proteoglycans and sulfated glycosaminoglycans. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar] [CrossRef]
- Laurent, T.C.; Laurent, U.B.; Fraser, J.R. The structure and function of hyaluronan: An overview. Immunol. Cell Biol. 1996, 74, A1–A7. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Mizumoto, S.; Fujita, M. Novel insight into glycosaminoglycan biosynthesis based on gene expression profiles. Front. Cell Dev. Biol. 2021, 9, 709018. [Google Scholar] [CrossRef]
- Mattson, J.M.; Turcotte, R.; Zhang, Y. Glycosaminoglycans contribute to extracellular matrix fiber recruitment and arterial wall mechanics. Biomech. Model. Mechanobiol. 2017, 16, 213–225. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.C.; Carvalho, M.S.; Han, X.; Xia, K.; Mikael, P.E.; Cabral, J.M.S.; Ferreira, F.C.; Linhardt, R.J. Compositional and structural analysis of glycosaminoglycans in cell-derived extracellular matrices. Glycoconj. J. 2019, 36, 141–154. [Google Scholar] [CrossRef]
- Puri, S.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Coulson-Thomas, V.J. Distribution and function of glycosaminoglycans and proteoglycans in the development, homeostasis and pathology of the ocular surface. Front. Cell Dev. Biol. 2020, 8, 731. [Google Scholar] [CrossRef] [PubMed]
- Rienks, M.; Papageorgiou, A.-P.; Frangogiannis, N.G.; Heymans, S. Myocardial extracellular matrix. Circ. Res. 2014, 114, 872–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricard-Blum, S. Glycosaminoglycans: Major biological players. Glycoconj. J. 2017, 34, 275–276. [Google Scholar] [CrossRef] [Green Version]
- Köwitsch, A.; Zhou, G.; Growth, T. Medical application of glycosaminoglycans: A review. J. Tissue Regen. Med. 2018, 12, e23–e41. [Google Scholar] [CrossRef] [PubMed]
- Banachowicz, P.; Buda, S. Gram-scale carbasugar synthesis via intramolecular seleno-Michael/aldol reaction. RSC Adv. 2019, 9, 12928. [Google Scholar] [CrossRef] [Green Version]
- Łopatkiewicz, G.; Buda, S.; Młynarski, J. Application of the EF and GH fragments to the synthesis of idraparinux. J. Org. Chem. 2017, 82, 12701–12714. [Google Scholar] [CrossRef]
- Mende, M.; Bednarek, C.; Wawryszyn, M.; Sauter, P.; Biskup, M.B.; Schepers, U.; Bräse, S. Chemical synthesis of glycosaminoglycans. Chem. Rev. 2016, 116, 8193–8255. [Google Scholar] [CrossRef]
- Miura, Y.; Fukuda, T.; Seto, H.; Hoshino, Y. Development of glycosaminoglycan mimetrics using glycopolymers. Polym. J. 2016, 48, 229–237. [Google Scholar] [CrossRef]
- Kalaska, B.; Kamiński, K.; Miklosz, J.; Nakai, K.; Yusa, S.-I.; Pawlak, D.; Nowakowska, M.; Mogielnicki, A.; Szczubiałka, K. Anticoagulant properties of poly(sodium 2-(acrylamido)-2-methylpropanesulfonate)-based di- and triblock polymers. Biomacromolecules 2018, 17, 3104–3118. [Google Scholar] [CrossRef]
- Gray, E.; Hogwood, J.; Mulloy, B. The anticoagulant and antithrombotic mechanisms of heparin. In Heparin—A Century of Progress. Handbook of Experimental Pharmacology; Lever, R., Mulloy, B., Page, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 207. [Google Scholar] [CrossRef]
- Sharma, S.K. Low molecular weight heparins. Med. J. Armed Forces India 1998, 54, 285–286. [Google Scholar] [CrossRef] [Green Version]
- Hirsh, J.; Anand, S.S.; Halperin, J.L.; Fuster, V. Mechanism of action and pharmacology of unfractionated heparin. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1094–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelberg, H. Heparin and the removal of triglyceride from the blood stream. Am. J. Cardiol. 1964, 14, 8–17. [Google Scholar] [CrossRef]
- Capila., I.; Linhardt, R. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 2002, 41, 391–412. [Google Scholar] [CrossRef]
- Soares da Costa, D.; Reis, R.L.; Pushkuleva, I. Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu. Rev. Biomed. Eng. 2017, 19, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariga, T.; Miyatake, T.; Yu, R.K. Role of proteoglycans and glycosaminoglycans in the pathogenesis of Alzheimer’s disease and related disorders: Amyloidogenesis and therapeutic strategies—A review. J Neurosci. Res. 2010, 88, 2303–20315. [Google Scholar] [CrossRef]
- Jorpes, J.E.; Gardell, S. On heparin monosulfuric acid. J. Biol. Chem. 1948, 176, 267–275. [Google Scholar] [CrossRef]
- Kalaska, B.; Miklosz, J.; Kamiński, K.; Musielak, B.; Yusa, S.-I.; Pawlak, D.; Nowakowska, M.; Szczubiałka, K.; Mogielnicki, A. The neutralization of heparan sulfate by heparin-binding copolymer as a potential therapeutic target. RSC Adv. 2019, 9, 3020–3029. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, J.T. Heparan sulfate: Growth control with a restricted sequence menu. J. Clin. Investig. 2001, 108, 357–361. [Google Scholar] [CrossRef]
- Li, J.-P.; Kusche-Gullberg, M. Heparan sulfate: Biosynthesis, structure, and function. Int. Rev. Cell Mol. Biol. 2016, 325, 215–273. [Google Scholar] [CrossRef]
- Gill, V.L.; Aich, U.; Rao, S.; Pohl, C.; Zaia, J. Disaccharide analysis of glycosaminoglycans using hydrophilic interaction chromatography and mass spectrometry. Anal. Chem. 2013, 15, 1138–1145. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, A.; Malvi, P.; Wajapeyee, N. Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front. Endocrinol. 2018, 9, 483. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Reis, C.A.; Vivès, R.R.; Magalhães, A. Heparan sulfate biosynthesis and sulfation profiles as modulators of cancer signalling and progression. Front. Oncol. 2021, 11, 778752. [Google Scholar] [CrossRef] [PubMed]
- Tekotte, H.; Engel, M.; Margolis, R.U.; Margolis, R.K. Disaccharide composition of heparan sulfates: Brain, nervous tissue storage organelles, kidney and lung. J. Neurochem. 1994, 62, 1126–1130. [Google Scholar] [CrossRef]
- Gordon, P.B.; Choi, H.U.; Conn, G.; Ahmed, A.; Ehrmann, B.; Rosenberg, L.; Hatcher, V.B. Extracellular matrix heparan sulfate proteoglycans modulate the mitogenic capacity of acidic fibroblast growth factor. J. Cell. Physiol. 1989, 140, 584–592. [Google Scholar] [CrossRef]
- Lindahl, U.; Kjellén, L. Pathophysiology of heparan sulfate: Many diseases, few drugs. J. Intern. Med. 2013, 273, 555–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoler-Barak, L.; Moussion, C.; Shezen, E.; Hatzav, M.; Sixt, M.; Alon, R. Blood vessels pattern heparan sulfate gradients between their apical and basolateral aspects. PLoS ONE 2014, 9, e85699. [Google Scholar] [CrossRef] [PubMed]
- Lindhal, B.; Lindhal, U. Amyloid-specific heparan sulfate from human liver and spleen. J. Biol. Chem. 1997, 272, 26091–26094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindhal, B.; Eriksson, L.; Lindhal, U. Structure of heparan sulfate from human brain, with special regard to Alzheimer’s disease. Biochem. J. 1995, 306, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Bernfield, M.; Götte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef]
- Masri, R.E.; Crétinon, Y.; Gout, E.; Vivès, R.R. HS and inflammation: A potential playground for the Sulfs? Front. Immunol. 2020, 11, 570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stringer, S.E.; Gallagher, J.T. Specific binding of the chemokine platelet factor 4 to heparan sulfate. J. Biol. Chem. 1997, 272, 20508–20514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Maguire, T.; Hileman, R.E.; Fromm, J.R.; Esko, J.D.; Linhardt, R.J.; Marks, R.M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 1997, 3, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Scarpellini, A.; Germack, R.; Lortat-Jacob, H.; Muramatsu, T.; Billett, E.; Johnson, T.; Verderio, E.A. Heparan sulfate proteoglycans are receptors for the cell-surface trafficking and biological activity of teansglutaminase-2. J. Biol. Chem. 2009, 284, 18411–18423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, A.D.; Selleck, S.B. The elusive functions of proteoglycans: In vivo veritas. J. Cell. Biol. 2000, 148, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Perrimon, N.; Bernfield, M. Specificities of heparan sulfate proteoglycans in development processes. Nature 2000, 404, 725–728. [Google Scholar] [CrossRef]
- Selleck, S.B. Proteoglycans and pattern formation: Sugar biochemistry meets development genetics. Trends Genet. 2000, 16, 206–212. [Google Scholar] [CrossRef]
- Onyeisi, J.O.S.; Ferreira, B.Z.F.; Nader, H.B.; Lopes, C.C. Heparan sulfate proteoglycans as targets for cancer therapy: A review. Cancer Biol. Ther. 2020, 21, 1087–1094. [Google Scholar] [CrossRef]
- Kresse, H.; Cantz, M.; von Figura, K.; Glössl, J.; Paschke, E. The Mucopolysaccharidoses: Biochemistry and clinical symptoms. Klin. Wochenschr. 1981, 59, 867–876. [Google Scholar] [CrossRef]
- Griffin, L.S.; Gloster, T.M. The enzymatic degradation of heparan sulfate. Protein Pept. Lett. 2017, 24, 710–722. [Google Scholar] [CrossRef]
- Bame, K.J. Heparanases: Endoglycosidases that degrade heparan sulfate proteoglycans. Glycobiology 2001, 11, 91R–98R. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valstar, M.J.; Ruijter, G.J.; van Diggelen, O.P.; Poorthuis, B.J.; Wijburg, F.A. Sanfilippo syndrome: A mini-review. J. Inherit. Metab. Dis. 2008, 31, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Kloska, A.; Tylki-Szymańska, A.; Węgrzyn, G. Mucopolysaccharidosis—Biochemical mechanisms of diseases and therapeutic possibilities. Postępy Biochem. 2011, 57, 133–147, (Article in Polish). [Google Scholar] [PubMed]
- Birrane, G.; Dassier, A.-L.; Romashko, A.; Lundberg, D.; Holmes, K.; Cottle, T.; Norton, A.W.; Zhang, B.; Concino, M.F.; Meiyappan, M. Structural characterization of the α-N-acetylglucosaminidase, a key enzyme in the pathogenesis of Sanfilippo syndrome B. J. Struct. Biol. 2019, 205, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Nidiffer, F.D.; Kelly, T.E. Developmental and degenerative patterns associated with cognitive, behavioral and motor difficulties in the Sanfilippo syndrome: An epidemiological study. J. Ment. Defic. Res. 1983, 27, 185–203. [Google Scholar] [CrossRef]
- Wraith, J.E.; Danks, D.M.; Rogers, J.G. Mild Sanfilippo syndrome: A further cause of hyperactivity and behavioral disturbance. Med. J. Aust. 1987, 147, 450–451. [Google Scholar] [CrossRef]
- Bax, M.C.; Colville, G.A. Behavior in mucopolysaccharide disorders. Arch. Dis. Child. 1995, 73, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Valstar, M.J.; Bruggenwirth, H.T.; Olmer, R.; Wevers, R.A.; Verheijen, F.W.; Poorthuis, B.J.; Halley, D.J.; Wijburg, F.A. Mucopolysaccharidosis type IIIB may predominantly present with an attenuated clinical phenotype. J. Inherit. Metab. Dis. 2010, 33, 759–767. [Google Scholar] [CrossRef] [Green Version]
- Malm, G.; Månsson, J.-E. Mucopolysaccharidosis type III (Sanfilippo disease) in Sweden: Clinical presentation of 322 children diagnosed during a 30-year period. Acta Paediatr. 2010, 99, 1253–1257. [Google Scholar] [CrossRef]
- White, K.K.; Karol, L.A.; White, D.R.; Hale, S. Musculoskeletal manifestations of Sanfilippo Syndrome (mucopolysaccharidosis type III). J. Pediatr. Orthop. 2011, 31, 594. [Google Scholar] [CrossRef]
- Van Schrojenstein-de Valk, H.M.; van de Kamp, J.J. Follow-up on seven adults patients with mild Sanfilippo B-disease. Am. J. Med. Genet. 1987, 28, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, W.M.A.; Csepán, R.; Marcelis, C.L.M.; Lefeber, D.J.; Egger, J.I.M.; Tuinier, S. Sanfilippo B in an elderly female psychiatric patient: A rare but relevant diagnosis in presenile dementia. Acta Psychiatr. Scand. 2009, 122, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Walkey, S.U. Secondary accumulation of gangliosides in lysosomal storage disorder. Semin. Cell Dev. Biol. 2004, 15, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.; Gargiulo, N.; Faraonio, R.; Castalodo, S.; Gonzales, Y.; Reyero, E.; Di Natale, P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J. Neurosci. Res. 2007, 85, 612–622. [Google Scholar] [CrossRef]
- Baydakova, G.; Iiyushkina, A.; Gaffke, L.; Pierzynowska, K.; Bychov, I.; Ługowska, A.; Węgrzyn, G.; Tylki-Szymanska, A.; Zakharova, E. Elevated LysoGb3 concentration in the neuronopathic forms of mucopolysaccharidoses. Diagnostics 2020, 10, 155. [Google Scholar] [CrossRef] [Green Version]
- Walkey, S.U.; Vanier, M.T. Pathomechanisms in lysosomal storage disorders. Biochim. Biophys. Acta 2009, 1793, 726–736. [Google Scholar] [CrossRef] [Green Version]
- Pearse, Y.; Iacovino, M. A cure for Sanfilippo syndrome? A summary of current therapeutic approaches and their promise. Med. Res. Arch. 2020, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Meikle, P.J.; Fuller, M.; Hopwood, J.J. Lysosomal Degradation of Heparin and Heparan Sulfate. In Chemistry and Biology of Heparin and Heparan Sulfate; Garg, H.G., Linhardt, R.J., Hales, C.A., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2005; pp. 285–311. [Google Scholar] [CrossRef]
- Stromberg, P.E.; Cumpstion, K.L. Sulfates. In Encyclopedia of Toxicology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 413–415. [Google Scholar] [CrossRef]
- Miller, C.G.; Schmidt, E.E. Sulfur metabolism under stress. Antioxid. Redox Sign. 2020, 33, 1158–1173. [Google Scholar] [CrossRef]
- Jonas, A.J.; Jobe, H. Sulfate transport by rat liver lysosomes. J. Biol. Chem. 1990, 265, 17545–17549. [Google Scholar] [CrossRef]
- Chou, H.F.; Passage, M.; Jonas, A.J. ATP stimulates lysosomal sulphate transport at neutral pH: Evidence for phosphorylation of the lysosomal sulphate carrier. Biochem. J. 1997, 327, 781–786. [Google Scholar] [CrossRef] [Green Version]
- Klaasen, C.D.; Boles, J.W. Sulfation and sulfotransferases 5: The importance of 3’-phosphoadenosine-5’-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997, 11, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Paganini, C.; Tota, C.G.; Superti-Furga, A.; Rossi, A. Skeletal dysplasias caused by sulfation defects. Int. J. Mol. Sci. 2020, 21, 2710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghiselli, G. Drug-mediated regulation of glycosaminoglycan biosynthesis. Med. Res. Rev. 2017, 37, 1051–1094. [Google Scholar] [CrossRef] [PubMed]
- Elgavish, A.; Meezan, E. Sulfation by human lung fibroblasts: SO4(2−) and sulfur-containing amino acids as a source for macromolecular sulfation. Am. J. Physiol.-Lung Cell. Mol. Physiol. 1991, 260, L450–L456. [Google Scholar] [CrossRef]
- Baranczyk-Kuzma, A.; Borchardt, R.T.; Schasteen, C.S.; Pinnick, C.L. Phenolsulfotransferase: Purification and characterization of the rat brain enzyme. Psychopharmacol. Bull. 1981, 17, 50–51. [Google Scholar]
- Hanna, E.; Ng, K.F.; MacRae, I.J.; Bley, C.J.; Fisher, A.J.; Segel, I.H. Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory domain. J. Bio. Chem. 2004, 279, 4415–4424. [Google Scholar] [CrossRef] [Green Version]
- Markovich, D. Physiological roles and regulation of mammalian sulfate transporters. Physiol. Rev. 2001, 81, 1499–1533. [Google Scholar] [CrossRef]
- Priyamvada, S.; Saksena, S.; Alrefai, W.A.; Dudeja, P.K. Intestinal anion absorption. In Physiology of the Gastrointestinal Track, 6th ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 1317–1362. [Google Scholar] [CrossRef]
- Langford, R.; Hurrion, E.; Dawson, P.A. Genetics and pathophysiology of mammalian sulfate biology. J. Genet. Genomics 2017, 44, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Dawson, J.R.; Norbeck, K.; Moldeus, P. The effectiveness of different sulfate precursors in supporting extrahepatic sulfate conjugation. Biochem. Pharm. 1983, 32, 1789–1791. [Google Scholar] [CrossRef]
- Stipanuk, M.H. Metabolism of sulfur-containing amino acids: How the body copes with excess methionine, cysteine and sulfide. J. Nutr. 2020, 150, 2494S–2505S. [Google Scholar] [CrossRef]
- Strott, C.A. Sulfonation and molecular action. Endocr. Rev. 2002, 23, 703–732. [Google Scholar] [CrossRef] [PubMed]
- Mosharov, E.; Cranfold, M.R.; Banerjee, R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by a transsulfuration pathway and its regulation by redox changes. Biochemistry 2000, 39, 13005–13011. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, N.; Sawada, N. The mercaptopyruvate pathway in cysteine metabolism: A physiologic role and related disease of the multifunctional 3-mercaptopyruvate sulfurtransferase. Curr. Med. Chem. 2006, 13, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, N.; Nagano, M.; Ito, T.; Suzuki, H. Redox regulation of mammalian 3-mercaptopyruvate sulfurtransferase. Meth. Enzymol. 2015, 554, 229–254. [Google Scholar] [CrossRef]
- Stipanuk, M.H. Sulfur amino acid metabolism: Pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 2004, 24, 539–577. [Google Scholar] [CrossRef]
- Kaczor-Kamińska, M.; Kamiński, K.; Wróbel, M. The expression and activity of rhodanese, 3-mercaptopyruvate sulfurtransferase, cystathionine γ-lyase in the most frequently chosen cellular research models. Biomolecules 2021, 11, 1859. [Google Scholar] [CrossRef]
- Wróbel, M.; Jurkowska, H.; Kaczor, M.; Uchacz, T. Rodanese and 3-mercaptopyruvate sulfurtransferase—Evolutionary related enzymes. KOSMOS 2012, 61, 233–244. [Google Scholar]
- Jurkowska, H.; Kaczor-Kamińska, M.; Bronowicka-Adamska, B.; Wróbel, M. Cystathionine γ-lyase. Adv. Hyg. Exp. Med. 2014, 68, 1–9. [Google Scholar] [CrossRef]
- Kaczor-Kamińska, M.; Sura, P.; Wróbel, M. Multidirectional changes in parameters related to sulfur metabolism in frog tissues exposed to heavy metal-related stress. Biomolecules 2020, 14, 574. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).