Diabetic Retinopathy: Are lncRNAs New Molecular Players and Targets?
Abstract
1. Introduction
2. Long Non Coding RNAs with Increased Expression in Diabetic Retinopathy
3. Long Non-Coding RNAs with Reduced Expression in Diabetic Retinopathy
4. The Deregulation of Small Nucleolar RNA Host Genes in Diabetic Retinopathy
| LncRNA | Chr | DR-Related Processes | Sponged miRNAs | Related Genes/Proteins | Ref. |
|---|---|---|---|---|---|
| SNHG1 | 11q12.3 | Inflammation and apoptosis. | Vimentin, α-SMA, IL-6, IL-1β, E-cadherin, ZO-1 | [184] | |
| SNHG16 | 17q.25.1 | Proliferation, migration, angiogenesis, apoptosis, oxidative stressand vessel-like formation. | miR-195, miR-146a-5p, miR-7-5p, miR-20a-5p | IRAK1, IRS1, NF-kB, PI3K/AKT, E2F1, mfn2 | [185,186,187] |
| GAS5 (SNHG2) | 1q25.1 | Apoptosis, oxidative stress and inflammation. | BCL2, BAD, BACX, SERCA2b | [188,189] | |
| SNHG4 | 5q31.2 | Apoptosis. | miR-200b | OXR1 | [190] |
| SNHG5 | 6q14.3 | Cell proliferation and angiogenesis. | VEGFA | [191] | |
| SNHG7 | 9q34.3 | Proliferation, migration and angiogenesis. | miR-543, miR-34a-5p | SIRT1 | [192,193] |
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; Available online: https://www.diabetesatlas.org (accessed on 30 June 2022).
- Reddy, M.A.; Zhang, E.; Natarajan, R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 2014, 58, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Fowler, M.J. Microvascular and Macrovascular Complications of Diabetes. Clin. Diabetes 2011, 29, 116–122. [Google Scholar] [CrossRef]
- Yau, J.W.Y.; Rogers, S.L.; Kawasaki, R.; Lamoureux, E.L.; Kowalski, J.W.; Bek, T.; Chen, S.-J.; Dekker, J.M.; Fletcher, A.; Grauslund, J.; et al. Global Prevalence and Major Risk Factors of Diabetic Retinopathy. Diabetes Care 2012, 35, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Fortun, J.A. Diabetic Macular Edema: Current Understanding, Pharmacologic Treatment Options, and Developing Therapies. Asia-Pacific J. Ophthalmol. 2019, 7, 28–35. [Google Scholar] [CrossRef]
- Kusuhara, S.; Fukushima, Y.; Ogura, S.; Inoue, N.; Uemura, A. Pathophysiology of Diabetic Retinopathy: The Old and the New. Diabetes Metab. J. 2018, 42, 364–376. [Google Scholar] [CrossRef]
- Shi, G.-J.; Shi, G.R.; Zhou, J.-Y.; Zhang, W.-J.; Gao, C.-Y.; Jiang, Y.-P.; Zi, Z.-G.; Zhao, H.-H.; Yang, Y.; Yu, J.-Q. Involvement of growth factors in diabetes mellitus and its complications: A general review. Biomed. Pharmacother. 2018, 101, 510–527. [Google Scholar] [CrossRef]
- Wang, W.; Lo, A.C.Y. Diabetic Retinopathy: Pathophysiology and Treatments. Int. J. Mol. Sci. 2018, 19, 1816. [Google Scholar] [CrossRef]
- Zhong, Q.; Kowluru, R.A. Epigenetic Modification of Sod2 in the Development of Diabetic Retinopathy and in the Metabolic Memory: Role of Histone Methylation. Investig. Ophthalmol. Vis. Sci. 2013, 54, 244–250. [Google Scholar] [CrossRef]
- Zhong, Q.; Kowluru, R.A. Regulation of Matrix Metalloproteinase-9 by Epigenetic Modifications and the Development of Diabetic Retinopathy. Diabetes 2013, 62, 2559–2568. [Google Scholar] [CrossRef]
- Tewari, S.; Santos, J.M.; Kowluru, R.A. Damaged Mitochondrial DNA Replication System and the Development of Diabetic Retinopathy. Antioxid. Redox Signal. 2012, 17, 492–504. [Google Scholar] [CrossRef]
- Tewari, S.; Zhong, Q.; Santos, J.M.; Kowluru, R.A. Mitochondria DNA Replication and DNA Methylation in the Metabolic Memory Associated with Continued Progression of Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4881–4888. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhou, X.; Liu, D.; Yun, L.; Zhang, L.; Chen, X.; Chai, Q.; Li, L. MicroRNA-29 regulates high-glucose-induced apoptosis in human retinal pigment epithelial cells through PTEN. In Vitro Cell Dev. Biol. Anim. 2016, 52, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dong, Y.; Wang, Y.; Gao, J.; Lv, J.; Sun, J.; Li, M.; Wang, M.; Zhao, Z.; Wang, J.; et al. Long non-codingRNAs in ocular diseases: New and potential therapeutic targets. FEBS J. 2019, 286, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qu, X. The roles of long non-coding RNAs in ocular diseases. Exp. Eye Res. 2021, 207, 108561. [Google Scholar] [CrossRef]
- Milluzzo, A.; Maugeri, A.; Barchitta, M.; Sciacca, L.; Agodi, A. Epigenetic Mechanisms in Type 2 Diabetes Retinopathy: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 10502. [Google Scholar] [CrossRef]
- Thomas, A.A.; Biswas, S.; Feng, B.; Chen, S.; Gonder, J.; Chakrabarti, S. lncRNA H19 prevents endothelial–mesenchymal transition in diabetic retinopathy. Diabetologia 2019, 62, 517–530. [Google Scholar] [CrossRef]
- Chen, Q.; Meng, X.; Liao, Q.; Chen, M. Versatile interactions and bioinformatics analysis of noncoding RNAs. Brief. Bioinform. 2018, 20, 1781–1794. [Google Scholar] [CrossRef]
- Aprile, M.; Katopodi, V.; Leucci, E.; Costa, V. LncRNAs in Cancer: From garbage to Junk. Cancers 2020, 12, 3220. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef]
- Pang, Y.; Mao, C.; Liu, S. Encoding activities of non-coding RNAs. Theranostics 2018, 8, 2496–2507. [Google Scholar] [CrossRef]
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-Encoded Peptide: Functions and Predicting Methods. Front. Oncol. 2021, 10, 622294. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, N.; De Crescenzo, A.; Mishra, K.; Perone, L.; Carella, M.; Palumbo, O.; Mussa, A.; Sparago, A.; Cerrato, F.; Russo, S.; et al. The KCNQ1OT1 imprinting control region and non-coding RNA: New properties derived from the study of Beckwith–Wiedemann syndrome and Silver–Russell syndrome cases. Hum. Mol. Genet. 2012, 21, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Akhade, V.S.; Pal, D.; Kanduri, C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv. Exp. Med. Biol. 2017, 1008, 47–74. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, J.-W.; Brüning, J.C. Regulation of metabolism by long, non-coding RNAs. Front. Genet. 2014, 5, 57. [Google Scholar] [CrossRef]
- Nam, J.-W.; Choi, S.-W.; You, B.-H. Incredible RNA: Dual Functions of Coding and Noncoding. Mol. Cells 2016, 39, 367–374. [Google Scholar] [CrossRef]
- Morlando, M.; Fatica, A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int. J. Mol. Sci. 2018, 19, 570. [Google Scholar] [CrossRef]
- Available online: https://pubmed.ncbi.nlm.nih.gov/?term=lncrna+and+diabetic+retinopathy&sort=date (accessed on 30 March 2022).
- Yan, B.; Tao, Z.-F.; Li, X.-M.; Zhang, H.; Yao, J.; Jiang, Q. Aberrant Expression of Long Noncoding RNAs in Early Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 941–951. [Google Scholar] [CrossRef]
- Sun, T.; Wei, C.; Wang, D.; Wang, X.; Wang, J.; Hu, Y.; Mao, X. The small RNA mascRNA differentially regulates TLR-induced proinflammatory and antiviral responses. JCI Insight 2021, 6, e150833. [Google Scholar] [CrossRef]
- Zhang, X.; Hamblin, M.H.; Yin, K.-J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Non-Coding RNA 2020, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag. Res. 2020, 12, 10181–10198. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.B.; Böing, S.; Zhou, X.Q.; Ji, P.; Dong, Y.; Yao, Q.; Müller-Tidow, C. Identification of metastasis-associated genes in early stage non-small cell lung cancer by subtractive hybridization. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 2002, 34, 273–278. [Google Scholar] [PubMed]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.; Goldstein, T.C.; et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.-C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2015, 30, 34–51. [Google Scholar] [CrossRef]
- Liu, J.-Y.; Yao, J.; Li, X.-M.; Song, Y.-C.; Wang, X.-Q.; Li, Y.-J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef]
- Yao, J.; Wang, X.-Q.; Li, Y.-J.; Shan, K.; Yang, H.; Wang, Y.-N.Z.; Yao, M.-D.; Liu, C.; Li, X.-M.; Shen, Y.; et al. Long non-coding RNA MALAT 1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol. Med. 2016, 8, 346–362. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Hu, H.-Y.; You, Z.-P.; Li, B.-Y.; Shi, K. Targeting long non-coding RNA MALAT1 alleviates retinal neurodegeneration in diabetic mice. Int. J. Ophthalmol. 2020, 13, 213–219. [Google Scholar] [CrossRef]
- Toraih, E.; El-Wazir, A.; Alghamdi, S.A.; Alhazmi, A.S.; El-Wazir, M.; Abdel-Daim, M.; Fawzy, M.S. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet. Mol. Biol. 2019, 42, 509–518. [Google Scholar] [CrossRef]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.-B.; Shi, J.-M.; Li, W.-J.; Tang, L.-S.; Zhu, X.-H.; Tong, P. LncRNA-MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR-125b/VE-cadherin axis. Biosci. Rep. 2019, 39, BSR20181469. [Google Scholar] [CrossRef]
- Yu, L.; Fu, J.; Yu, N.; Wu, Y.; Han, N. Long noncoding RNA MALAT1 participates in the pathological angiogenesis of diabetic retinopathy in an oxygen-induced retinopathy mouse model by sponging miR-203a-3p. Can. J. Physiol. Pharmacol. 2020, 98, 219–227. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kowluru, R.A. Long Noncoding RNA MALAT1 and Regulation of the Antioxidant Defense System in Diabetic Retinopathy. Diabetes 2020, 70, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Thomas, A.A.; Chen, S.; Aref-Eshghi, E.; Feng, B.; Gonder, J.; Sadikovic, B.; Chakrabarti, S. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci. Rep. 2018, 8, 6526. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Guo, H.; Peng, Y.; Nie, D.; Mo, J.; Ye, L. Knockdown of MALAT1 attenuates high-glucose-induced angiogenesis and inflammation via endoplasmic reticulum stress in human retinal vascular endothelial cells. Biomed. Pharmacother. 2020, 124, 109699. [Google Scholar] [CrossRef]
- Park, Y.; Kim, H.-L.; Lee, S.; Zhang, Y.; Kim, I.-B. Expression of the Endoplasmic Reticulum Stress Marker GRP78 in the Normal Retina and Retinal Degeneration Induced by Blue LED Stimuli in Mice. Cells 2021, 10, 995. [Google Scholar] [CrossRef]
- Dong, N.; Xu, B.; Shi, H. Long noncoding RNA MALAT1 acts as a competing endogenous RNA to regulate Amadori-glycated albumin-induced MCP-1 expression in retinal microglia by a microRNA-124-dependent mechanism. Inflamm. Res. 2018, 67, 913–925. [Google Scholar] [CrossRef]
- Shaker, O.G.; Abdelaleem, O.O.; Mahmoud, R.H.; Abdelghaffar, N.K.; Ahmed, T.I.; Said, O.; Zaki, O.M. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life 2018, 71, 310–320. [Google Scholar] [CrossRef]
- Han, N.; Tian, W.; Yu, N.; Yu, L. YAP1 is required for the angiogenesis in retinal microvascular endothelial cells via the inhibition of MALAT1-mediated miR-200b-3p in high glucose-induced diabetic retinopathy. J. Cell. Physiol. 2019, 235, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yang, J.; Gao, Y.; Jiang, S.; Li, Z.; Wang, Y.; Hu, Z.; Han, F.; Ni, N. LncRNA MALAT1 aggravates the retinal angiogenesis via miR-320a/HIF-1α axis in diabetic retinopathy. Exp. Eye Res. 2022, 218, 108984. [Google Scholar] [CrossRef]
- Tan, A.; Li, T.; Ruan, L.; Yang, J.; Luo, Y.; Li, L.; Wu, X. Knockdown of Malat1 alleviates high-glucose-induced angiogenesis through regulating miR-205-5p/VEGF-A axis. Exp. Eye Res. 2021, 207, 108585. [Google Scholar] [CrossRef] [PubMed]
- Li, X. lncRNA MALAT1 promotes diabetic retinopathy by upregulating PDE6G via miR-378a-3p. Arch. Physiol. Biochem. 2021, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- You, Z.-P.; Zhang, Y.-L.; Li, B.-Y.; Zhu, X.-G.; Shi, K. Bioinformatics Analysis of Weighted Genes in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5558–5563. [Google Scholar] [CrossRef]
- Mohammad, G.; Kowluru, R.A. Nuclear Genome-Encoded Long Noncoding RNAs and Mitochondrial Damage in Diabetic Retinopathy. Cells 2021, 10, 3271. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.-Y.; Li, X.-M.; Wang, X.-Q.; Li, Y.-J.; Tao, Z.-F.; Song, Y.-C.; Yu-Chen, S.; Jiang, Q. lncRNA-MIAT Regulates Microvascular Dysfunction by Functioning as a Competing Endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Li, Q.; Pang, L.; Yang, W.; Liu, X.; Su, G.; Dong, Y. Long Non-Coding RNA of Myocardial Infarction Associated Transcript (LncRNA-MIAT) Promotes Diabetic Retinopathy by Upregulating Transforming Growth Factor-β1 (TGF-β1) Signaling. Med. Sci. Monit. 2018, 24, 9497–9503. [Google Scholar] [CrossRef]
- Sathishkumar, C.; Prabu, P.; Mohan, V.; Balasubramanyam, M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genom. 2018, 12, 41. [Google Scholar] [CrossRef]
- Toraih, E.A.; Abd Elghany, A.A.; Abd El Fadeal, N.M.; Al Ageeli, E.; Fawzy, M.S. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch. Clin. Exp. Ophthalmol. 2019, 257, 1897–1913. [Google Scholar] [CrossRef]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Aprea, J.; Prenninger, S.; Dori, M.; Ghosh, T.; Monasor, L.S.; Wessendorf, E.; Zocher, S.; Massalini, S.; Alexopoulou, D.; Lesche, M.; et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013, 32, 3145–3160. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, A.; Hasegawa, Y.; Ishida, K.; Yanaka, K.; Nakagawa, S. Formation of nuclear bodies by the lnc RNA Gomafu-associating proteins Celf3 and SF 1. Genes Cells 2014, 19, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2013, 19, 486–494. [Google Scholar] [CrossRef]
- Zhu, M.; Li, N.; Luo, P.; Jing, W.; Wen, X.; Liang, C.; Tu, J. Peripheral Blood Leukocyte Expression of lncRNA MIAT and Its Diagnostic and Prognostic Value in Ischemic Stroke. J. Stroke Cerebrovasc. Dis. 2018, 27, 326–337. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Chen, J.; Lin, S.; Cai, D.; Chen, C.; Chen, Z. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci. Rep. 2017, 37, BSR20170036. [Google Scholar] [CrossRef]
- Yu, C.; Yang, K.; Meng, X.; Cao, B.; Wang, F. Downregulation of Long Noncoding RNA MIAT in the Retina of Diabetic Rats with Tail-vein Injection of Human Umbilical-cord Mesenchymal Stem Cells. Int. J. Med. Sci. 2020, 17, 591–598. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, C.; Wu, L.; Wang, Q.; Chen, J.; Zhang, S.; Chen, Z. C-myc contributes to the release of Müller cells-derived proinflammatory cytokines by regulating lncRNA MIAT/XNIP pathway. Int. J. Biochem. Cell Biol. 2019, 114, 105574. [Google Scholar] [CrossRef]
- Yu, X.; Ma, X.; Lin, W.; Xu, Q.; Zhou, H.; Kuang, H. Long noncoding RNA MIAT regulates primary human retinal pericyte pyroptosis by modulating miR-342–3p targeting of CASP1 in diabetic retinopathy. Exp. Eye Res. 2020, 202, 108300. [Google Scholar] [CrossRef]
- Burd, C.E.; Jeck, W.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N. Expression of Linear and Novel Circular Forms of an INK4/ARF-Associated Non-Coding RNA Correlates with Atherosclerosis Risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Razeghian-Jahromi, I.; Akhormeh, A.K.; Zibaeenezhad, M.J. The Role of ANRIL in Atherosclerosis. Dis. Markers 2022, 2022, 8859677. [Google Scholar] [CrossRef] [PubMed]
- De Martino, M.; Esposito, F.; Pallante, P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl. Cancer Res. 2021, 10, 3140–3157. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, W.; Li, C.; Wang, X.; Luo, J.; Cheng, B. LncRNA ANRIL Facilitates Vascular Smooth Muscle Cell Proliferation and Suppresses Apoptosis via Modulation of miR-7/FGF2 Pathway in Intracranial Aneurysms. Neurocritical Care 2022, 36, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, A.; Ada, A.O. The roles of ANRIL polymorphisms in periodontitis: A systematic review and meta-analysis. Clin. Oral Investig. 2022, 26, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Safari, M.; Taheri, M.; Samadian, M. Expression of Linear and Circular lncRNAs in Alzheimer’s Disease. J. Mol. Neurosci. 2021, 72, 187–200. [Google Scholar] [CrossRef]
- Nakaoka, H.; Gurumurthy, A.; Hayano, T.; Ahmadloo, S.; Omer, W.H.; Yoshihara, K.; Yamamoto, A.; Kurose, K.; Enomoto, T.; Akira, S.; et al. Allelic Imbalance in Regulation of ANRIL through Chromatin Interaction at 9p21 Endometriosis Risk Locus. PLoS Genet. 2016, 12, e1005893. [Google Scholar] [CrossRef]
- Huang, G.; Liang, D.; Luo, L.; Lan, C.; Luo, C.; Xu, H.; Lai, J. Significance of the lncRNAs MALAT1 and ANRIL in occurrence and development of glaucoma. J. Clin. Lab. Anal. 2022, 36, e24215. [Google Scholar] [CrossRef]
- Dieter, C.; Lemos, N.E.; Corrêa, N.R.d.F.; Assmann, T.S.; Crispim, D. The Impact of lncRNAs in Diabetes Mellitus: A Systematic Review and In Silico Analyses. Front. Endocrinol. 2021, 12, 602597. [Google Scholar] [CrossRef]
- Wei, J.C.; Shi, Y.L.; Wang, Q. LncRNA ANRIL knockdown ameliorates retinopathy in diabetic rats by inhibiting the NF-κB pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7732–7739. [Google Scholar] [CrossRef]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhong, H.; Wang, Y.; Wang, Z.; Liang, X.; Li, S.; Li, Z.; Yu, Z.; Li, L.; Yi, G.; et al. The clinical significance of long non-coding RNA ANRIL level in diabetic retinopathy. Acta Diabetol. 2019, 57, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Pisani, G.; Baron, B. NEAT1 and Paraspeckles in Cancer Development and Chemoresistance. Non-Coding RNA 2020, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, K.; Huang, W. Long non-coding RNA NEAT1-centric gene regulation. Cell Mol. Life Sci. 2020, 77, 3769–3779. [Google Scholar] [CrossRef]
- Li, X.-J. Long non-coding RNA nuclear paraspeckle assembly transcript 1 inhibits the apoptosis of retina Müller cells after diabetic retinopathy through regulating miR-497/brain-derived neurotrophic factor axis. Diabetes Vasc. Dis. Res. 2018, 15, 204–213. [Google Scholar] [CrossRef]
- Shao, K.; Xi, L.; Cang, Z.; Chen, C.; Huang, S. Knockdown of NEAT1 exerts suppressive effects on diabetic retinopathy progression via inactivating TGF-β1 and VEGF signaling pathways. J. Cell. Physiol. 2020, 235, 9361–9369. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, J.; Li, W.H.; Zhou, Z.X.; Xia, X.B. LncRNA NEAT1 regulated diabetic retinal epithelial-mesenchymal transition through regulating miR-204/SOX4 axis. PeerJ 2021, 9, e11817. [Google Scholar] [CrossRef]
- Cantile, M.; Di Bonito, M.; Cerrone, M.; Collina, F.; De Laurentiis, M.; Botti, G. Long Non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers 2020, 12, 1197. [Google Scholar] [CrossRef]
- Yuan, C.; Ning, Y.; Pan, Y. Emerging roles of HOTAIR in human cancer. J. Cell. Biochem. 2020, 121, 3235–3247. [Google Scholar] [CrossRef]
- Biswas, S.; Feng, B.; Chen, S.; Liu, J.; Aref-Eshghi, E.; Gonder, J.; Ngo, V.; Sadikovic, B.; Chakrabarti, S. The Long Non-Coding RNA HOTAIR Is a Critical Epigenetic Mediator of Angiogenesis in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2021, 62, 20. [Google Scholar] [CrossRef]
- Zhao, D.; Zhao, Y.; Wang, J.; Wu, L.; Liu, Y.; Zhao, S.; Guo, F.; Ma, X.; Zhang, H.; Li, Z.; et al. Long noncoding RNA Hotair facilitates retinal endothelial cell dysfunction in diabetic retinopathy. Clin. Sci. 2020, 134, 2419–2434. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Murakami, K.; Meguro, M.; Soejima, H.; Higashimoto, K.; Urano, T.; Kugoh, H.; Mukai, T.; Ikeguchi, M.; Oshimura, M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006, 97, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Cagle, P.; Qi, Q.; Niture, S.; Kumar, D. KCNQ1OT1: An Oncogenic Long Noncoding RNA. Biomolecules 2021, 11, 1602. [Google Scholar] [CrossRef]
- Jin, X.; Jin, H.; Shi, Y.; Guo, Y.; Zhang, H. Long Non-Coding RNA KCNQ1OT1 Promotes Cataractogenesis via miR-214 and Activation of the Caspase-1 Pathway. Cell. Physiol. Biochem. 2017, 42, 295–305. [Google Scholar] [CrossRef]
- Chen, B.; Ma, J.; Li, C.; Wang, Y. Long noncoding RNA KCNQ1OT1 promotes proliferation and epithelial-mesenchymal transition by regulation of SMAD4 expression in lens epithelial cells. Mol. Med. Rep. 2018, 18, 16–24. [Google Scholar] [CrossRef]
- Shao, J.; Pan, X.; Yin, X.; Fan, G.; Tan, C.; Yao, Y.; Xin, Y.; Sun, C. KCNQ1OT1 affects the progression of diabetic retinopathy by regulating miR-1470 and epidermal growth factor receptor. J. Cell. Physiol. 2019, 234, 17269–17279. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.L.; Ceresa, B.P. Epidermal Growth Factor Receptor Expression in the Corneal Epithelium. Cells 2021, 10, 2409. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, Z.; Li, X.; Xu, S.; Zhou, S.; Jin, X.; Zhang, H. Long noncoding RNA KCNQ1OT1 induces pyroptosis in diabetic corneal endothelial keratopathy. Am. J. Physiol. Cell. Physiol. 2020, 318, C346–C359. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Ni, M.-M. Long non-coding RNA HEIH: A novel tumor activator in multiple cancers. Cancer Cell Int. 2021, 21, 558. [Google Scholar] [CrossRef]
- Zhao, C.; Fei, X.; Xu, B.; Lu, Y.; Zhang, Q. Long non-coding RNA HEIH contributes to diabetic retinopathy by regulating miR-939/VEGF axis. Int. J. Clin. Exp. Pathol. 2019, 12, 2022–2033. [Google Scholar]
- Shan, K.; Jiang, Q.; Wang, X.Q.; Wang, Y.N.Z.; Yang, H.; Yao, M.D.; Liu, C.; Li, X.M.; Yao, J.; Liu, B.; et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016, 7, e2248. [Google Scholar] [CrossRef] [PubMed]
- Shan, K.; Li, C.-P.; Liu, C.; Liu, X.; Yan, B. RNCR3: A regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 482, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, C.-P.; Wang, J.-J.; Shan, K.; Liu, X.; Yan, B. RNCR3 knockdown inhibits diabetes mellitus-induced retinal reactive gliosis. Biochem. Biophys. Res. Commun. 2016, 479, 198–203. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Song, G.; Zhang, X.; Gao, S.; Liu, H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019, 450, 14–21. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.-X. LncRNA HOTTIP improves diabetic retinopathy by regulating the p38-MAPK pathway. Eur. Rev. Med Pharmacol. Sci. 2018, 22, 2941–2948. [Google Scholar]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Ghanbari, M. A concise review on the role of BDNF-AS in human disorders. Biomed. Pharmacother. 2021, 142, 112051. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Z.; Xie, T.; Zhang, X.; Dai, T. Inhibition of BDNF-AS Provides Neuroprotection for Retinal Ganglion Cells against Ischemic Injury. PLoS ONE 2016, 11, e0164941. [Google Scholar] [CrossRef]
- Li, Y.; Xu, F.; Xiao, H.; Han, F. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J. Cell. Biochem. 2017, 119, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, C.; Xu, Y.; Liu, Y.; Zhang, H.; Liu, Y. LncRNA FENDRR promotes high-glucose-induced proliferation and angiogenesis of human retinal endothelial cells. Biosci. Biotechnol. Biochem. 2019, 83, 869–875. [Google Scholar] [CrossRef]
- Kun-Peng, Z.; Chun-Lin, Z.; Xiao-Long, M. Antisense lncRNA FOXF1-AS1 Promotes Migration and Invasion of Osteosarcoma Cells Through the FOXF1/MMP-2/-9 Pathway. Int. J. Biol. Sci. 2017, 13, 1180–1191. [Google Scholar] [CrossRef]
- Ren, X.; Ustiyan, V.; Pradhan, A.; Cai, Y.; Havrilak, J.; Bolte, C.S.; Shannon, J.M.; Kalin, T.V.; Kalinichenko, V.V. FOXF1 Transcription Factor Is Required for Formation of Embryonic Vasculature by Regulating VEGF Signaling in Endothelial Cells. Circ. Res. 2014, 115, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Luo, Y.; Chen, G.; Liu, H.; Tian, N.; Zen, X.; Liu, Q. Long noncoding RNA IGF2AS regulates high-glucose induced apoptosis in human retinal pigment epithelial cells. IUBMB Life 2019, 71, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Yang, H.; Zeng, Y. Long non-coding RNA Arid2-IR affects advanced glycation end products-induced human retinal endothelial cell injury by binding to Smad3. Int. Ophthalmol. 2020, 40, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Dong, W.; Fan, Y.; Chen, F.; Bian, X.; Xu, X.; Qian, T.; Yu, P. LncRNA TDRG1-Mediated Overexpression of VEGF Aggravated Retinal Microvascular Endothelial Cell Dysfunction in Diabetic Retinopathy. Front. Pharmacol. 2020, 10, 1703. [Google Scholar] [CrossRef]
- Li, X.; Zhu, J.; Zhong, Y.; Liu, C.; Yao, M.; Sun, Y.; Yao, W.; Ni, X.; Zhou, F.; Yao, J.; et al. Targeting long noncoding RNA-AQP4-AS1 for the treatment of retinal neurovascular dysfunction in diabetes mellitus. eBioMedicine 2022, 77, 103857. [Google Scholar] [CrossRef]
- Fiedler, J.; Breckwoldt, K.; Remmele, C.W.; Hartmann, D.; Dittrich, M.; Pfanne, A.; Just, A.; Xiao, K.; Kunz, M.; Müller, T.; et al. Development of Long Noncoding RNA-Based Strategies to Modulate Tissue Vascularization. J. Am. Coll. Cardiol. 2015, 66, 2005–2015. [Google Scholar] [CrossRef]
- Atef, M.M.; Shafik, N.M.; Hafez, Y.M.; Watany, M.M.; Selim, A.; Shafik, H.M.; El-Deeb, O.S. The evolving role of long noncoding RNA HIF1A-AS2 in diabetic retinopathy: A cross-link axis between hypoxia, oxidative stress and angiogenesis via MAPK/VEGF-dependent pathway. Redox Rep. 2022, 27, 70–78. [Google Scholar] [CrossRef]
- Wang, J.-J.; Wu, K.-F.; Wang, D.-D. A novel regulatory network of linc00174/miR-150-5p/VEGFA modulates pathological angiogenesis in diabetic retinopathy. Can. J. Physiol. Pharmacol. 2021, 99, 1175–1183. [Google Scholar] [CrossRef]
- Shi, Q.; Tang, J.; Wang, M.; Xu, L.; Shi, L. Knockdown of Long Non-coding RNA TUG1 Suppresses Migration and Tube Formation in High Glucose-Stimulated Human Retinal Microvascular Endothelial Cells by Sponging miRNA-145. Mol. Biotechnol. 2021, 64, 171–177. [Google Scholar] [CrossRef]
- Yan, H.; Yao, P.; Hu, K.; Li, X.; Li, H. Long non-coding ribonucleic acid urothelial carcinoma-associated 1 promotes high glucose-induced human retinal endothelial cells angiogenesis through regulating micro-ribonucleic acid-624-3p/vascular endothelial growth factor C. J. Diabetes Investig. 2021, 12, 1948–1957. [Google Scholar] [CrossRef]
- Zhang, R.; Niu, C.; Guan, Y.; Wu, J.; Hu, L. LINC00963 silencing inhibits the proliferation and migration of high glucose-induced retinal endothelial cells via targeting miR-27b. Exp. Ther. Med. 2021, 22, 1274. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Yang, J.; Yan, X.; Cao, Y.; Liu, Y.; Lei, Y.; Lv, H. Knockdown of lncRNA TUG1 alleviates diabetic retinal vascular dysfunction through regulating miR-524-5p/FGFR2. Bioengineered 2022, 13, 12661–12672. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, W.; Lao, W.; Chen, Y. Silencing LncRNA PVT1 Reverses High Glucose-Induced Regulation of the High Expression of PVT1 in HRMECs by Targeting miR-128-3p. Horm. Metab. Res. 2022, 54, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Zheng, Y.; Sun, Y.; Lai, M.; Qiu, J.; Gui, F.; Zeng, Q.; Liu, F. Suppressing long noncoding RNA OGRU ameliorates diabetic retinopathy by inhibition of oxidative stress and inflammation via miR-320/USP14 axis. Free Radic. Biol. Med. 2021, 169, 361–381. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef]
- Sellers, Z.P.; Schneider, G.; Maj, M.; Ratajczak, M.Z. Analysis of the Paternally-Imprinted DLK1–MEG3 and IGF2–H19 Tandem Gene Loci in NT2 Embryonal Carcinoma Cells Identifies DLK1 as a Potential Therapeutic Target. Stem Cell Rev. Rep. 2018, 14, 823–836. [Google Scholar] [CrossRef]
- He, C.; Yang, W.; Yang, J.; Ding, J.; Li, S.; Wu, H.; Zhou, F.; Jiang, Y.; Teng, L.; Yang, J. Long Noncoding RNAMEG3Negatively Regulates Proliferation and Angiogenesis in Vascular Endothelial Cells. DNA Cell Biol. 2017, 36, 475–481. [Google Scholar] [CrossRef]
- Ruan, W.; Zhao, F.; Zhao, S.; Zhang, L.; Shi, L.; Pang, T. Knockdown of long noncoding RNA MEG3 impairs VEGF-stimulated endothelial sprouting angiogenesis via modulating VEGFR2 expression in human umbilical vein endothelial cells. Gene 2018, 649, 32–39. [Google Scholar] [CrossRef]
- Zhang, D.; Qin, H.; Leng, Y.; Li, X.; Zhang, L.; Bai, D.; Meng, Y.; Wang, J. LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF-β1 and VEGF. Exp. Ther. Med. 2018, 16, 2337–2342. [Google Scholar] [CrossRef]
- Luo, R.; Jin, H.; Li, L.; Hu, Y.-X.; Xiao, F. Long Noncoding RNA MEG3 Inhibits Apoptosis of Retinal Pigment Epithelium Cells Induced by High Glucose via the miR-93/Nrf2 Axis. Am. J. Pathol. 2020, 190, 1813–1822. [Google Scholar] [CrossRef]
- Chen, J.; Liao, L.; Xu, H.; Zhang, Z.; Zhang, J. Long non-coding RNA MEG3 inhibits neovascularization in diabetic retinopathy by regulating microRNA miR-6720-5p and cytochrome B5 reductase 2. Bioengineered 2021, 12, 11872–11884. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.-Z.; Tian, W.; Fu, H.-T.; Li, C.-P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Dan, Y.; Gao, X.; Huang, L.; Lv, H.; Chen, J. DNMT1-mediated lncRNA MEG3 methylation accelerates endothelial-mesenchymal transition in diabetic retinopathy through the PI3K/Akt/mTOR signaling pathway. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E598–E608. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Peng, Q.-H.; Gu, L.-M.; Xie, W.-W.; Li, W.-J. LncRNA-MEG3 alleviates high glucose induced inflammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis. Exp. Mol. Pathol. 2018, 107, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, X.; Tong, X.-L. Effect of lncRNA MEG3 on retinopathy in diabetic rats through regulating Fox01 expression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9163–9170. [Google Scholar]
- Fan, G.; Gu, Y.; Zhang, J.; Xin, Y.; Shao, J.; Giampieri, F.; Battino, M. Transthyretin Upregulates Long Non-Coding RNA MEG3 by Affecting PABPC1 in Diabetic Retinopathy. Int. J. Mol. Sci. 2019, 20, 6313. [Google Scholar] [CrossRef]
- Xiao, F.; Li, L.; Fu, J.-S.; Hu, Y.-X.; Luo, R. Regulation of the miR-19b-mediated SOCS6-JAK2/STAT3 pathway by lncRNA MEG3 is involved in high glucose-induced apoptosis in hRMECs. Biosci. Rep. 2020, 40, BSR20194370. [Google Scholar] [CrossRef]
- Tu, Y.; Zhu, M.; Wang, Z.; Wang, K.; Chen, L.; Liu, W.; Shi, Q.; Zhao, Q.; Sun, Y.; Wang, X.; et al. Melatonin inhibits Müller cell activation and pro-inflammatory cytokine production via upregulating the MEG3/miR-204/Sirt1 axis in experimental diabetic retinopathy. J. Cell. Physiol. 2020, 235, 8724–8735. [Google Scholar] [CrossRef]
- Elahian, F.; Sepehrizadeh, Z.; Moghimi, B.; Mirzaei, S.A. Human cytochrome b5 reductase: Structure, function, and potential applications. Crit. Rev. Biotechnol. 2012, 34, 134–143. [Google Scholar] [CrossRef]
- Ming, H.; Lan, Y.; He, F.; Xiao, X.; Zhou, X.; Zhang, Z.; Li, P.; Huang, G. Cytochrome b5 reductase 2 suppresses tumor formation in nasopharyngeal carcinoma by attenuating angiogenesis. Chin. J. Cancer 2015, 34, 459–467. [Google Scholar] [CrossRef]
- Nie, Q.-Z.; Di, Y.; Wang, Y.; Wang, X.; Ma, Y. Maternally expressed gene 3 regulates retinal neovascularization in retinopathy of prematurity. Neural Regen. Res. 2022, 17, 1364–1368. [Google Scholar] [CrossRef] [PubMed]
- Galupa, R.; Heard, E. X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annu. Rev. Genet. 2018, 52, 535–566. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.; Ballabio, A.; Rupert, J.L.; LaFreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef]
- Dong, Y.; Wan, G.; Peng, G.; Yan, P.; Qian, C.; Li, F. Long non-coding RNA XIST regulates hyperglycemia-associated apoptosis and migration in human retinal pigment epithelial cells. Biomed. Pharmacother. 2020, 125, 109959. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, C.; Zhang, S.; Chen, J.; Wu, L.; Chen, Z. LncRNA XIST restrains the activation of Müller cells and inflammation in diabetic retinopathy via stabilizing SIRT1. Autoimmunity 2021, 54, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Kamiya, S.; Ishiwata, T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front. Biosci. 2018, 23, 614–625. [Google Scholar] [CrossRef]
- Alipoor, B.; Parvar, S.N.; Sabati, Z.; Ghaedi, H.; Ghasemi, H. An updated review of the H19 lncRNA in human cancer: Molecular mechanism and diagnostic and therapeutic importance. Mol. Biol. Rep. 2020, 47, 6357–6374. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 2012, 50, 171–179. [Google Scholar] [CrossRef]
- Riccio, A.; Sparago, A.; Verde, G.; De Crescenzo, A.; Citro, V.; Cubellis, M.V.; Ferrero, G.B.; Silengo, M.C.; Russo, S.; Larizza, L.; et al. Inherited and Sporadic Epimutations at the IGF2-H19 Locus in Beckwith-Wiedemann Syndrome and Wilms’ Tumor. Endocr. Dev. 2009, 14, 1–9. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Selvatici, R.; Di Domenico, M.; Marci, R.; Vesce, F.; Tognon, M.; Martini, F. Methylation loss atH19imprinted gene correlates with methylenetetrahydrofolate reductasegene promoter hypermethylation in semen samples from infertile males. Epigenetics 2013, 8, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shen, A.; Liu, A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull. Cancer 2019, 106, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Jiang, R.; Lin, X.; Shao, M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget 2016, 8, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Xiao, F.; Wang, P.; Hu, Y.-X. lncRNA H19 sponging miR-93 to regulate inflammation in retinal epithelial cells under hyperglycemia via XBP1s. Inflamm. Res. 2020, 69, 255–265. [Google Scholar] [CrossRef]
- Luo, R.; Li, L.; Hu, Y.; Xiao, F. LncRNA H19 inhibits high glucose-induced inflammatory responses of human retinal epithelial cells by targeting miR-19b to increase SIRT1 expression. Kaohsiung J. Med. Sci. 2020, 37, 101–110. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Abdelghany, A.A.; Toraih, E.A.; Mohamed, A.M. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn. J. Basic Med. Sci. 2020, 20, 365–371. [Google Scholar] [CrossRef]
- Zha, T.; Su, F.; Liu, X.; Yang, C.; Liu, L. Role of Long Non-Coding RNA (LncRNA) LINC-PINT Downregulation in Cardiomyopathy and Retinopathy Progression Among Patients with Type 2 Diabetes. Med. Sci. Monit. 2019, 25, 8509–8514. [Google Scholar] [CrossRef]
- Marín-Béjar, O.; Marchese, F.P.; Athie, A.; Sánchez, Y.; González, J.; Segura, V.; Huang, L.; Moreno, I.; Navarro, A.; Monzó, M.; et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013, 14, R104. [Google Scholar] [CrossRef]
- Zeng, F.; Luo, G.; Lu, Y.; Zhang, Z.; Zhou, Y.; Chen, Y.; Zhou, Z. Long non-coding RNA VIM Antisense RNA 1 (VIM-AS1) sponges microRNA-29 to participate in diabetic retinopathy. Acta Diabetol. 2020, 57, 1111–1116. [Google Scholar] [CrossRef]
- Bozgeyik, E.; Ege, B.; Koparal, M.; Ceylan, O. Clinical significance of Vimentin Antisense RNA 1 and its correlation with other epithelial to mesenchymal transition markers in oral cancers. Pathol.-Res. Pract. 2022, 232, 153807. [Google Scholar] [CrossRef]
- Zhang, X.F.; Zhang, Y.; Shen, Z.; Yang, G.G.; Wang, H.D.; Li, L.F.; Liu, D.C.; Qiu, J.M. LncRNA LUADT1 is overexpressed in colorectal cancer and its expression level is related to clinicopathology. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2282–2286. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-H.; Zhao, W.-Y.; Fang, Q.-Q.; Wang, X.-F.; Zhang, D.-D.; Hu, Y.-Y.; Zheng, B.; Tan, W.-Q. Long Noncoding RNA LUADT1 Is Upregulated in Melanoma and May Sponge miR-28-5p to Upregulate RAP1B. Cancer Biother. Radiopharm. 2020, 35, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Xu, Y.; Wang, J.; Zhang, E.; Sun, M.; Zheng, Y.; Li, M.; Xia, W.; Feng, D.; Yin, R.; et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015, 6, e1858. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.; Sun, Z.; Qian, Y.; Zhang, B.; Han, Y.; Deng, G. LncRNA LUADT1 inhibits cell apoptosis in diabetic retinopathy by regulating miR-383/peroxiredoxin 3 axis. Arch. Physiol. Biochem. 2020, 128, 637–642. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, E.; Yang, L.; Fu, W.; Hu, F.; Zhou, X. LncRNA AK077216 is downregulated in diabetic retinopathy and inhibited the apoptosis of retinal pigment epithelial cells by downregulating miR-383. Endocr. J. 2019, 66, 1011–1016. [Google Scholar] [CrossRef]
- Niu, T.; An, Y.; Lv, T.; Liu, D. Long non-coding RNA RPSAP52 upregulates Timp3 by serving as the endogenous sponge of microRNA-365 in diabetic retinopathy. Exp. Ther. Med. 2020, 20, 246. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, X. Long non-coding ribonucleic acid ATP2B1-AS1 modulates endothelial permeability through regulating the miR-4729–IQGAP2 axis in diabetic retinopathy. J. Diabetes Investig. 2021, 13, 443–452. [Google Scholar] [CrossRef]
- Li, C.-P.; Wang, S.-H.; Wang, W.-Q.; Song, S.-G.; Liu, X.-M. Long Noncoding RNA-Sox2OT Knockdown Alleviates Diabetes Mellitus-Induced Retinal Ganglion Cell (RGC) injury. Cell. Mol. Neurobiol. 2016, 37, 361–369. [Google Scholar] [CrossRef]
- Shao, J.; Zhang, Y.; Fan, G.; Xin, Y.; Yao, Y. Transcriptome analysis identified a novel 3-LncRNA regulatory network of transthyretin attenuating glucose induced hRECs dysfunction in diabetic retinopathy. BMC Med. Genom. 2019, 12, 134. [Google Scholar] [CrossRef]
- Yang, J.; Yang, F.-J.; Wang, Y.-G.; Su, G.-F.; Miao, X. LncRNA MIR497HG inhibits proliferation and migration of retinal endothelial cells under high-level glucose treatment via miRNA-128-3p/SIRT1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5871–5877. [Google Scholar]
- Sun, X.; Lu, Y.; Lei, T. TPTEP1 suppresses high glucose-induced dysfunction in retinal vascular endothelial cells by interacting with STAT3 and targeting VEGFA. Acta Diabetol. 2021, 58, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Yang, C.; Yang, X.; Wu, S.; Feng, Z.; Qu, L.; Chen, X.; Liu, L.; Ma, Y. LncRNA MCM3AP-AS1 is downregulated in diabetic retinopathy and promotes cell apoptosis by regulating miR-211/SIRT1. Diabetol. Metab. Syndr. 2022, 14, 73. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhu, Y.; Ma, L. LncRNA LINC00673 is Downregulated in Diabetic Retinopathy and Regulates the Apoptosis of Retinal Pigment Epithelial Cells via Negatively Regulating p53. Diabetes Metab. Syndr. Obes. 2021, 14, 4233–4240. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, P.; Mathew, S.; Sivadas, A.; Ray, A.; Tanwar, J.; Vishwakarma, S.; Ranjan, G.; Shamsudheen, K.V.; Bhoyar, R.C.; Pateria, A.; et al. LncRNA VEAL2 regulates PRKCB2 to modulate endothelial permeability in diabetic retinopathy. EMBO J. 2021, 40, e107134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, X.; Li, Y.; Wang, Y. Downregulation of lncRNA BANCR participates in the development of retinopathy among diabetic patients. Exp. Ther. Med. 2019, 17, 4132–4138. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Sun, Z.; Ren, Q.; Su, X.; Zhang, D. Long Non-Coding RNA BANCR Is Overexpressed in Patients with Diabetic Retinopathy and Promotes Apoptosis of Retinal Pigment Epithelial Cells. Med Sci. Monit. 2019, 25, 2845–2851. [Google Scholar] [CrossRef]
- Awata, T.; Yamashita, H.; Kurihara, S.; Morita-Ohkubo, T.; Miyashita, Y.; Katayama, S.; Mori, K.; Yoneya, S.; Kohda, M.; Okazaki, Y.; et al. A Genome-Wide Association Study for Diabetic Retinopathy in a Japanese Population: Potential Association with a Long Intergenic Non-Coding RNA. PLoS ONE 2014, 9, e111715. [Google Scholar] [CrossRef]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef]
- Ojha, S.; Malla, S.; Lyons, S.M. snoRNPs: Functions in Ribosome Biogenesis. Biomolecules 2020, 10, 783. [Google Scholar] [CrossRef]
- Qin, Y.; Sun, W.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers. OncoTargets Ther. 2020, 13, 7699–7717. [Google Scholar] [CrossRef]
- Biagioni, A.; Tavakol, S.; Ahmadirad, N.; Zahmatkeshan, M.; Magnelli, L.; Mandegary, A.; Fekri, H.S.; Asadi, M.H.; Mohammadinejad, R.; Ahn, K.S. Small nucleolar RNA host genes promoting epithelial–mesenchymal transition lead cancer progression and metastasis. IUBMB Life 2021, 73, 825–842. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, K.; Meng, X.; Liu, P.; Fu, Y.; Wang, Y. Silenced SNHG1 Inhibited Epithelial-Mesenchymal Transition and Inflammatory Response of ARPE-19 Cells Induced by High Glucose. J. Inflamm. Res. 2021, 14, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Jiang, H.; Li, Y.; Li, Q.; Yang, C. Upregulation of long non-coding RNA SNHG16 promotes diabetes-related RMEC dysfunction via activating NF-κB and PI3K/AKT pathways. Mol. Ther.-Nucleic Acids 2021, 24, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, C.; Chen, Y.; Yu, F. Long non-coding RNA SNHG16 regulates E2F1 expression by sponging miR-20a-5p and aggravating proliferative diabetic retinopathy. Can. J. Physiol. Pharmacol. 2021, 99, 1207–1216. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, X.; Jiang, L.; Xia, W.; Li, H.; Zhao, N.; Cui, X.; Zhang, N.; Zhou, H.; Xu, S. Decreased lncRNA SNHG16 Accelerates Oxidative Stress Induced Pathological Angiogenesis in Human Retinal Microvascular Endothelial Cells by Regulating miR-195/mfn2 Axis. Curr. Pharm. Des. 2021, 27, 3047–3060. [Google Scholar] [CrossRef]
- Li, Z.; Yu, Z.; Meng, X.; Zhou, S.; Xiao, S.; Li, X.; Liu, S.; Yu, P. Long noncoding RNA GAS5 impairs the proliferation and invasion of endometrial carcinoma induced by high glucose via targeting miR-222-3p/p27. Am. J. Transl. Res. 2019, 11, 2413–2421. [Google Scholar]
- Jiang, L.; Wang, C.; Shen, X. LncRNA GAS5 suppresses ER stress-induced apoptosis and inflammation by regulating SERCA2b in HG-treated retinal epithelial cell. Mol. Med. Rep. 2020, 22, 1072–1080. [Google Scholar] [CrossRef]
- Yu, J.; Qin, M.; Li, J.; Cui, S. LncRNA SNHG4 sponges miR-200b to inhibit cell apoptosis in diabetic retinopathy. Arch. Physiol. Biochem. 2021, 6, 1–6. [Google Scholar] [CrossRef]
- He, J.; Rui, Z.; Gao, J.; Chen, Y.; Li, Y.; Xu, T.; Wang, S. Expression of Long Non-Coding RNA (lncRNA) SNHG5 in Patients with Refractory Diabetic Macular Edema and Its Regulatory Mechanism. Med. Sci. Monit. 2021, 27, e932996. [Google Scholar] [CrossRef]
- Ke, N.; Pi, L.-H.; Liu, Q.; Chen, L. Long noncoding RNA SNHG7 inhibits high glucose-induced human retinal endothelial cells angiogenesis by regulating miR-543/SIRT1 axis. Biochem. Biophys. Res. Commun. 2019, 514, 503–509. [Google Scholar] [CrossRef]
- Cao, X.; Xue, L.-D.; Di, Y.; Li, T.; Tian, Y.-J.; Song, Y. MSC-derived exosomal lncRNA SNHG7 suppresses endothelial-mesenchymal transition and tube formation in diabetic retinopathy via miR-34a-5p/XBP1 axis. Life Sci. 2021, 272, 119232. [Google Scholar] [CrossRef] [PubMed]
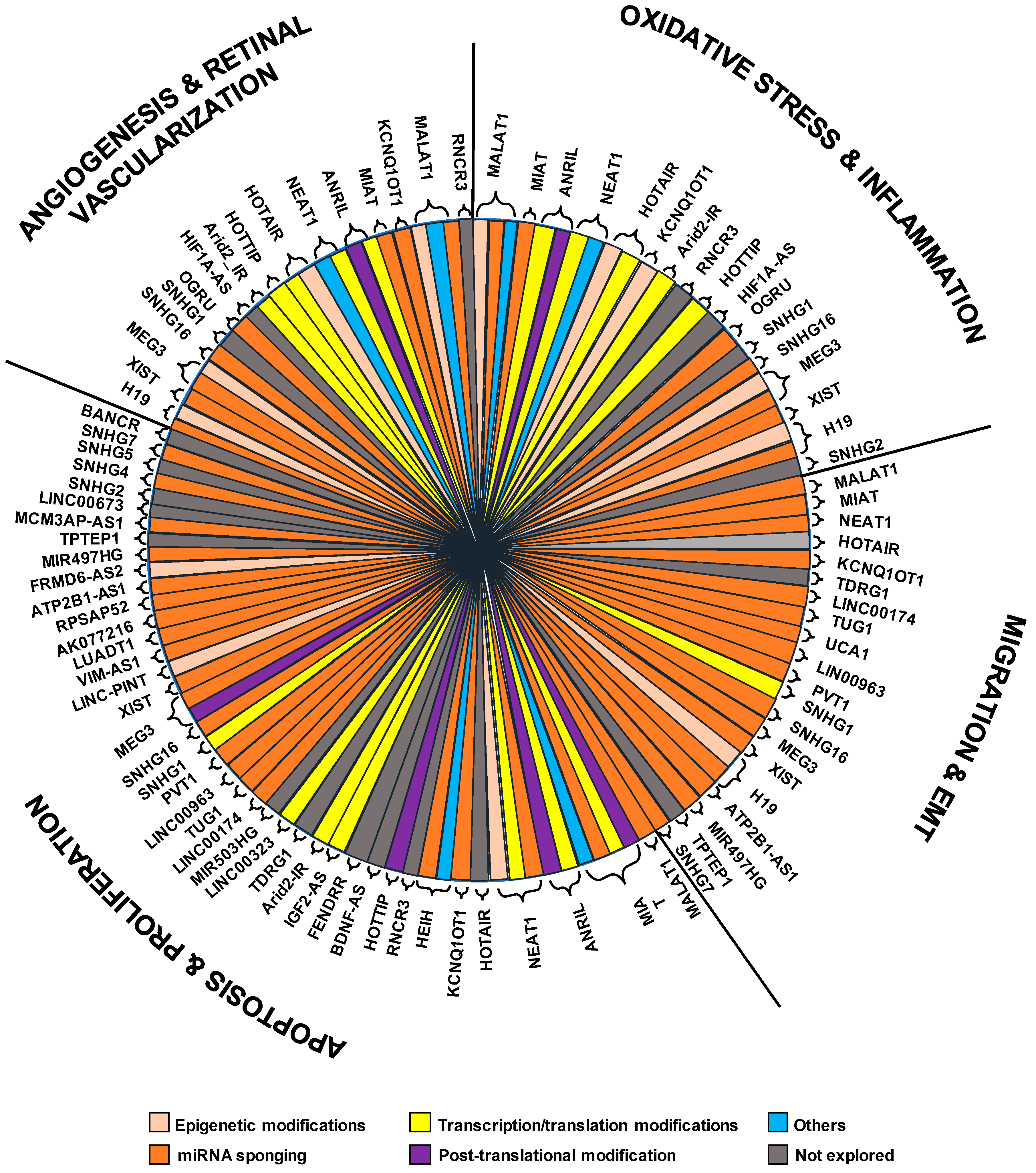
| LncRNA | Chr | DR-Related Processes | Sponged miRNAs | Related Genes/Proteins | Ref. |
|---|---|---|---|---|---|
| MALAT1 | 11q13.1 | Cell proliferation, migration, tube formation, vascular permeability, retinal vascularization, pericytes loss, capillary degeneration, microvascular leakage, oxidative stress, inflammation | miR-124, miR-20b, miR-17-3p, miR-124-3p, miR-125b-5p, miR-200b-3p, miR-203a-3p, miR-205-5p, miR-378a-3p, miR-320a | CREB, p38 MAPK, PP2A, NRF2, KEAP1, TNF-α, IL-6, SAA3, GRP78, CHOP, PRC2 complex, VE-cadherin/β-catenin, CDH5, YAP1, VEGFA, HIF-1α, Pde6g, Guca1a, Rho, Sag, Prph2, MCP-1 | [30,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58] |
| MIAT | 22q12.1 | Cell proliferation, migration, tube formation, cell viability, apoptosis, microvascular permeability, vascular leakage, inflammation, pyroptosis | miR-150-5p, miR-29b, miR-342–3p | Casp3, AKT1, VEGF, NF-κB, IL-6, IL1B, TXNIP, CASP1, TGFB1 | [59,60,61,62,63,64,65,66,67,68,69,70,71,72] |
| ANRIL | 9p21.3 | Regulates VEGF expression, inflammation, apoptosis | miR-200b | VEGF, Bax, P65, Bcl-2, IL-1, IL-10, MCP-1, p300, EZH2 (PRC2 complex) | [62,82,83,84] |
| NEAT1 | 11q13.1 | Cell proliferation, cell apoptosis, epithelial-mesenchymal transition, oxidative stress, inflammation | miR-497, miR-204 | BCL2, BAX, SOX4, COX2, IL-6, TNF-α, VEGFA, TGF-β1, BDNF | [58,87,88,89] |
| HOTAIR | 12q13.13 | Angiogenesis, oxidative damage, mitochondrial alterations, retinal acellular capillaries, vascular leakage, proliferation, invasion, migration, permeability | miR-20b, miR-17-3p | LSD1, VE-cadherin, H3K4me3, HIF1α, VEGFA | [52,92,93] |
| KCNQ1OT1 | 11p15.5 | Proliferation, apotosis, epithelial-mesenchymal transition | miR-214, miR-1470 | CASP1, EGFR, IL-1β | [98,99,100] |
| HEIH | 5q35.3 | Apoptosis and cell damage | miR-939 | VEGF, PI3K/AKT1, CASP3, CYPC | [101,102] |
| RNCR3 | 8p23.1 | Retinal vascular functions, cytokines release | miR-185-5p | Klf2 | [103,104,105] |
| HOTTIP | 7p15.2 | Cell viability, apoptosis | VEGF, ICAM-1, P38/MAPK | [107] | |
| BDNF-AS | 11p14.1 | Apoptosis | BDNF | [108,109,110] | |
| FENDRR | 16q24.1 | Proliferation, angiogenesis | FOX1, VEGF | [111,112,113] | |
| IGF2AS | 11p15.5 | Apoptosis | IGF2/AKT, CASP-9 | [114] | |
| Arid2-IR | 12q12 | inflammation, oxidative stress, apoptosis | Smad-3, Bax, Bcl2 | [115] | |
| TDRG1 | 6p21.2 | Proliferation, permeability, migration, tube formation | VEGF | [116] | |
| AQP4-AS1 | 18q11.2 | Retinal neurodegeneration, capillary degeneration | AQP4 | [117] | |
| LINC00323 | 21q22.2 | Proliferation, angiogenesis | GATA2 | [118] | |
| MIR503HG | Xq26.3 | Proliferation, angiogenesis. | GATA2 | [118] | |
| HIF1A-AS2 | 14q23.2 | Hypoxia, oxidative stress, angiogenesis | HIFα, VEGF, MAPK | [119] | |
| LINC00174 | 7q11.21 | Proliferation, migration, angiogenesis | miR-150-5p | VEGFA | [120] |
| TUG1 | 22q12.2 | Proliferation, migration, tube formation | miR-145, miR-524-5p | VEGFA, FGFR2 | [121,124] |
| UCA1 | 19p13.12 | Epithelial-mesenchymal transition | miR-624-3p | VEGFC | [122] |
| LINC00963 | 9q34.11 | Proliferation, invasion, migration | miR-27b | [123] | |
| PVT1 | 8q24.21 | Proliferation, migration | miR-128-3p | [62,125] | |
| OGRU | chr9qA4 | Inflammation, oxidative stress | miR-320 | TGF-β1, USP14 | [126] |
| LncRNA | Chr | DR-Related Processes | Sponged miRNAs | Related Genes/Proteins | Ref. |
|---|---|---|---|---|---|
| MEG3 | 14q32.2 | Proliferation, migration, angiogenesis, oxidative stress, inflammation, neovascolarization | miR-34a, miR-223-3p, miR-204, miR-93, miR-19b, miR-6720-5p | NF-kB, DNMT1, PI3K, Akt, mTOR, IL-1β, VEGF, NRF2, SOCS6, CYB5R2, Sirt1, Notch1 | [129,130,131,132,133,134,135,136,137,138,139,140] |
| XIST | Xq13.2 | Apoptosis, migration, inflammation | miR-21-5p | VEGF, SIRT1 | [146,147] |
| H19 | 11p15.5 | Endothelial–mesenchymal transition, vascular leakage, inflammation | miR-675-3p, miR-675-5p, miR-200b, miR-93, miR-19b | XBP1, SIRT1, TGF-β1, Smad | [17,149,150,151,152,153,154,155,156,157,158] |
| LINC-PINT | 7q32.3 | Cell viability | P53 | [159,160] | |
| VIM-AS1 | 10p13 | Apoptosis | miR-29 | [161] | |
| LUADT1 | 6q24.3 | Apoptosis | miR-383 | PRX3 | [166] |
| AK077216 | 8p23.2 | Apoptosis | miR-383 | [167] | |
| RPSAP52 | 12q14.3 | Apoptosis | miR-365 | TIMP3 | [168] |
| ATP2B1-AS1 | 12q21.33 | Proliferation, migration, angiogenesis, permeability | miR-4729 | IQGAP2 | [167,169] |
| SOX2OT | 3q26.33 | Retinal neurodegeneration | [170] | ||
| FRMD6-AS2 | 14q22.1 | Proliferation, neovascularization | PBRM1, PPP2R5C, ASB | [171] | |
| MIR497HG | 17p13.1 | Proliferation, migration | miR-128-3p | SIRT1 | [172,173] |
| TPTEP1 | 22q.11.1 | Proliferation, migration | STAT3, VEGFA | [173] | |
| MCM3AP-AS1 | 21q22.3 | Apoptosis | miR-211 | SIRT1 | [174] |
| LINC00673 | 17q24.3 | Regulation of P53, apoptosis | P53 | [175] | |
| VEAL2 | 16p12.2 | Endothelial permeability | PRKCB | [176] | |
| BANCR | 9q21.11-q21.12 | Apoptosis | [177,178] | ||
| RP1-90L14.1 | 6q14.3 | Ciliary function | CE162 | [179] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cataldi, S.; Tramontano, M.; Costa, V.; Aprile, M.; Ciccodicola, A. Diabetic Retinopathy: Are lncRNAs New Molecular Players and Targets? Antioxidants 2022, 11, 2021. https://doi.org/10.3390/antiox11102021
Cataldi S, Tramontano M, Costa V, Aprile M, Ciccodicola A. Diabetic Retinopathy: Are lncRNAs New Molecular Players and Targets? Antioxidants. 2022; 11(10):2021. https://doi.org/10.3390/antiox11102021
Chicago/Turabian StyleCataldi, Simona, Mariagiovanna Tramontano, Valerio Costa, Marianna Aprile, and Alfredo Ciccodicola. 2022. "Diabetic Retinopathy: Are lncRNAs New Molecular Players and Targets?" Antioxidants 11, no. 10: 2021. https://doi.org/10.3390/antiox11102021
APA StyleCataldi, S., Tramontano, M., Costa, V., Aprile, M., & Ciccodicola, A. (2022). Diabetic Retinopathy: Are lncRNAs New Molecular Players and Targets? Antioxidants, 11(10), 2021. https://doi.org/10.3390/antiox11102021










