Peroxisomal PEX7 Receptor Affects Cadmium-Induced ROS and Auxin Homeostasis in Arabidopsis Root System
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. pex7-1 Mutant Screening Procedure
2.3. Root and Shoot Cd Determination by ICP-OES
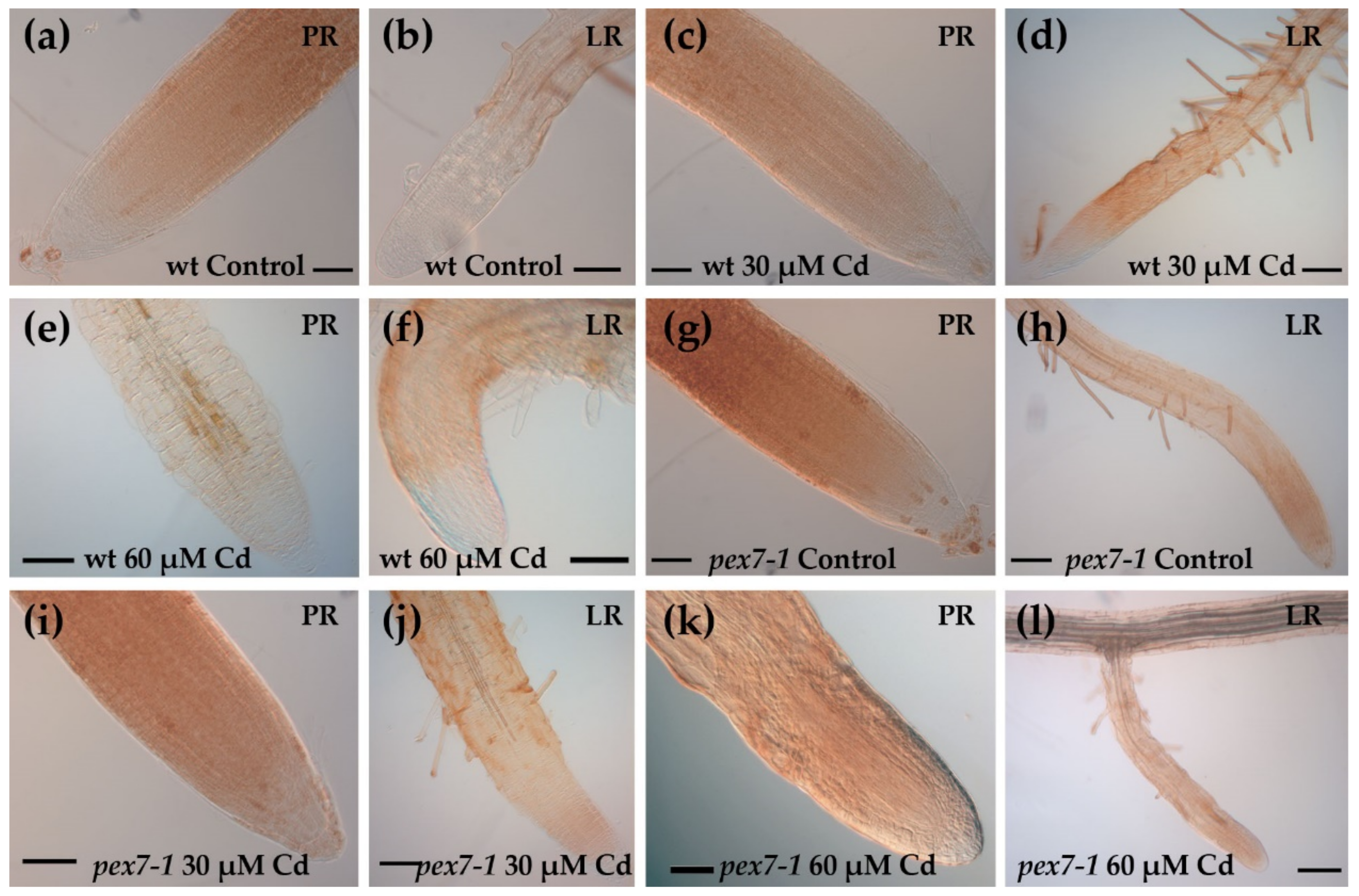
2.4. Root Morphological Analysis
2.5. Auxin Quantification
2.6. Root Protoplast Isolation and N-BODIPY Fluorescence Assay
2.7. In Situ Root ROS and RNS Visualization
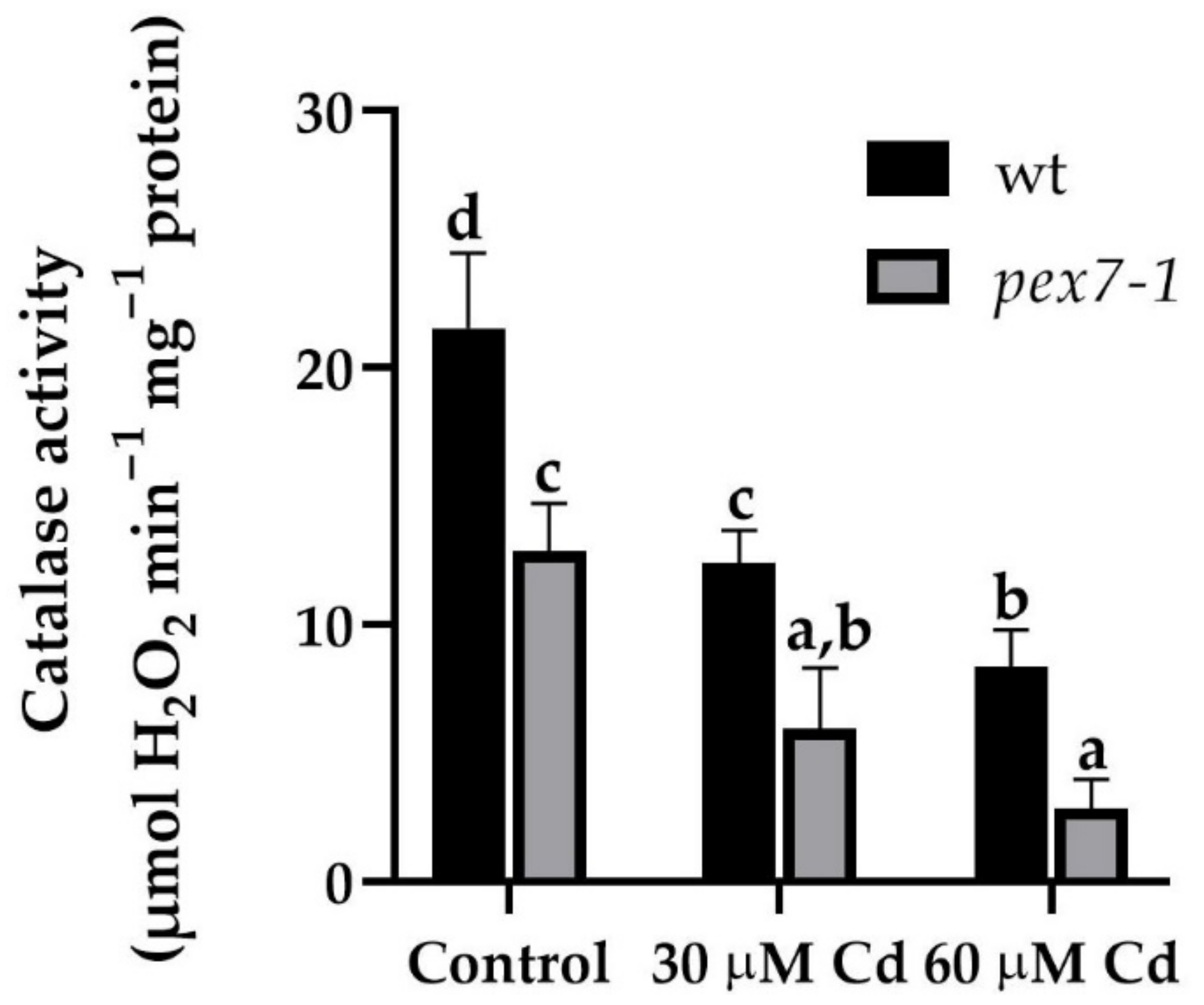
2.8. Catalase (CAT) Activity Assays
2.9. Statistical Analysis
3. Results
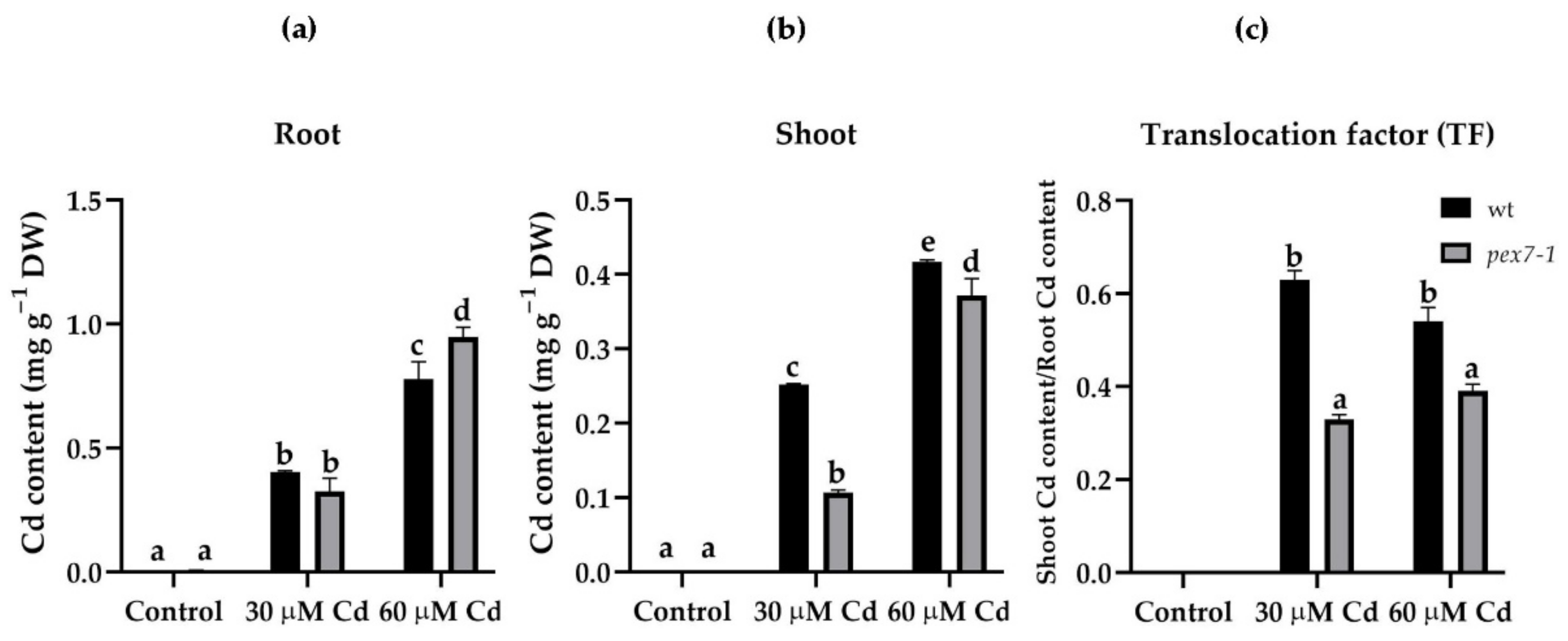
3.1. pex7-1 Mutation Reduces Cd Translocation from Root to Shoot
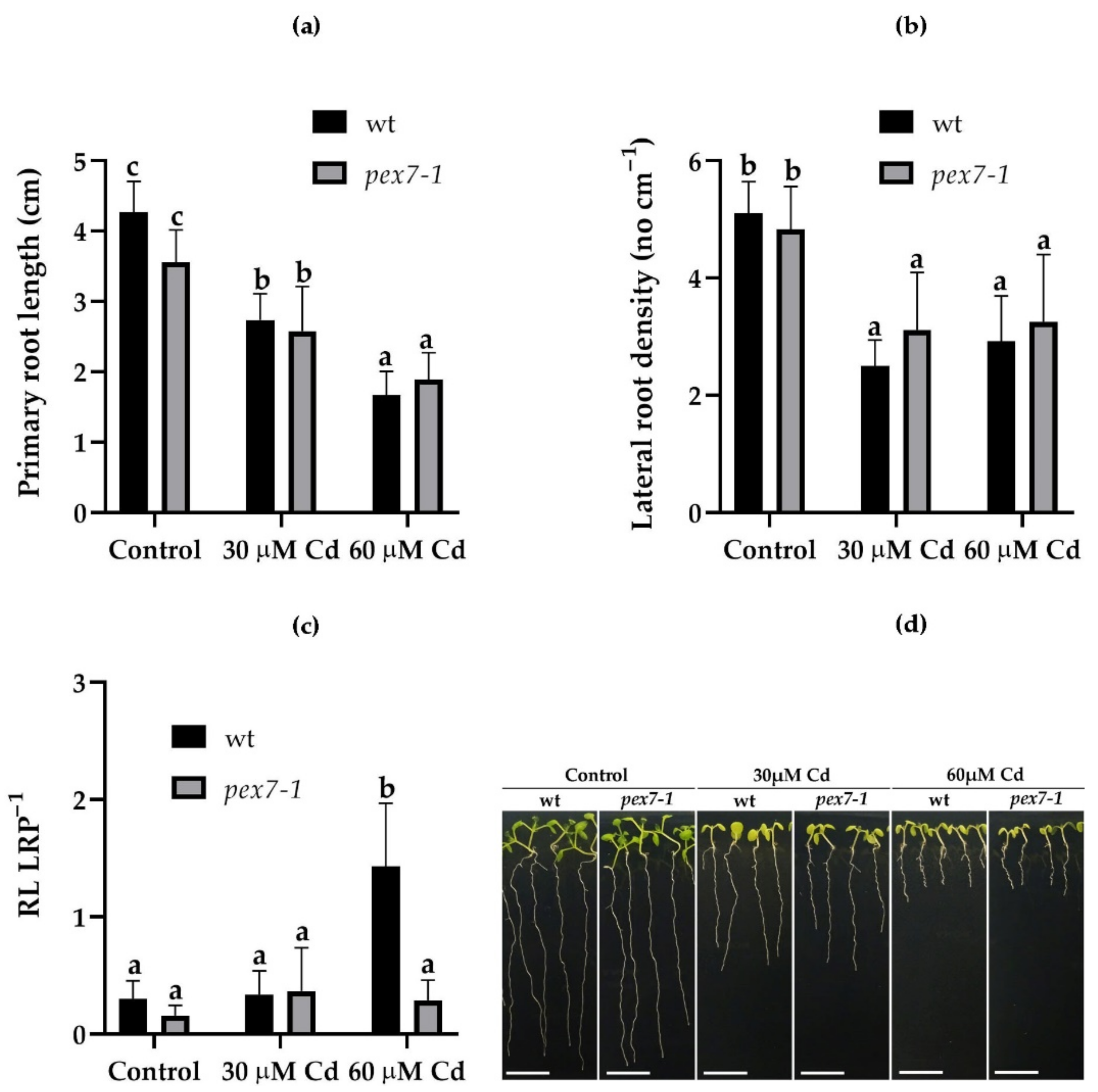
3.2. pex7-1 Mutation Reduces LR Elongation in Cd Presence
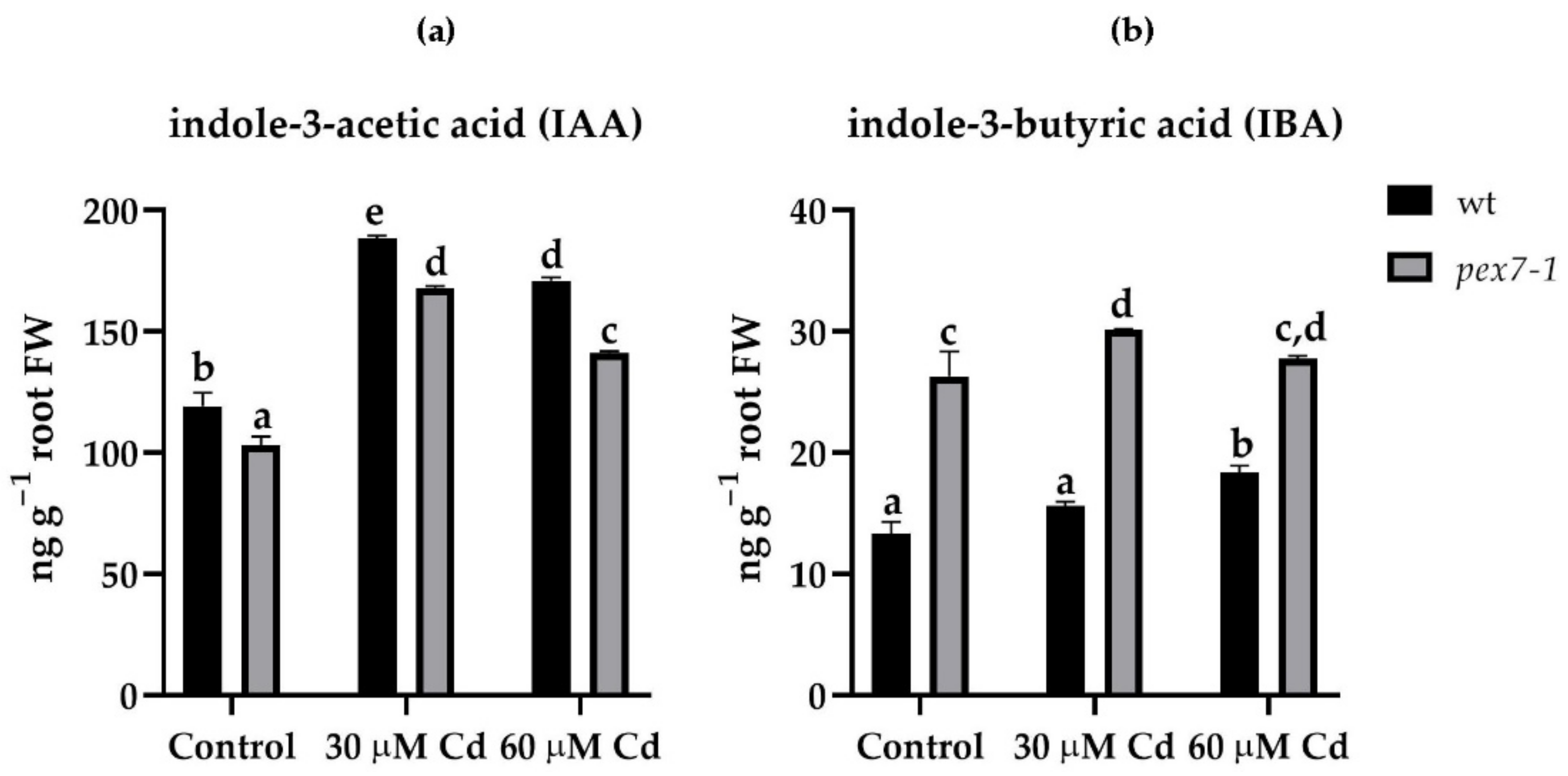
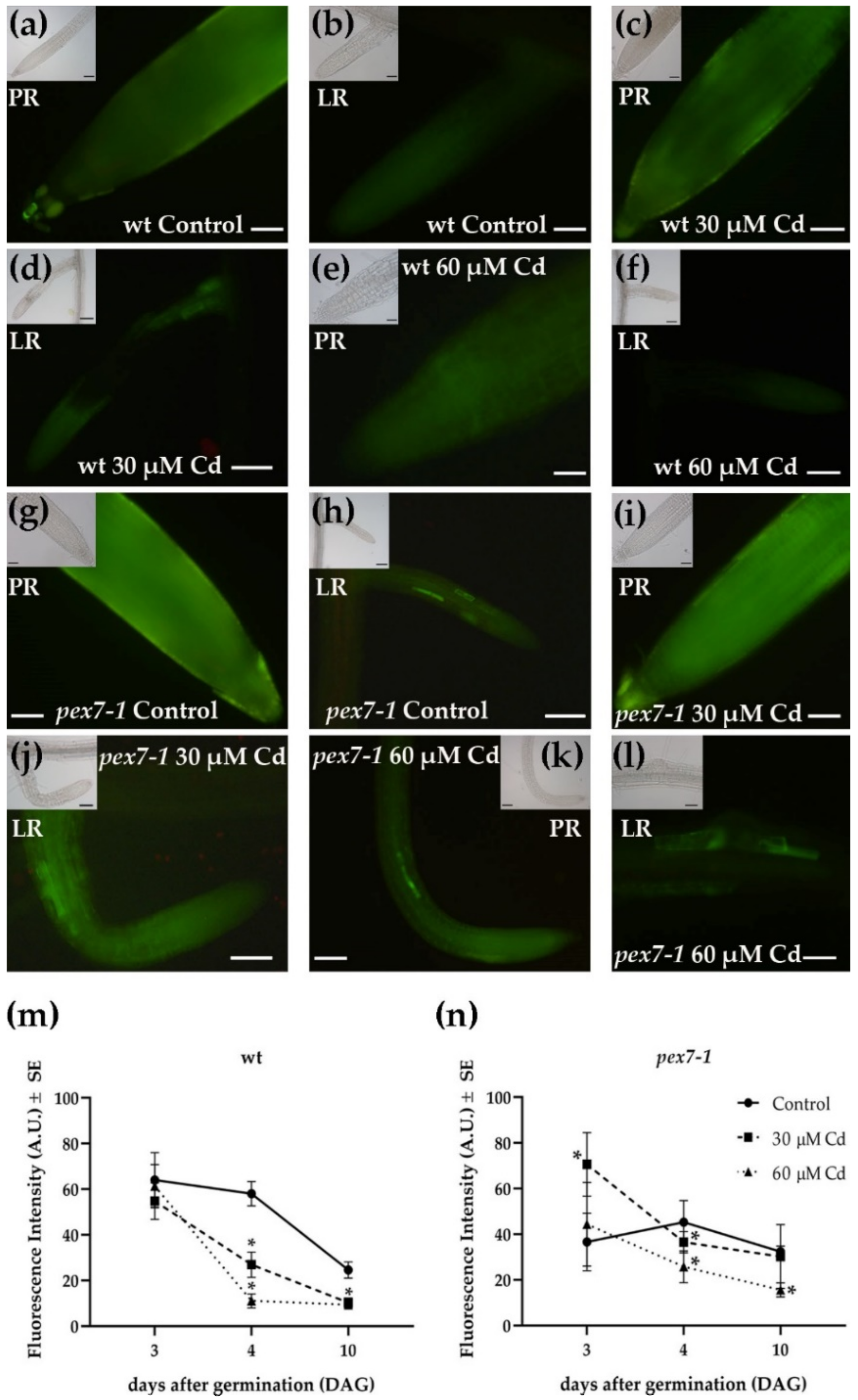
3.3. pex7-1 Mutation Reduces IBA to IAA Conversion
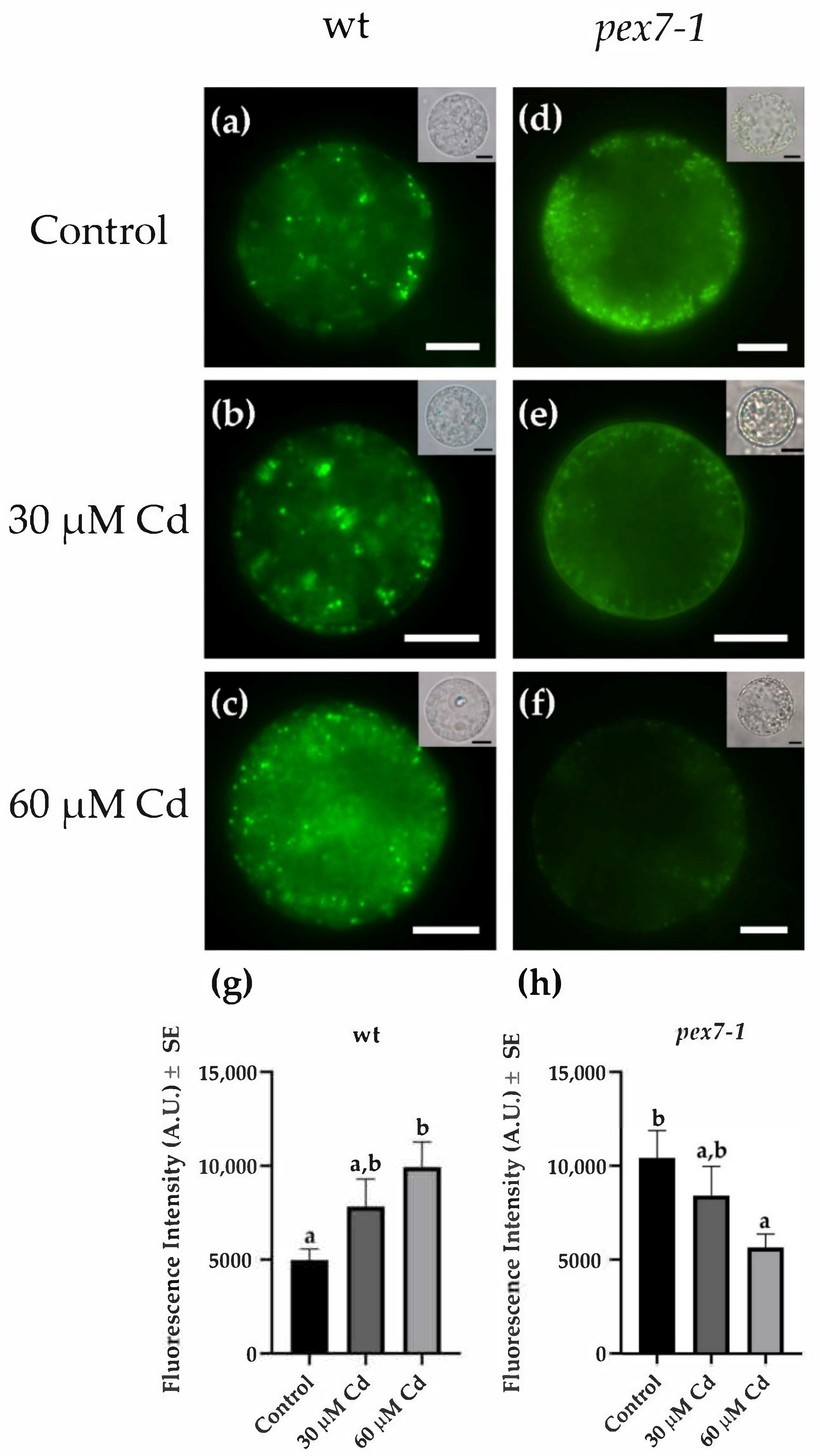
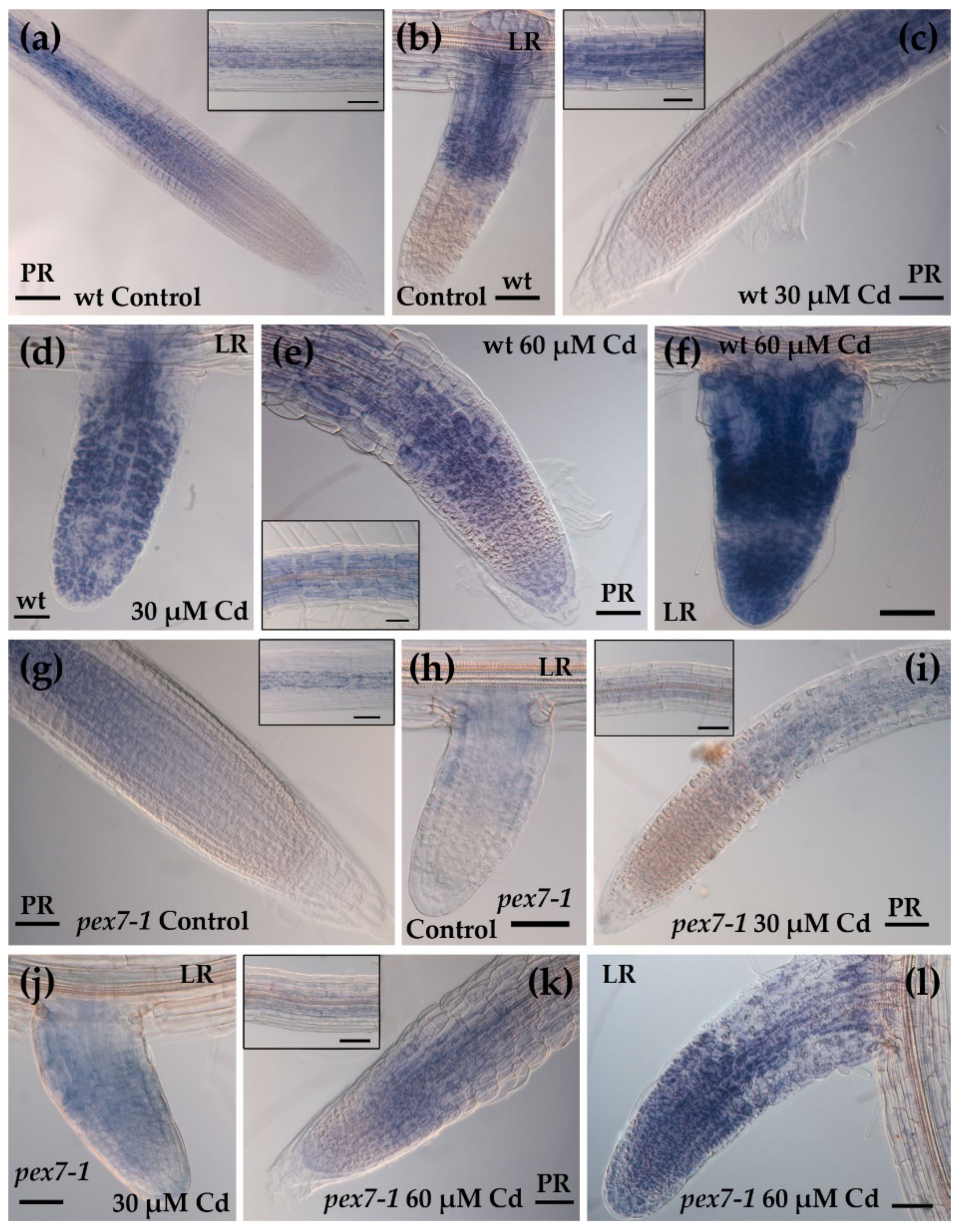
3.4. pex7-1 Mutation Reduces Cd-Induced Root Peroxisomal Signal
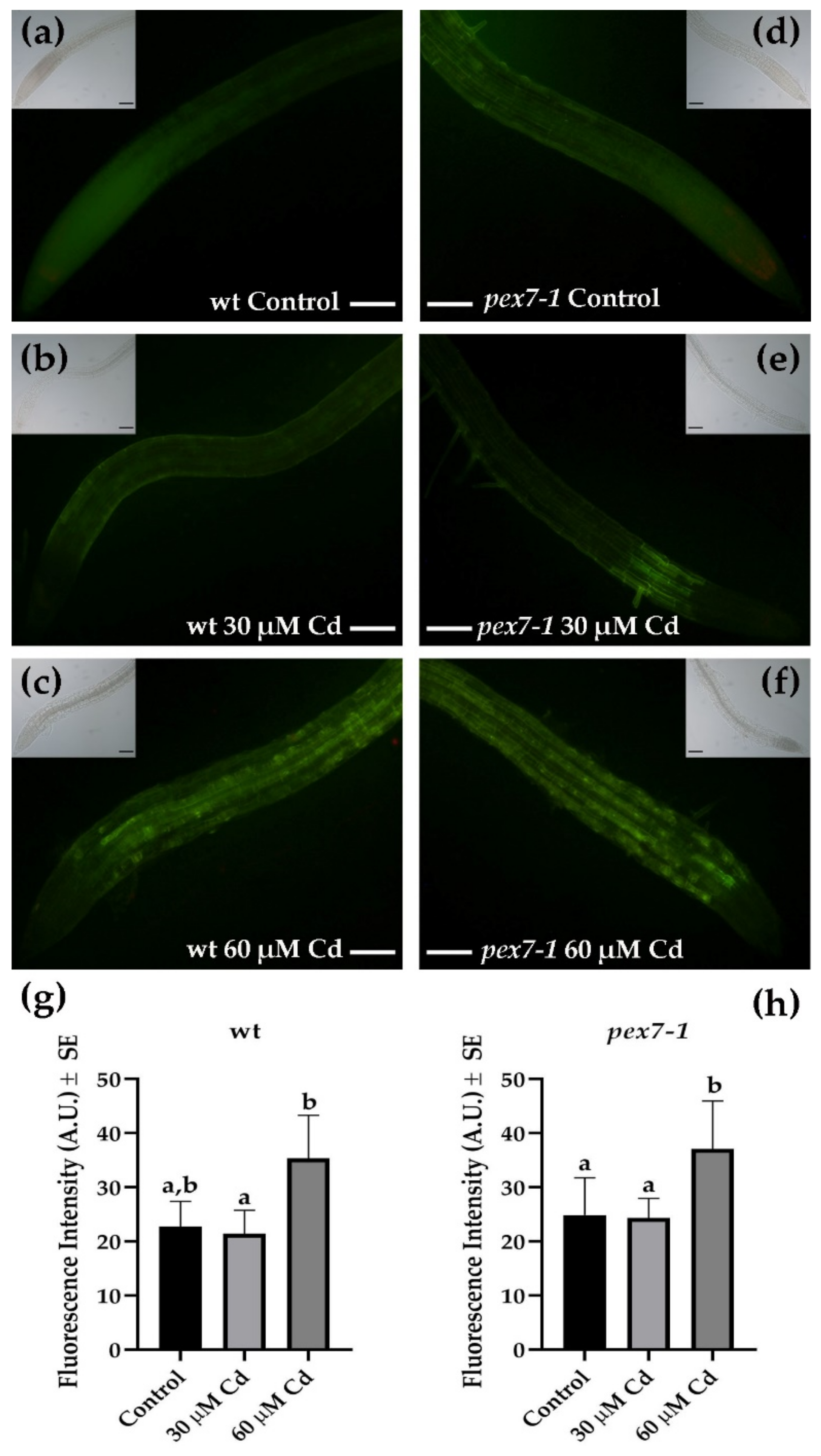
3.5. Cd-Induced Nitric Oxide and Peroxynitrite Levels Do Not Change in pex7-1 Roots Compared to wt
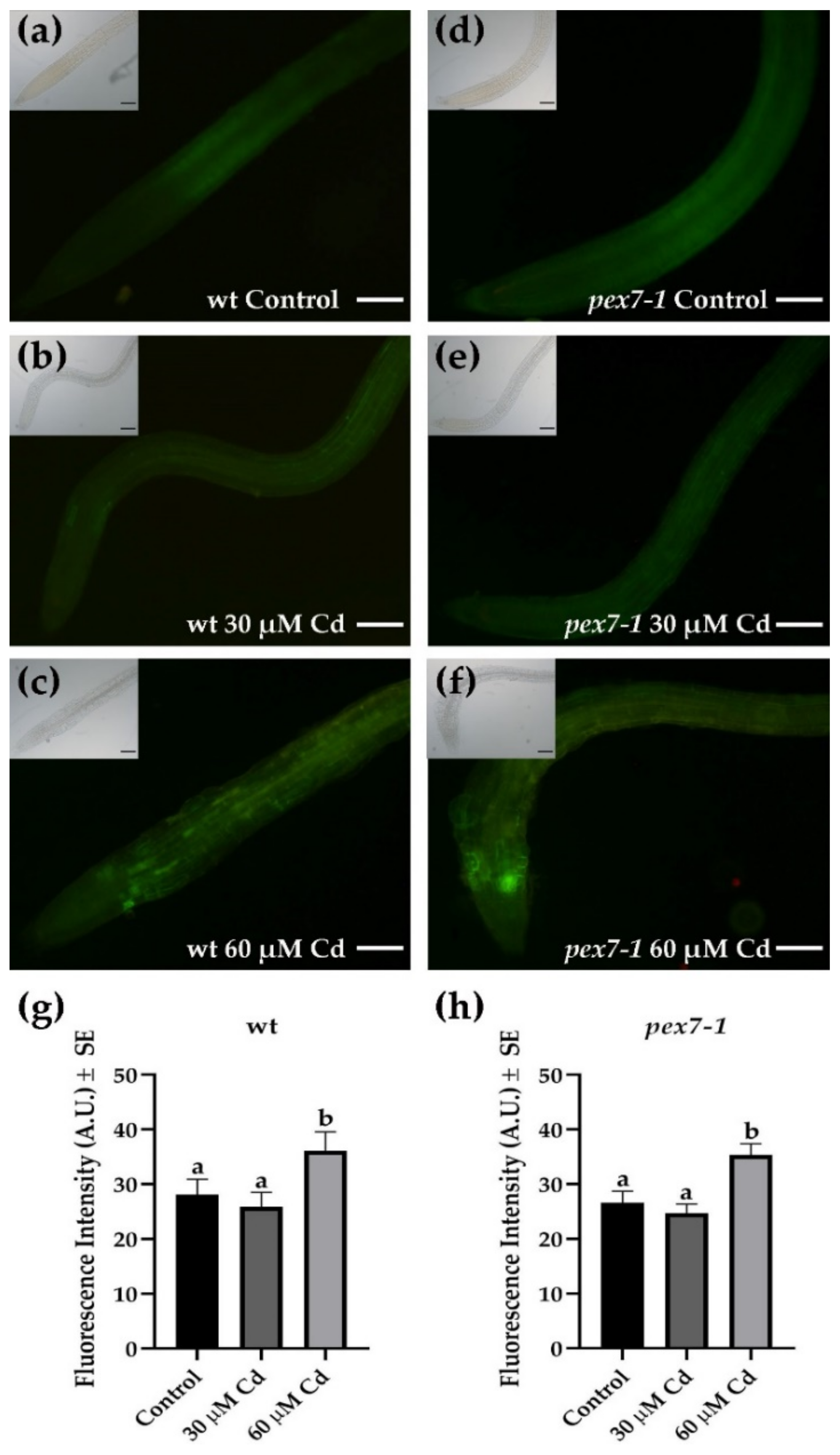
3.6. Cd-Induced Superoxide Anion is Affected by pex7-1 Mutation
3.7. pex7-1 Mutation Delays H2O2 Scavenging in Cd-Exposed Roots
4. Discussion
4.1. PEX7 Has a Morphogenic Role in the Root System in Response to Cd
4.2. Endogenous IAA and IBA Protect the Root system from Cd Stress
4.3. pex7-1 Mutation Negatively Affects Cellular Homeostasis of ROS, But Does Not Affect RNS Homeostasis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebeed, H.T.; Stevenson, S.R.; Cuming, A.C.; Baker, A. Conserved and Differential Transcriptional Responses of Peroxisome Associated Pathways to Drought, Dehydration and ABA. J. Exp. Bot. 2018, 69, 4971–4985. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Liu, J.; Hu, J. Peroxisomes in Plant Reproduction and Seed-Related Development. J. Integr. Plant Biol. 2019, 61, 784–802. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxisomal Plant Metabolism—An Update on Nitric Oxide, Ca2+ and the NADPH Recycling Network. J. Cell Sci. 2018, 131, jcs202978. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Peláez-Vico, M.A.; Molina-Moya, E.; Romero-Puertas, M.C. Peroxisomes as Redox-Signaling Nodes in Intracellular Communication and Stress Responses. Plant Physiol. 2021, 186, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Liu, J.; Wang, S.; Hu, J. Peroxisomes: Versatile Organelles with Diverse Roles in Plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef] [PubMed]
- Cross, L.L.; Ebeed, H.T.; Baker, A. Peroxisome Biogenesis, Protein Targeting Mechanisms and PEX Gene Functions in Plants. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 850–862. [Google Scholar] [CrossRef]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant Peroxisomes: Biogenesis and Function. Plant Cell 2012, 24, 2279–2303. [Google Scholar] [CrossRef]
- Kao, Y.-T.; Gonzalez, K.L.; Bartel, B. Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef]
- Reumann, S.; Chowdhary, G.; Lingner, T. Characterization, Prediction and Evolution of Plant Peroxisomal Targeting Signals Type 1 (PTS1s). Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 790–803. [Google Scholar] [CrossRef]
- Distel, B.; Erdmann, R.; Gould, S.J.; Blobel, G.; Crane, D.I.; Cregg, J.M.; Dodt, G.; Fujiki, Y.; Goodman, J.M.; Just, W.W.; et al. A Unified Nomenclature for Peroxisome Biogenesis Factors. J. Cell Biol. 1996, 135, 1–3. [Google Scholar] [CrossRef]
- Reumann, S.; Bartel, B. Plant Peroxisomes: Recent Discoveries in Functional Complexity, Organelle Homeostasis, and Morphological Dynamics. Curr. Opin. Plant Biol. 2016, 34, 17–26. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Sanz-Fernández, M.; Hu, J.; Sandalio, L.M. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016, 171, 1665–1674. [Google Scholar] [CrossRef]
- Cui, P.; Liu, H.; Islam, F.; Li, L.; Farooq, M.A.; Ruan, S.; Zhou, W. OsPEX11, a Peroxisomal Biogenesis Factor 11, Contributes to Salt Stress Tolerance in Oryza Sativa. Front. Plant Sci. 2016, 7, 1357. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. The Arabidopsis Peroxisomal Targeting Signal Type 2 Receptor PEX7 Is Necessary for Peroxisome Function and Dependent on PEX5. Mol. Biol. Cell 2005, 16, 573–583. [Google Scholar] [CrossRef]
- Zolman, B.K.; Martinez, N.; Millius, A.; Adham, A.R.; Bartel, B. Identification and Characterization of Arabidopsis Indole-3-Butyric Acid Response Mutants Defective in Novel Peroxisomal Enzymes. Genetics 2008, 180, 237–251. [Google Scholar] [CrossRef]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of Peroxisome Homeostasis and Its Role in Stress Response and Signaling in Plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Fukao, Y.; Hayashi, M.; Hara-Nishimura, I.; Nishimura, M. Novel Glyoxysomal Protein Kinase, GPK1, Identified by Proteomic Analysis of Glyoxysomes in Etiolated Cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 1002–1012. [Google Scholar] [CrossRef]
- Reumann, S.; Quan, S.; Aung, K.; Yang, P.; Manandhar-Shrestha, K.; Holbrook, D.; Linka, N.; Switzenberg, R.; Wilkerson, C.G.; Weber, A.P.M.; et al. In-Depth Proteome Analysis of Arabidopsis Leaf Peroxisomes Combined with in Vivo Subcellular Targeting Verification Indicates Novel Metabolic and Regulatory Functions of Peroxisomes. Plant Physiol. 2009, 150, 125–143. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin Biosynthesis and Storage Forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Strader, L.C.; Bartel, B. Transport and Metabolism of the Endogenous Auxin Precursor Indole-3-Butyric Acid. Mol. Plant 2011, 4, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Robert, S.; Kleine-Vehn, J. Auxin: Simply Complicated. J. Exp. Bot. 2013, 64, 2565–2577. [Google Scholar] [CrossRef]
- Veloccia, A.; Fattorini, L.; Della Rovere, F.; Sofo, A.; D’Angeli, S.; Betti, C.; Falasca, G.; Altamura, M.M. Ethylene and Auxin Interaction in the Control of Adventitious Rooting in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6445–6458. [Google Scholar] [CrossRef]
- Fattorini, L.; Veloccia, A.; Della Rovere, F.; D’Angeli, S.; Falasca, G.; Altamura, M.M. Indole-3-Butyric Acid Promotes Adventitious Rooting in Arabidopsis thaliana Thin Cell Layers by Conversion into Indole-3-Acetic Acid and Stimulation of Anthranilate Synthase Activity. BMC Plant Biol. 2017, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Della Rovere, F.; Fattorini, L.; Ronzan, M.; Falasca, G.; Altamura, M.M.; Betti, C. Jasmonic Acid Methyl Ester Induces Xylogenesis and Modulates Auxin-Induced Xylary Cell Identity with NO Involvement. Int. J. Mol. Sci. 2019, 20, 4469. [Google Scholar] [CrossRef]
- Fattorini, L.; Ronzan, M.; Piacentini, D.; Della Rovere, F.; De Virgilio, C.; Sofo, A.; Altamura, M.M.; Falasca, G. Cadmium and Arsenic Affect Quiescent Centre Formation and Maintenance in Arabidopsis thaliana Post-Embryonic Roots Disrupting Auxin Biosynthesis and Transport. Environ. Exp. Bot. 2017, 144, 37–48. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; Proia, A.; De Paolis, A.; Falasca, G.; Altamura, M.M.; Sanità di Toppi, L.; Costantino, P.; Cardarelli, M. Cadmium Tolerance and Phytochelatin Content of Arabidopsis Seedlings Over-Expressing the Phytochelatin Synthase Gene AtPCS1. J. Exp. Bot. 2011, 62, 5509–5519. [Google Scholar] [CrossRef]
- Bruno, L.; Pacenza, M.; Forgione, I.; Lamerton, L.R.; Greco, M.; Chiappetta, A.; Bitonti, M.B. In Arabidopsis thaliana Cadmium Impact on the Growth of Primary Root by Altering SCR Expression and Auxin-Cytokinin Cross-Talk. Front. Plant Sci. 2017, 8, 1323. [Google Scholar] [CrossRef]
- Piacentini, D.; Ronzan, M.; Fattorini, L.; Della Rovere, F.; Massimi, L.; Altamura, M.M.; Falasca, G. Nitric Oxide Alleviates Cadmium- but Not Arsenic-Induced Damages in Rice Roots. Plant Physiol. Biochem. 2020, 151, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Piacentini, D.; Della Rovere, F.; Sofo, A.; Fattorini, L.; Falasca, G.; Altamura, M.M. Nitric Oxide Cooperates With Auxin to Mitigate the Alterations in the Root System Caused by Cadmium and Arsenic. Front. Plant Sci. 2020, 11, 1182. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxynitrite (ONOO−) Is Endogenously Produced in Arabidopsis Peroxisomes and Is Overproduced under Cadmium Stress. Ann. Bot. 2014, 113, 87–96. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Zeiner, A.; Šamaj, J.; Takáč, T. Signaling Toward Reactive Oxygen Species-Scavenging Enzymes in Plants. Front. Plant Sci. 2021, 11, 618835. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; Van Breusegem, F.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-Mediated ROS Production Facilitates Lateral Root Emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef]
- Lv, B.; Tian, H.; Zhang, F.; Liu, J.; Lu, S.; Bai, M.; Li, C.; Ding, Z. Brassinosteroids Regulate Root Growth by Controlling Reactive Oxygen Species Homeostasis and Dual Effect on Ethylene Synthesis in Arabidopsis. PLoS Genet. 2018, 14, e1007144. [Google Scholar] [CrossRef] [PubMed]
- Del Río, L.A.; López-Huertas, E. ROS Generation in Peroxisomes and Its Role in Cell Signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Betti, C.; Della Rovere, F.; Piacentini, D.; Fattorini, L.; Falasca, G.; Altamura, M.M. Jasmonates, Ethylene and Brassinosteroids Control Adventitious and Lateral Rooting as Stress Avoidance Responses to Heavy Metals and Metalloids. Biomolecules 2021, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Piacentini, D.; Corpas, F.J.; D’Angeli, S.; Altamura, M.M.; Falasca, G. Cadmium and Arsenic-Induced-Stress Differentially Modulates Arabidopsis Root Architecture, Peroxisome Distribution, Enzymatic Activities and Their Nitric Oxide Content. Plant Physiol. Biochem. 2020, 148, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-C.; Zheng, S.-Q.; Yu, Z.-D.; Gao, X.; Shen, R.; Lu, Y.-T. WD40-REPEAT 5a Represses Root Meristem Growth by Suppressing Auxin Synthesis through Changes of Nitric Oxide Accumulation in Arabidopsis. Plant J. 2018, 93, 883–893. [Google Scholar] [CrossRef]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant Catalases: Peroxisomal Redox Guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Nazir, F.; Hussain, A.; Fariduddin, Q. Hydrogen Peroxide Modulate Photosynthesis and Antioxidant Systems in Tomato (Solanum lycopersicum L.) Plants under Copper Stress. Chemosphere 2019, 230, 544–558. [Google Scholar] [CrossRef]
- Nazir, F.; Hussain, A.; Fariduddin, Q. Interactive Role of Epibrassinolide and Hydrogen Peroxide in Regulating Stomatal Physiology, Root Morphology, Photosynthetic and Growth Traits in Solanum lycopersicum L. under Nickel Stress. Environ. Exp. Bot. 2019, 162, 479–495. [Google Scholar] [CrossRef]
- Nazir, F.; Fariduddin, Q.; Khan, T.A. Hydrogen Peroxide as a Signalling Molecule in Plants and Its Crosstalk with Other Plant Growth Regulators under Heavy Metal Stress. Chemosphere 2020, 252, 126486. [Google Scholar] [CrossRef]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakière, B.; Vanacker, H.; Miginiac-Maslow, M.; Breusegem, F.V.; Noctor, G. Conditional Oxidative Stress Responses in the Arabidopsis Photorespiratory Mutant Cat2 Demonstrate That Redox State Is a Key Modulator of Daylength-Dependent Gene Expression, and Define Photoperiod as a Crucial Factor in the Regulation of H2O2-Induced Cell Death. Plant J. 2007, 52, 640–657. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Gill, S.S.; Alharby, H.F.; Razafindrabe, B.H.N.; Fujita, M. Hydrogen Peroxide Pretreatment Mitigates Cadmium-Induced Oxidative Stress in Brassica napus L.: An Intrinsic Study on Antioxidant Defense and Glyoxalase Systems. Front. Plant Sci. 2017, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Yang, L.; Xu, C.; Shi, Z.; Shao, J.; Xian, M.; Chen, J. Cadmium Disrupts the Balance between Hydrogen Peroxide and Superoxide Radical by Regulating Endogenous Hydrogen Sulfide in the Root Tip of Brassica rapa. Front. Plant Sci. 2017, 8, 232. [Google Scholar] [CrossRef]
- Dunand, C.; Crèvecoeur, M.; Penel, C. Distribution of Superoxide and Hydrogen Peroxide in Arabidopsis Root and Their Influence on Root Development: Possible Interaction with Peroxidases. New Phytol. 2007, 174, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Heyno, E.; Klose, C.; Krieger-Liszkay, A. Origin of Cadmium-Induced Reactive Oxygen Species Production: Mitochondrial Electron Transfer versus Plasma Membrane NADPH Oxidase. New Phytol. 2008, 179, 687–699. [Google Scholar] [CrossRef]
- Manzano, C.; Pallero-Baena, M.; Casimiro, I.; De Rybel, B.; Orman-Ligeza, B.; Van Isterdael, G.; Beeckman, T.; Draye, X.; Casero, P.; del Pozo, J.C. The Emerging Role of Reactive Oxygen Species Signaling during Lateral Root Development. Plant Physiol. 2014, 165, 1105–1119. [Google Scholar] [CrossRef]
- Mullen, R.T.; Lee, M.S.; Trelease, R.N. Identification of the Peroxisomal Targeting Signal for Cottonseed Catalase. Plant J. 1997, 12, 313–322. [Google Scholar] [CrossRef]
- Del Río, L.A.; Sandalio, L.M.; Corpas, F.J.; Palma, J.M.; Barroso, J.B. Reactive Oxygen Species and Reactive Nitrogen Species in Peroxisomes. Production, Scavenging, and Role in Cell Signaling. Plant Physiol. 2006, 141, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Wang, P.; Li, H.; Zhao, Y.; Lu, Y.; Dai, P.; Ren, T.; Wang, X.; Li, X.; Shao, Q.; et al. The Arabidopsis Catalase Triple Mutant Reveals Important Roles of Catalases and Peroxisome-Derived Signaling in Plant Development. J. Integr. Plant Biol. 2018, 60, 591–607. [Google Scholar] [CrossRef]
- Palma, J.M.; Gómez, M.; Yáñez, J.; Del Río, L.A. Increased Levels of Peroxisomal Active Oxygen-Related Enzymes in Copper-Tolerant Pea Plants 1. Plant Physiol. 1987, 85, 570–574. [Google Scholar] [CrossRef]
- Zong, H.; Liu, S.; Xing, R.; Chen, X.; Li, P. Protective Effect of Chitosan on Photosynthesis and Antioxidative Defense System in Edible Rape (Brassica rapa L.) in the Presence of Cadmium. Ecotoxicol. Environ. Saf. 2017, 138, 271–278. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase Function in Plants: A Focus on Arabidopsis Mutants as Stress-Mimic Models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Jia, W.; Zhang, J. AtMEK1 Mediates Stress-Induced Gene Expression of CAT1 Catalase by Triggering H2O2 Production in Arabidopsis. J. Exp. Bot. 2007, 58, 2969–2981. [Google Scholar] [CrossRef]
- Zou, J.-J.; Li, X.-D.; Ratnasekera, D.; Wang, C.; Liu, W.-X.; Song, L.-F.; Zhang, W.-Z.; Wu, W.-H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 Function in Abscisic Acid-Mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Isono, K.; Sakata, Y.; Taji, T. CATALASE2 Plays a Crucial Role in Long-Term Heat Tolerance of Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2021, 534, 747–751. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Lead-Induced Stress, Which Triggers the Production of Nitric Oxide (NO) and Superoxide Anion (O2−) in Arabidopsis Peroxisomes, Affects Catalase Activity. Nitric Oxide 2017, 68, 103–110. [Google Scholar] [CrossRef]
- Bueso, E.; Alejandro, S.; Carbonell, P.; Perez-Amador, M.A.; Fayos, J.; Bellés, J.M.; Rodriguez, P.L.; Serrano, R. The Lithium Tolerance of the Arabidopsis Cat2 Mutant Reveals a Cross-Talk between Oxidative Stress and Ethylene. Plant J. 2007, 52, 1052–1065. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J.; et al. Lack of GLYCOLATE OXIDASE1, but Not GLYCOLATE OXIDASE2, Attenuates the Photorespiratory Phenotype of CATALASE2-Deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Kerchev, P.I.; Mühlenbock, P.; Hoeberichts, F.A.; Van Der Kelen, K.; Mhamdi, A.; Willems, P.; Denecker, J.; Kumpf, R.P.; Noctor, G.; et al. SHORT-ROOT Deficiency Alleviates the Cell Death Phenotype of the Arabidopsis Catalase2 Mutant under Photorespiration-Promoting Conditions. Plant Cell 2016, 28, 1844–1859. [Google Scholar] [CrossRef]
- Yang, Z.; Mhamdi, A.; Noctor, G. Analysis of Catalase Mutants Underscores the Essential Role of CATALASE2 for Plant Growth and Day Length-Dependent Oxidative Signalling. Plant Cell Environ. 2019, 42, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Kessel-Vigelius, S.K.; Wiese, J.; Schroers, M.G.; Wrobel, T.J.; Hahn, F.; Linka, N. An Engineered Plant Peroxisome and Its Application in Biotechnology. Plant Sci. Int. J. Exp. Plant Biol. 2013, 210, 232–240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-Inducible Expression of the ABC-Type Transporter AtABCC3 Increases Phytochelatin-Mediated Cadmium Tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [PubMed]
- Canepari, S.; Cardarelli, E.; Pietrodangelo, A.; Strincone, M. Determination of Metals, Metalloids and Non-Volatile Ions in Airborne Particulate Matter by a New Two-Step Sequential Leaching Procedure: Part B: Validation on Equivalent Real Samples. Talanta 2006, 69, 588–595. [Google Scholar] [CrossRef]
- Rezvani, M.; Zaefarian, F. Bioaccumulation and Translocation Factors of Cadmium and Lead in “Aeluropus Littoralis”. Aust. J. Agric. Eng. 2011, 2, 114–119. [Google Scholar] [CrossRef]
- Lindberg, S.; Landberg, T.; Greger, M. A New Method to Detect Cadmium Uptake in Protoplasts. Planta 2004, 219, 526–532. [Google Scholar] [CrossRef]
- Fahy, D.; Sanad, M.N.M.E.; Duscha, K.; Lyons, M.; Liu, F.; Bozhkov, P.; Kunz, H.-H.; Hu, J.; Neuhaus, H.E.; Steel, P.G.; et al. Impact of Salt Stress, Cell Death, and Autophagy on Peroxisomes: Quantitative and Morphological Analyses Using Small Fluorescent Probe N-BODIPY. Sci. Rep. 2017, 7, 39069. [Google Scholar] [CrossRef]
- Piacentini, D.; Falasca, G.; Canepari, S.; Massimi, L. Potential of PM-Selected Components to Induce Oxidative Stress and Root System Alteration in a Plant Model Organism. Environ. Int. 2019, 132, 105094. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Oxygen Radicals in Biological Systems; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Tran, T.A.; Popova, L.P. Functions and Toxicity of Cadmium in Plants: Recent Advances and Future Prospects. Turk. J. Bot. 2013, 37, 1–13. [Google Scholar]
- Kubier, A.; Wilkin, R.T.; Pichler, T. Cadmium in Soils and Groundwater: A Review. Appl. Geochem. 2019, 108, 104388. [Google Scholar] [CrossRef]
- Chaffei, C.; Pageau, K.; Suzuki, A.; Gouia, H.; Ghorbel, M.H.; Masclaux-Daubresse, C. Cadmium Toxicity Induced Changes in Nitrogen Management in Lycopersicon Esculentum Leading to a Metabolic Safeguard Through an Amino Acid Storage Strategy. Plant Cell Physiol. 2004, 45, 1681–1693. [Google Scholar] [CrossRef]
- Ramón, N.M.; Bartel, B. Interdependence of the Peroxisome-Targeting Receptors in Arabidopsis thaliana: PEX7 Facilitates PEX5 Accumulation and Import of PTS1 Cargo into Peroxisomes. Mol. Biol. Cell 2010, 21, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Nakamori, C.; Nito, K.; Kondo, M.; Nishimura, M. The Arabidopsis Pex12 and Pex13 Mutants Are Defective in Both PTS1- and PTS2-Dependent Protein Transport to Peroxisomes. Plant J. 2006, 47, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Frick, E.M.; Strader, L.C. Roles for IBA-Derived Auxin in Plant Development. J. Exp. Bot. 2018, 69, 169–177. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, J.; Zheng, L.; Wang, Y.; Luo, L.; Ma, M.; Zhang, C.; Han, Y.; Beeckman, T.; Xu, G.; et al. Cadmium Stress Suppresses Lateral Root Formation by Interfering with the Root Clock. Plant Cell Environ. 2019, 42, 3182–3196. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, M.V.; Salguero, J.; Lloret, P.G. Auxin Modulated Initiation of Lateral Roots Is Linked to Pericycle Cell Length in Maize. Front. Plant Sci. 2019, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Rolli, E.; Brunoni, F.; Dramis, L.; Sacco, E.; Fattorini, L.; Ruffoni, B.; Díaz-Sala, C.; Altamura, M.M. 1,3-Di(Benzo[d]Oxazol-5-Yl)Urea Acts as Either Adventitious Rooting Adjuvant or Xylogenesis Enhancer in Carob and Pine Microcuttings Depending on the Presence/Absence of Exogenous Indole-3-Butyric Acid. Plant Cell Tissue Organ Cult. PCTOC 2016, 126, 411–427. [Google Scholar] [CrossRef]
- Zolman, B.K.; Yoder, A.; Bartel, B. Genetic Analysis of Indole-3-Butyric Acid Responses in Arabidopsis thaliana Reveals Four Mutant Classes. Genetics 2000, 156, 1323–1337. [Google Scholar] [CrossRef]
- Strader, L.C.; Culler, A.H.; Cohen, J.D.; Bartel, B. Conversion of Endogenous Indole-3-Butyric Acid to Indole-3-Acetic Acid Drives Cell Expansion in Arabidopsis Seedlings. Plant Physiol. 2010, 153, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, L.; Hause, B.; Gutierrez, L.; Veloccia, A.; Della Rovere, F.; Piacentini, D.; Falasca, G.; Altamura, M.M. Jasmonate Promotes Auxin-Induced Adventitious Rooting in Dark-Grown Arabidopsis thaliana Seedlings and Stem Thin Cell Layers by a Cross-Talk with Ethylene Signalling and a Modulation of Xylogenesis. BMC Plant Biol. 2018, 18, 182. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Schubert, B.; Pieper, K. Regulation of IBA Synthetase from Maize (Zea mays L.) by Drought Stress and ABA. J. Exp. Bot. 1995, 46, 423–432. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Ronzan, M.; Piacentini, D.; Fattorini, L.; Federica, D.R.; Caboni, E.; Eiche, E.; Ziegler, J.; Hause, B.; Riemann, M.; Betti, C.; et al. Auxin-Jasmonate Crosstalk in Oryza sativa L. Root System Formation after Cadmium and/or Arsenic Exposure. Environ. Exp. Bot. 2019, 165, 59–69. [Google Scholar] [CrossRef]
- Bashri, G.; Singh, S.; Prasad, S.M.; Ansari, M.J.; Usmani, S.; Alfarraj, S.; Alharbi, S.A.; Brestic, M. Kinetin Mitigates Cd-Induced Damages to Growth, Photosynthesis and PS II Photochemistry of Trigonella Seedlings by up-Regulating Ascorbate-Glutathione Cycle. PLoS ONE 2021, 16, e0249230. [Google Scholar] [CrossRef] [PubMed]
- Demecsová, L.; Tamás, L. Reactive Oxygen Species, Auxin and Nitric Oxide in Metal-Stressed Roots: Toxicity or Defence. BioMetals 2019, 32, 717–744. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-M.; Huang, X. Inhibition of Root Meristem Growth by Cadmium Involves Nitric Oxide-Mediated Repression of Auxin Accumulation and Signalling in Arabidopsis. Plant Cell Environ. 2016, 39, 120–135. [Google Scholar] [CrossRef] [PubMed]
- Terrón-Camero, L.C.; Rodríguez-Serrano, M.; Sandalio, L.M.; Romero-Puertas, M.C. Nitric Oxide Is Essential for Cadmium-Induced Peroxule Formation and Peroxisome Proliferation. Plant Cell Environ. 2020, 43, 2492–2507. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Terrón-Camero, L.C.; Peláez-Vico, M.Á.; Olmedilla, A.; Sandalio, L.M. Reactive Oxygen and Nitrogen Species as Key Indicators of Plant Responses to Cd Stress. Environ. Exp. Bot. 2019, 161, 107–119. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxisomal Plant Nitric Oxide Synthase (NOS) Protein Is Imported by Peroxisomal Targeting Signal Type 2 (PTS2) in a Process That Depends on the Cytosolic Receptor PEX7 and Calmodulin. FEBS Lett. 2014, 588, 2049–2054. [Google Scholar] [CrossRef] [PubMed]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J. A Physiological Perspective on Targets of Nitration in NO-Based Signaling Networks in Plants. J. Exp. Bot. 2019, 70, 4379–4389. [Google Scholar] [CrossRef] [PubMed]
- Abozeid, A.; Ying, Z.; Lin, Y.; Liu, J.; Zhang, Z.; Tang, Z. Ethylene Improves Root System Development under Cadmium Stress by Modulating Superoxide Anion Concentration in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Chaca, M.V.; Rodríguez-Serrano, M.; Molina, A.S.; Pedranzani, H.E.; Zirulnik, F.; Sandalio, L.M.; Romero-Puertas, M.C. Cadmium Induces Two Waves of Reactive Oxygen Species in Glycine max (L.) Roots. Plant Cell Environ. 2014, 37, 1672–1687. [Google Scholar] [CrossRef] [PubMed]
- Nyathi, Y.; Baker, A. Plant Peroxisomes as a Source of Signalling Molecules. Biochim. Biophys. Acta BBA Mol. Cell Res. 2006, 1763, 1478–1495. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Stress-Triggered Redox Signalling: What’s in PROSpect? Plant Cell Environ. 2016, 39, 951–964. [Google Scholar] [CrossRef]
- Reumann, S.; Ma, C.; Lemke, S.; Babujee, L. AraPerox. A Database of Putative Arabidopsis Proteins from Plant Peroxisomes. Plant Physiol. 2004, 136, 2587–2608. [Google Scholar] [CrossRef]
- Kamigaki, A.; Mano, S.; Terauchi, K.; Nishi, Y.; Tachibe-Kinoshita, Y.; Nito, K.; Kondo, M.; Hayashi, M.; Nishimura, M.; Esaka, M. Identification of Peroxisomal Targeting Signal of Pumpkin Catalase and the Binding Analysis with PTS1 Receptor. Plant J. 2003, 33, 161–175. [Google Scholar] [CrossRef]
- Walton, P.A.; Brees, C.; Lismont, C.; Apanasets, O.; Fransen, M. The Peroxisomal Import Receptor PEX5 Functions as a Stress Sensor, Retaining Catalase in the Cytosol in Times of Oxidative Stress. Biochim. Biophys. Acta BBA Mol. Cell Res. 2017, 1864, 1833–1843. [Google Scholar] [CrossRef]
- Cho, S.-C.; Chao, Y.-Y.; Hong, C.-Y.; Kao, C.H. The Role of Hydrogen Peroxide in Cadmium-Inhibited Root Growth of Rice Seedlings. Plant Growth Regul. 2012, 66, 27–35. [Google Scholar] [CrossRef]
- Okumoto, K.; El Shermely, M.; Natsui, M.; Kosako, H.; Natsuyama, R.; Marutani, T.; Fujiki, Y. The Peroxisome Counteracts Oxidative Stresses by Suppressing Catalase Import via Pex14 Phosphorylation. eLife 2020, 9, e55896. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piacentini, D.; Della Rovere, F.; Bertoldi, I.; Massimi, L.; Sofo, A.; Altamura, M.M.; Falasca, G. Peroxisomal PEX7 Receptor Affects Cadmium-Induced ROS and Auxin Homeostasis in Arabidopsis Root System. Antioxidants 2021, 10, 1494. https://doi.org/10.3390/antiox10091494
Piacentini D, Della Rovere F, Bertoldi I, Massimi L, Sofo A, Altamura MM, Falasca G. Peroxisomal PEX7 Receptor Affects Cadmium-Induced ROS and Auxin Homeostasis in Arabidopsis Root System. Antioxidants. 2021; 10(9):1494. https://doi.org/10.3390/antiox10091494
Chicago/Turabian StylePiacentini, Diego, Federica Della Rovere, Ilaria Bertoldi, Lorenzo Massimi, Adriano Sofo, Maria Maddalena Altamura, and Giuseppina Falasca. 2021. "Peroxisomal PEX7 Receptor Affects Cadmium-Induced ROS and Auxin Homeostasis in Arabidopsis Root System" Antioxidants 10, no. 9: 1494. https://doi.org/10.3390/antiox10091494
APA StylePiacentini, D., Della Rovere, F., Bertoldi, I., Massimi, L., Sofo, A., Altamura, M. M., & Falasca, G. (2021). Peroxisomal PEX7 Receptor Affects Cadmium-Induced ROS and Auxin Homeostasis in Arabidopsis Root System. Antioxidants, 10(9), 1494. https://doi.org/10.3390/antiox10091494










