H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Expression and Purification
2.3. H2S Production Assays
2.4. Measurement of CSE Activity Using S-Methylcysteine as a Substrate
2.5. Mass Spectrometry
2.6. Labeling of Unpaired Cysteine Thiols in CSE
2.7. Determination of Unpaired Cysteine Thiols in CSE
2.8. Molecular Dynamics (MD) Simulations of the Reduced CSE and the Oxidized CSE
2.9. Molecular Docking
2.10. Images of Endogenous H2S in Live Cells
3. Results
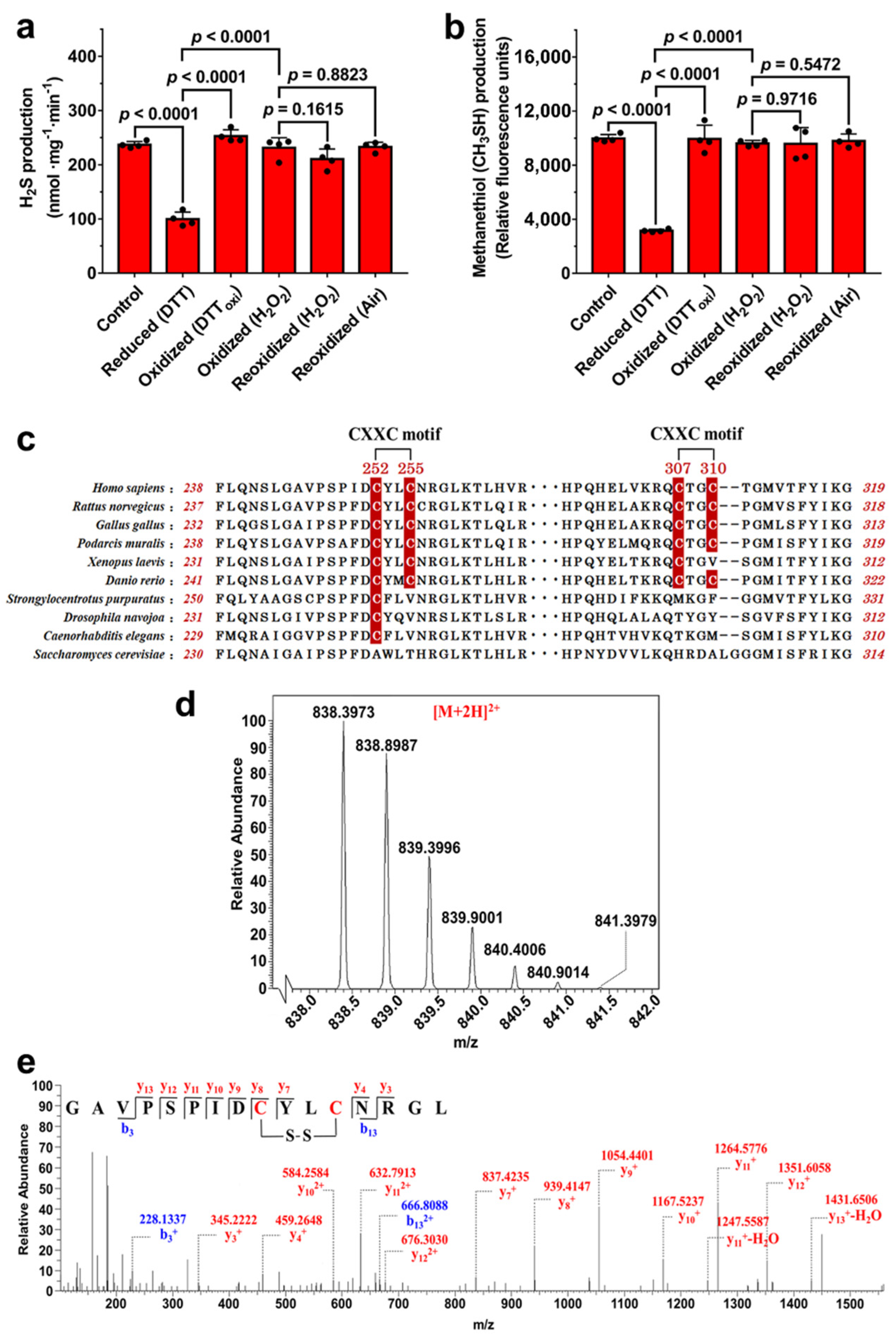
3.1. Potential Disulfide Bond Redox Switch Regulates the Activity of CSE
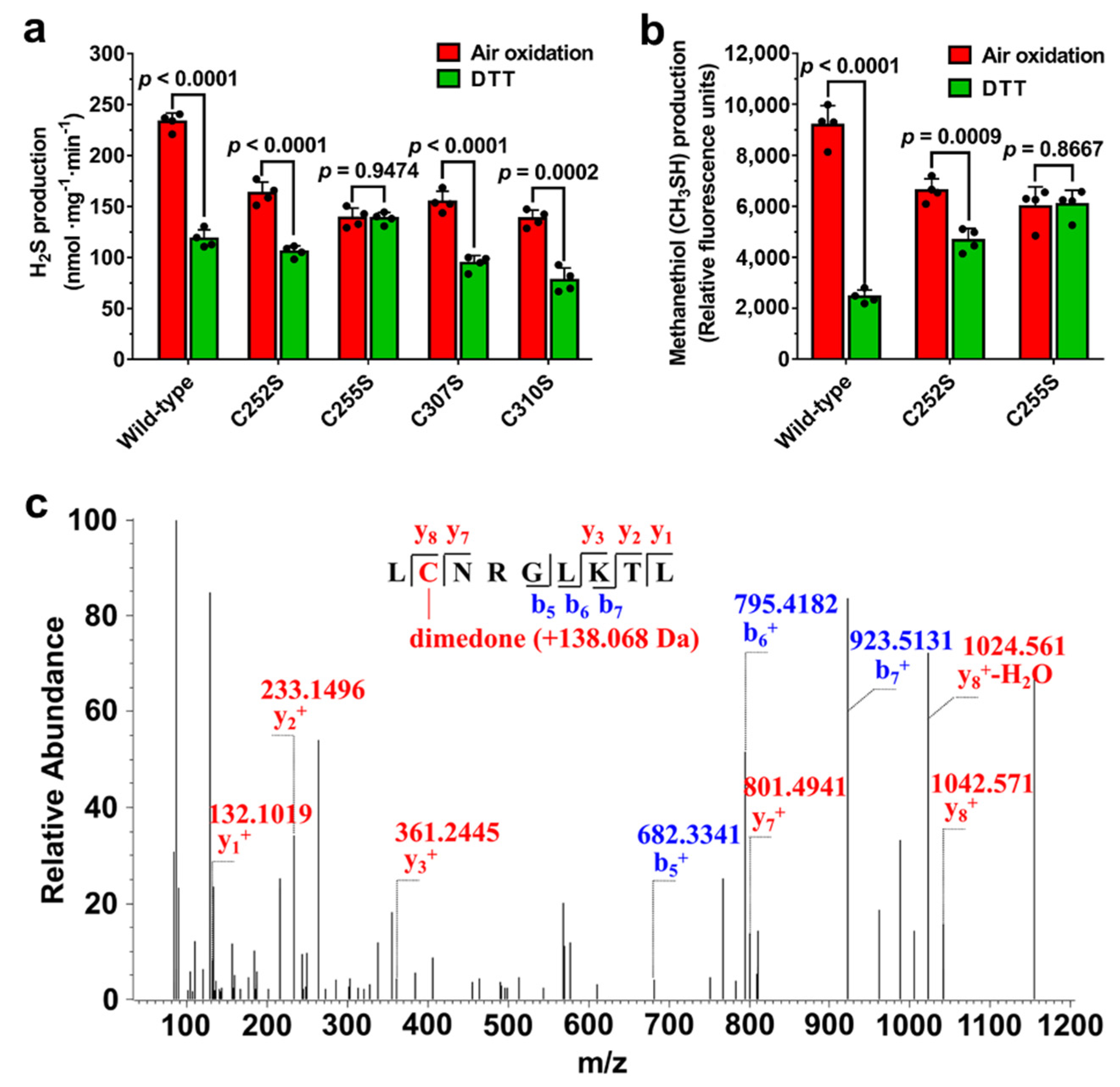
3.2. Disulfide Bond Formation between Cys252 and Cys255 in CXXC Motif
3.3. Residue Cys255 Is Crucial for Oxidation Sensing
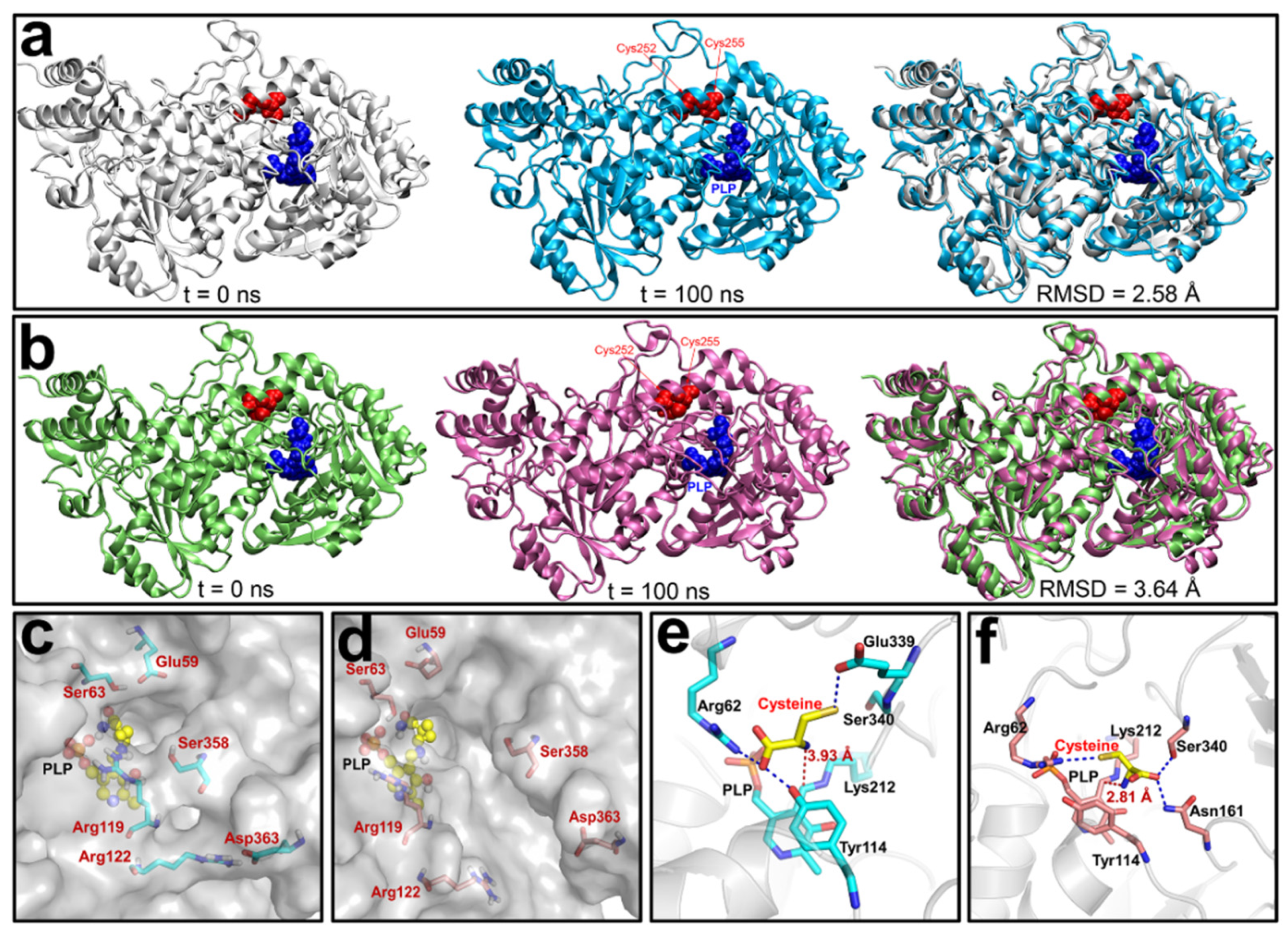
3.4. Molecular Dynamics (MD) Simulations of the Reduced and Oxidized CSE
3.5. Oxidative Stress Stimulates H2S Production via Enhancement of CSE Activity in HA-VSMCs
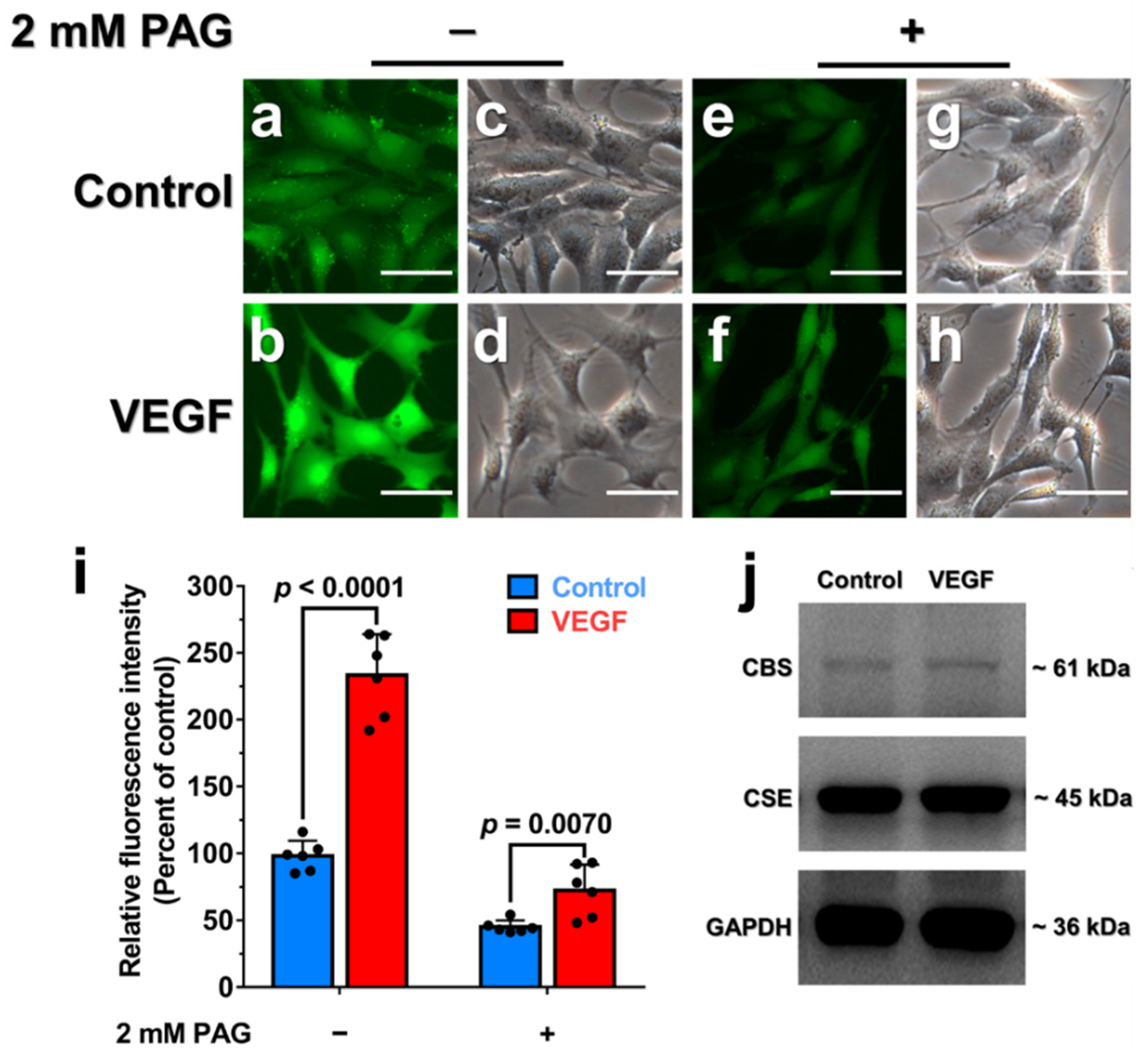
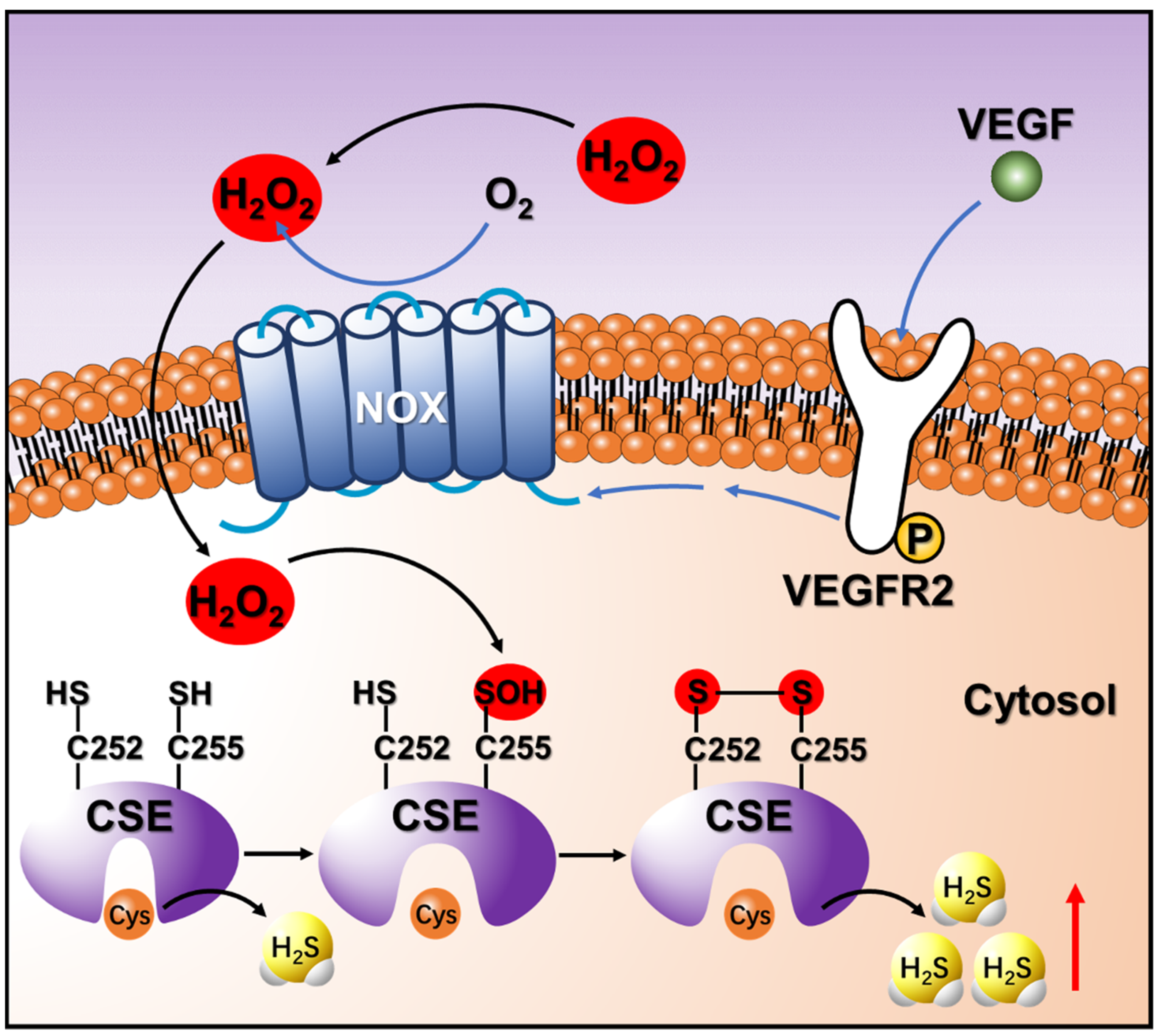
3.6. Endogenous H2O2 Triggered by VEGF Promotes Cellular H2S Production via the Enhancement of CSE Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kabil, O.; Banerjee, R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Sign. 2014, 20, 770–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, H. Hydrogen sulfide: From brain to gut. Antioxid. Redox Sign. 2010, 12, 1111–1123. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, R. H2S and blood vessels: An overview. Chem. Biochem. Pharmacol. Hydrog. Sulfide 2015, 230, 85–110. [Google Scholar]
- Pan, L.L.; Liu, X.H.; Gong, Q.H.; Yang, H.B.; Zhu, Y.Z. Role of cystathionine γ-lyase/hydrogen sulfide pathway in cardiovascular disease: A novel therapeutic strategy? Antioxid. Redox Sign. 2012, 17, 106–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, A.; Bailey, S.M. Redox biology of hydrogen sulfide: Implications for physiology, pathophysiology, and pharmacology. Redox Biol. 2013, 1, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Meng, G.; Ma, Y.; Xie, L.; Ferro, A.; Ji, Y. Emerging role of hydrogen sulfide in hypertension and related cardiovascular diseases. Br. J. Pharmacol. 2015, 172, 5501–5511. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, J.; Lu, Y.; Wang, R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO J. 2001, 20, 6008–6016. [Google Scholar] [CrossRef] [Green Version]
- Kraus, J.P.; Hasek, J.; Kozich, V.; Collard, R.; Venezia, S.; Janosíková, B.; Wang, J.; Stabler, S.P.; Allen, R.H.; Jakobs, C.; et al. Cystathionine γ-lyase: Clinical, metabolic, genetic, and structural studies. Mol. Genet. Metab. 2009, 97, 250–259. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Collins, R.; Huang, S.; Holmberg-Schiavone, L.; Anand, G.S.; Tan, C.H.; van-den-Berg, S.; Deng, L.W.; Moore, P.K.; Karlberg, T.; et al. Structural basis for the inhibition mechanism of human cystathionine γ-lyase, an enzyme responsible for the production of H2S. J. Biol. Chem. 2009, 284, 3076–3085. [Google Scholar] [CrossRef] [Green Version]
- Papapetropoulos, A.; Pyriochou, A.; Altaany, Z.; Yang, G.; Marazioti, A.; Zhou, Z.; Jeschke, M.G.; Branski, L.K.; Herndon, D.N.; Wang, R.; et al. Hydrogen sulfide is an endogenous stimulator of angiogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 21972–21977. [Google Scholar] [CrossRef] [Green Version]
- Lin, V.S.; Lippert, A.R.; Chang, C.J. Cell-trappable fluorescent probes for endogenous hydrogen sulfide signaling and imaging H2O2-dependent H2S production. Proc. Natl. Acad. Sci. USA 2013, 110, 7131–7135. [Google Scholar] [CrossRef] [Green Version]
- Niu, W.; Wu, P.; Chen, F.; Wang, J.; Shang, X.; Xu, C. Discovery of selective cystathionine beta-synthase inhibitors by high-throughput screening with a fluorescent thiol probe. Medchemcomm 2017, 8, 198–201. [Google Scholar] [CrossRef]
- Niu, W.; Yadav, P.K.; Adamec, J.; Banerjee, R. S-glutathionylation enhances human cystathionine β-synthase activity under oxidative stress conditions. Antioxid. Redox Sign. 2015, 22, 350–361. [Google Scholar] [CrossRef] [Green Version]
- Chiku, T.; Padovani, D.; Zhu, W.; Singh, S.; Vitvitsky, V.; Banerjee, R. H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 2009, 284, 11601–11612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Guo, X.; Li, H.; Qi, H.; Qian, J.; Yan, S.; Shi, J.; Niu, W. Hydrogen sulfide from cysteine desulfurase, not 3-mercaptopyruvate sulfurtransferase, contributes to sustaining cell growth and bioenergetics in E. coli under anaerobic conditions. Front. Microbiol. 2019, 10, 2357. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, J.; Lindsay, R.H. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman’s reagent. Anal. Biochem. 1968, 25, 192–205. [Google Scholar] [CrossRef]
- Niu, W.; Wang, J.; Qian, J.; Wang, M.; Wu, P.; Chen, F.; Yan, S. Allosteric control of human cystathionine β-synthase activity by a redox active disulfide bond. J. Biol. Chem. 2018, 293, 2523–2533. [Google Scholar] [CrossRef] [Green Version]
- Botello-Morte, L.; Bes, M.T.; Heras, B.; Fernάndez-Otal, A.; Peleato, M.L.; Fillat, M.F. Unraveling the redox properties of the global regulator FurA from Anabaena sp. PCC 7120: Disulfide reductase activity based on its CXXC motifs. Antioxid. Redox Sign. 2014, 20, 1396–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. Gromacs: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh ewald: An N·Log(N) method for ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Martonak, R.; Laio, A.; Parrinello, M. Predicting crystal structures: The parrinello-rahman method revisited. Phys. Rev. Lett. 2003, 90, 075503. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.J.P. MOPAC: A semiempirical molecular orbital program. J. Comput. Aid. Mol. Des. 1990, 4, 1–105. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods. III extension of PM3 to Be, Mg, Zn, Ga, Ge, As, Se, Cd, In, Sn, Sb, Te, Hg, Tl, Pb, and Bi. J. Comput. Chem. 1991, 12, 320–341. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model 1999, 17, 57–61. [Google Scholar] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehder, D.S.; Borges, C.R. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 2010, 49, 7748–7755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Cheng, Z.; Reddie, K.; Carroll, K.; Hammad, L.A.; Karty, J.A.; Bauer, C.E. RegB kinase activity is repressed by oxidative formation of cysteine sulfenic acid. J. Biol. Chem. 2013, 288, 4755–4762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballal, S.; Alvarez, B.; Turell, L.; Botti, H.; Freeman, B.A.; Radi, R. Sulfenic acid in human serum albumin. Amino Acids 2007, 32, 543–551. [Google Scholar] [CrossRef]
- Cheng, Z.; Wu, J.; Setterdahl, A.; Reddie, K.; Carroll, K.; Hammad, L.A.; Karty, J.A.; Bauer, C.E. Activity of the tetrapyrrole regulator CrtJ is controlled by oxidation of a redox active cysteine located in the DNA binding domain. Mol. Microbiol. 2012, 85, 734–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.Y.; Ma, G.; Su, Z. Native PAGE eliminates the problem of PEG-SDS interaction in SDS-PAGE and provides an alternative to HPLC in characterization of protein PEGylation. Electrophoresis 2007, 28, 2801–2807. [Google Scholar] [CrossRef] [PubMed]
- Tetsch, L.; Koller, C.; Dönhöfer, A.; Jung, K. Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 2011, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Zavaczki, E.; Jeney, V.; Agarwal, A.; Zarjou, A.; Oros, M.; Katko, M.; Varga, Z.; Balla, G.; Balla, J. Hydrogen sulfide inhibits the calcification and osteoblastic differentiation of vascular smooth muscle cells. Kidney Int. 2011, 80, 731–739. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.W.; Wang, X.Y.; Liang, X.H.; Wu, J.C.; Dong, S.Y.; Li, H.Z.; Jin, M.L.; Sun, D.J.; Zhang, W.H.; Zhong, X. Inhibition of hydrogen sulfide on the proliferation of vascular smooth muscle cells involved in the modulation of calcium sensing receptor in high homocysteine. Exp. Cell Res. 2016, 347, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.R.; Spokes, K.C.; Shih, S.C.; Aird, W.C. NADPH oxidase activity selectively modulates vascular endothelial growth factor signaling pathways. J. Biol. Chem. 2007, 282, 35373–35385. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.R.; Kwon, K.S.; Kim, S.R.; Rhee, S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998, 273, 15366–15372. [Google Scholar] [CrossRef] [Green Version]
- Meng, T.C.; Fukada, T.; Tonks, N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 2002, 9, 387–399. [Google Scholar] [CrossRef]
- Sbodio, J.I.; Snyder, S.H.; Paul, B.D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 2019, 176, 583–593. [Google Scholar] [CrossRef]
- Singh, S.; Padovani, D.; Leslie, R.A.; Chiku, T.; Banerjee, R. Relative contributions of cystathionine β-synthase and γ-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 2009, 284, 22457–22466. [Google Scholar] [CrossRef] [Green Version]
- Kabil, O.; Banerjee, R. Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 2010, 285, 21903–21907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Ding, L.; Xie, Z.Z.; Yang, Y.; Whiteman, M.; Moore, P.K.; Bian, J.S. A review of hydrogen sulfide synthesis, metabolism, and measurement: Is modulation of hydrogen sulfide a novel therapeutic for cancer? Antioxid. Redox Sign. 2019, 31, 1–38. [Google Scholar] [CrossRef]
- Smith, S. Exploring the Roles of the CXXC Motifs in Cystathionine γ-Lyase. Master’s Thesis, The University of Windsor, Windsor, ON, Canada, 2019. [Google Scholar]
- Poole, L.B.; Karplus, P.A.; Claiborne, A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 325–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Roos, G.; Messens, J. Protein sulfenic acid formation: From cellular damage to redox regulation. Free Radical Biol. Med. 2011, 51, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, L.; Montaut, S.; Yang, G. Hydrogen Sulfide Signaling Axis as a Target for Prostate Cancer Therapeutics. Prostate Cancer 2016, 2016, 8108549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, M.E.; Vranes, D.; Youssef, S.; Stacker, S.A.; Cox, A.J.; Rizkalla, B.; Casley, D.J.; Bach, L.A.; Kelly, D.J.; Gilbert, R.E. Increased renal expression of vascular endothelial growth factor (VEGF) and its receptor VEGFR-2 in experimental diabetes. Diabetes 1999, 48, 2229–2239. [Google Scholar] [CrossRef]
- Murphy, J.F.; Fitzgerald, D.J. Vascular endothelial cell growth factor (VEGF) induces cyclooxygenase (COX)-dependent proliferation of endothelial cells (EC) via the VEGF-2 receptor. FASEB J. 2001, 15, 1667–1669. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.D.; Wang, H.; Kho, S.H.; Rinkiko, S.; Sheng, X.; Shen, H.M.; Zhu, Y.Z. Hydrogen sulfide protects HUVECs against hydrogen peroxide induced mitochondrial dysfunction and oxidative stress. PLoS ONE 2013, 8, e53147. [Google Scholar] [CrossRef] [Green Version]
- Cheung, S.H.; Lau, J.Y.W. Hydrogen sulfide mediates athero-protection against oxidative stress via S-sulfhydration. PLoS ONE 2018, 13, e0194176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, N.; Moshal, K.S.; Sen, U.; Vacek, T.P.; Kumar, M.; Hughes, W.M.; Kundu, S.; Tyagi, S.C. H2S protects against methionine-induced oxidative stress in brain endothelial cells. Antioxid. Redox Sign. 2009, 11, 25–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, G.; Zhao, S.; Xie, L.; Han, Y.; Ji, Y. Protein S-sulfhydration by hydrogen sulfide in cardiovascular system. Br. J. Pharmacol. 2018, 175, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Fu, M.; Stokes, E.; Wu, L.; Yang, G. H2S-mediated protein S-sulfhydration: A prediction for its formation and regulation. Molecules 2017, 22, 1334. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Jia, G.; Li, H.; Yan, S.; Qian, J.; Guo, X.; Li, G.; Qi, H.; Zhu, Z.; Wu, Y.; et al. H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate. Antioxidants 2021, 10, 1488. https://doi.org/10.3390/antiox10091488
Wang J, Jia G, Li H, Yan S, Qian J, Guo X, Li G, Qi H, Zhu Z, Wu Y, et al. H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate. Antioxidants. 2021; 10(9):1488. https://doi.org/10.3390/antiox10091488
Chicago/Turabian StyleWang, Jun, Guanya Jia, Heng Li, Shasha Yan, Jing Qian, Xin Guo, Ge Li, Haizhen Qi, Zhilong Zhu, Yanjun Wu, and et al. 2021. "H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate" Antioxidants 10, no. 9: 1488. https://doi.org/10.3390/antiox10091488
APA StyleWang, J., Jia, G., Li, H., Yan, S., Qian, J., Guo, X., Li, G., Qi, H., Zhu, Z., Wu, Y., He, W., & Niu, W. (2021). H2O2-Mediated Oxidative Stress Enhances Cystathionine γ-Lyase-Derived H2S Synthesis via a Sulfenic Acid Intermediate. Antioxidants, 10(9), 1488. https://doi.org/10.3390/antiox10091488




