Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization
Abstract
1. Introduction
2. Materials and Methods
2.1. E. histolytica Culture
2.2. Adaptation of E. histolytica Trophozoites to AF
2.3. Growth Rate of WT Trophozoites and AFAT
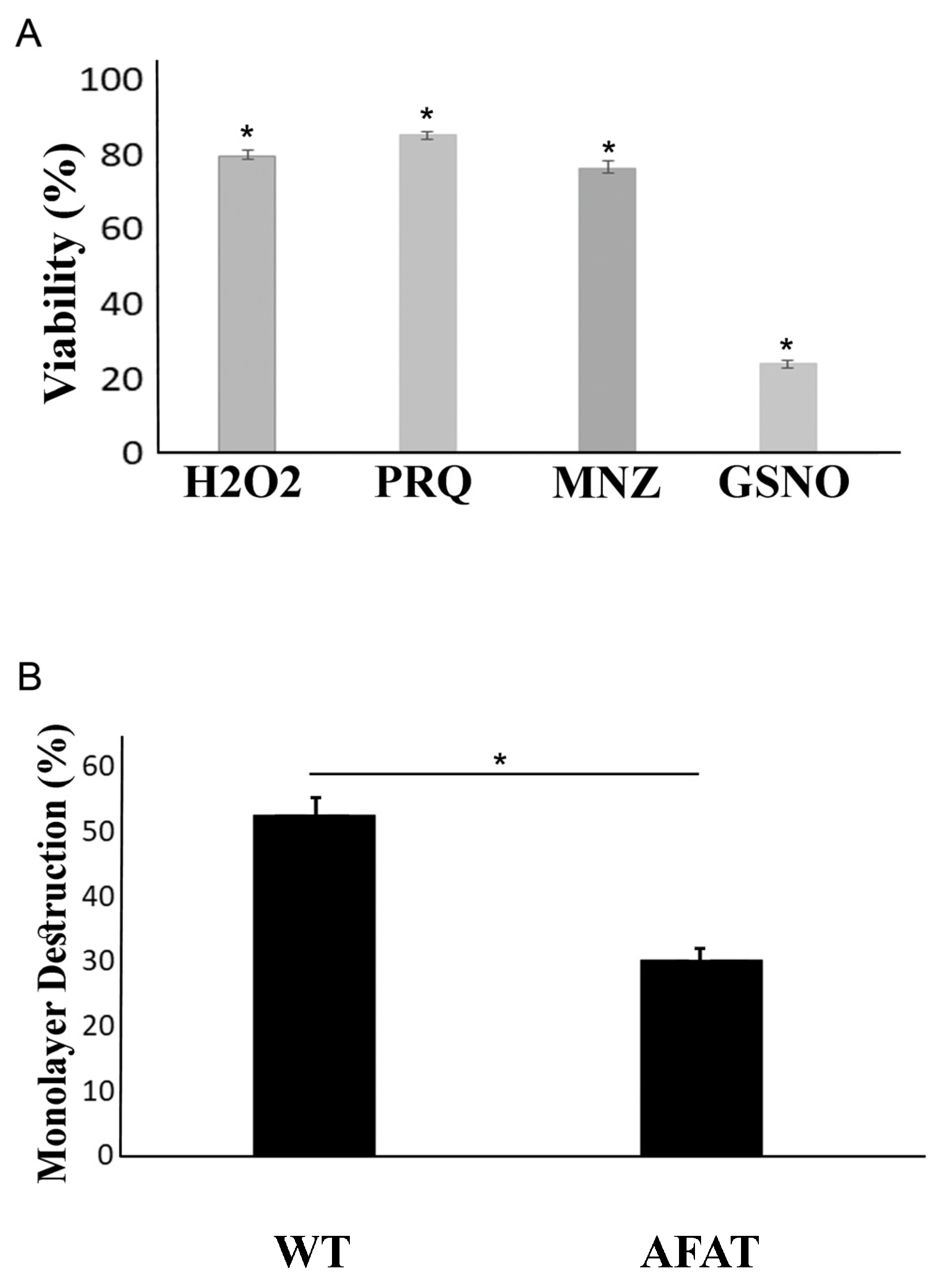
2.4. Viability of AFAT Exposed to H2O2, Paraquat, MNZ or GSNO
2.5. Measurement of Cytopathic Activity
2.6. RNA Extraction
2.7. RNA Sequencing (RNAseq): Library Preparation and Data Generation
2.8. Descriptive Analysis
2.9. Differential Expression Analysis
2.10. Availability of Data
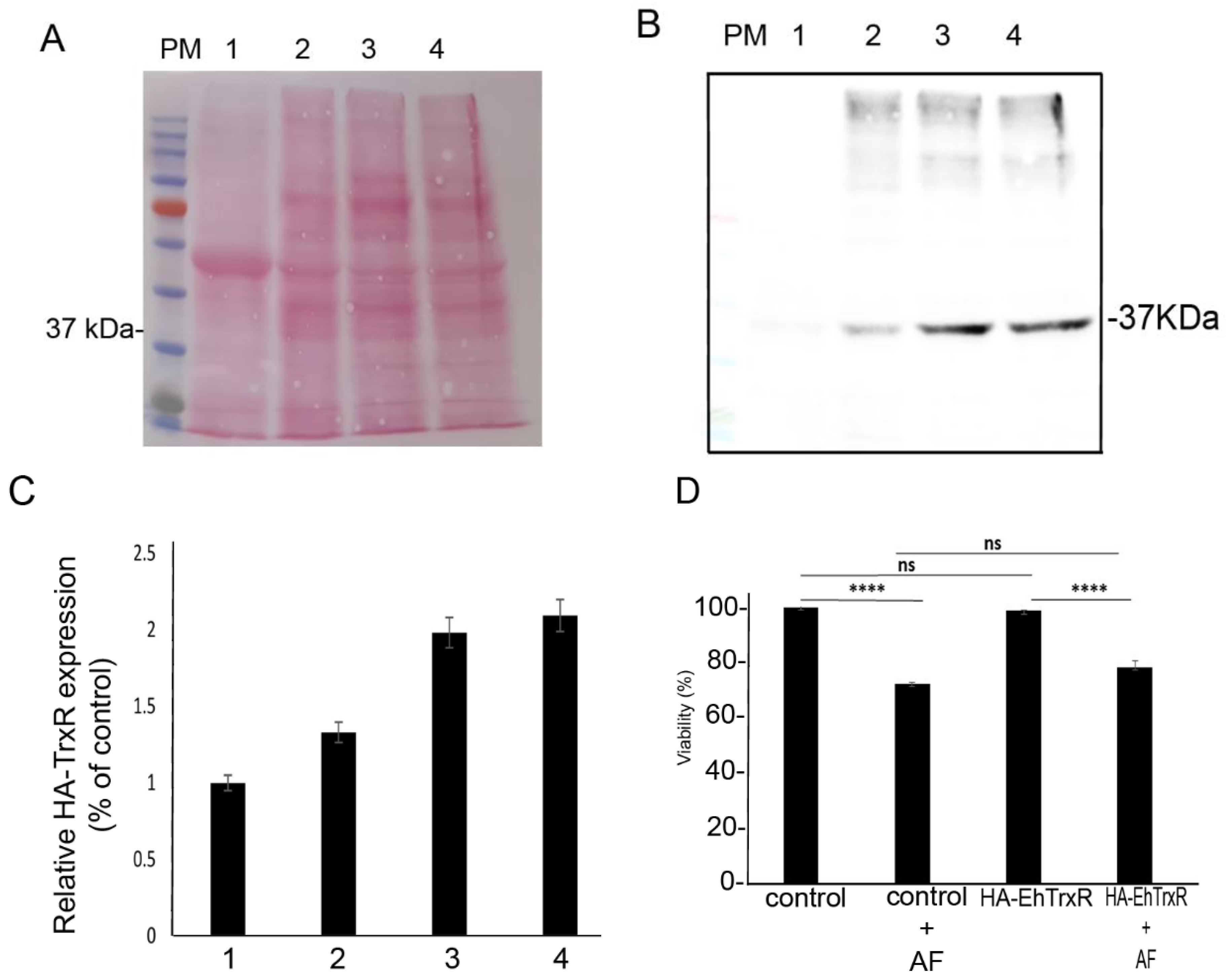
2.11. Construction of HA-Tagged EhTrxR Trophozoites
2.12. Immunodetection of (HA)-Tagged EhTrxR
2.13. Viability Assay
2.14. Detection of ROS
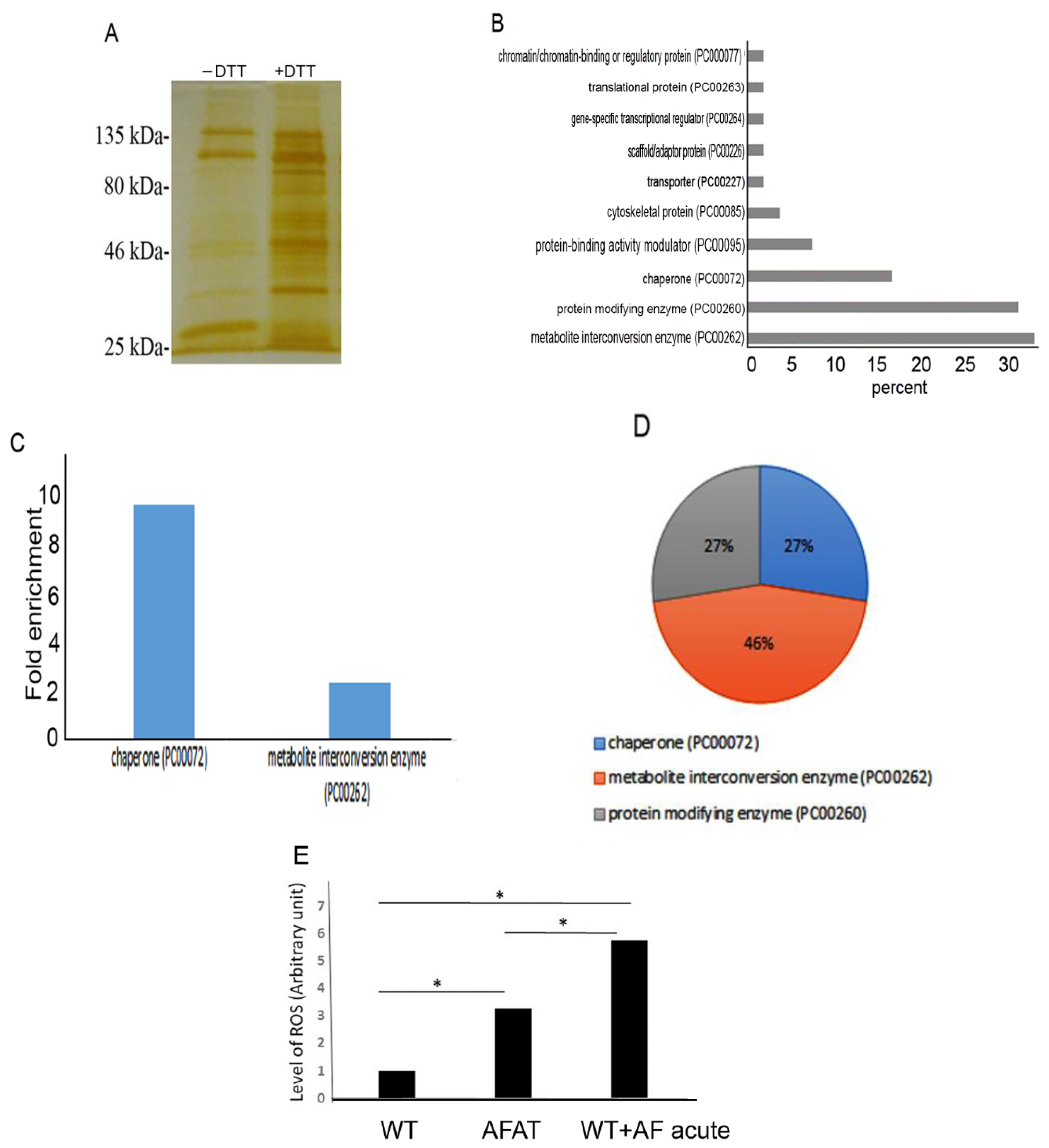
2.15. Detection of OXs by RAC (OX-RAC)
2.16. In-Gel Proteolysis and MS Analysis
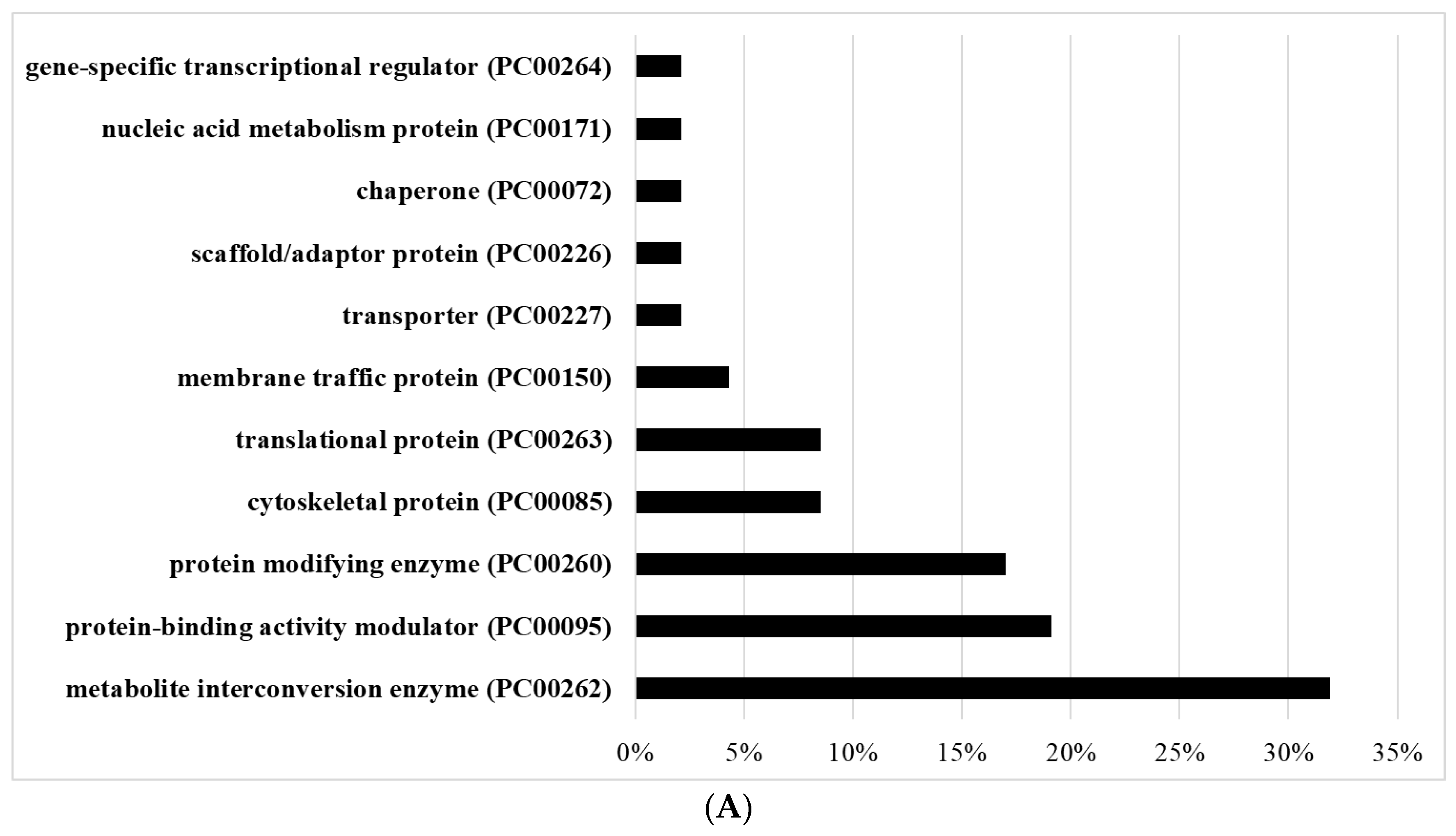
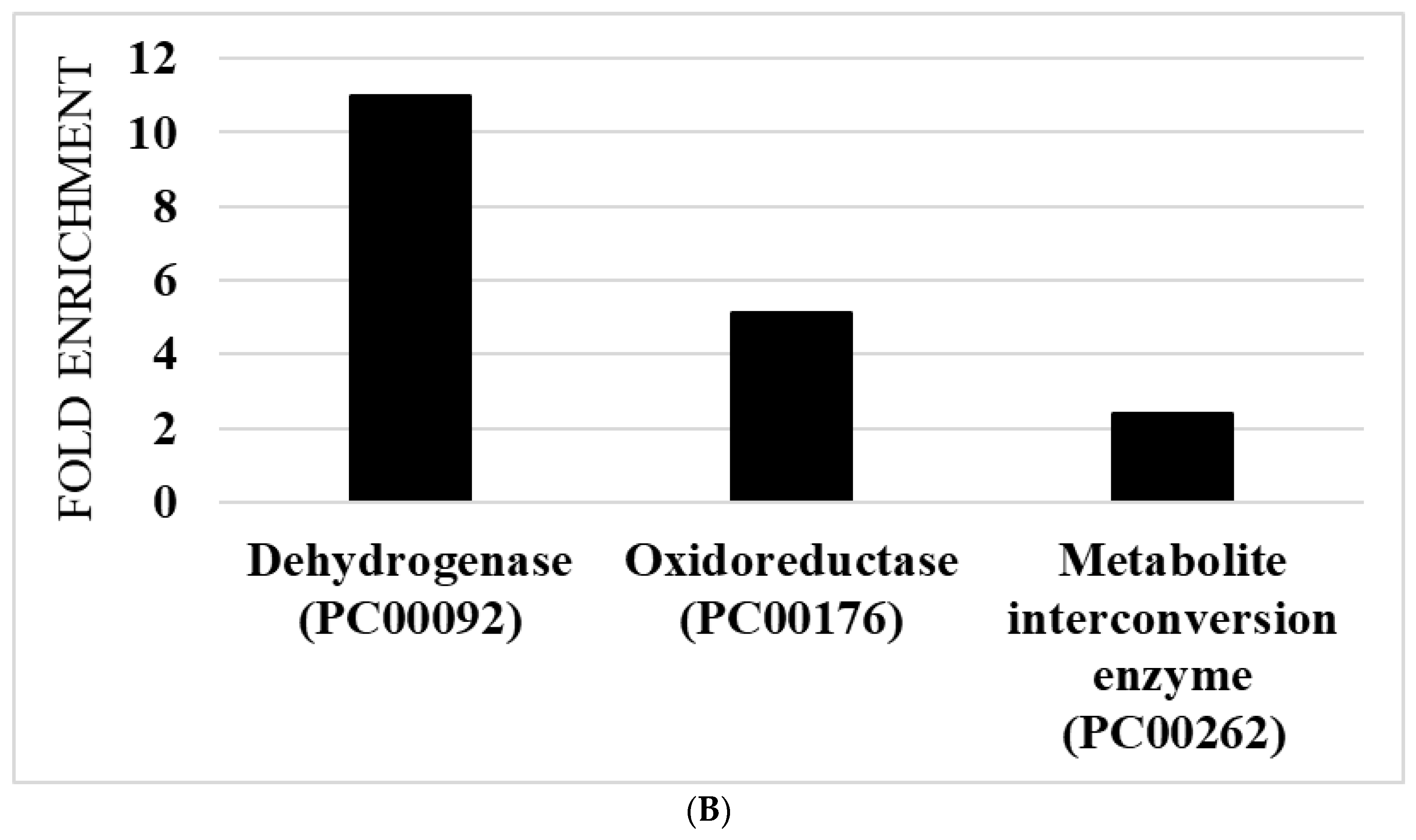
2.17. Classification of OXs According to Their Protein Class
2.18. Immunofluorescence Microscopy Analysis
3. Results
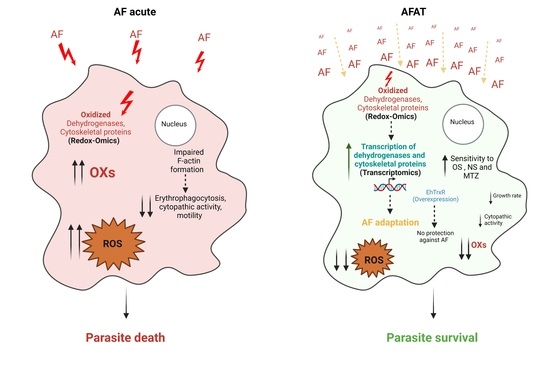
3.1. Generation of AFAT
3.2. Reponse of AFAT to OS, NS, and Cytopathic Activity
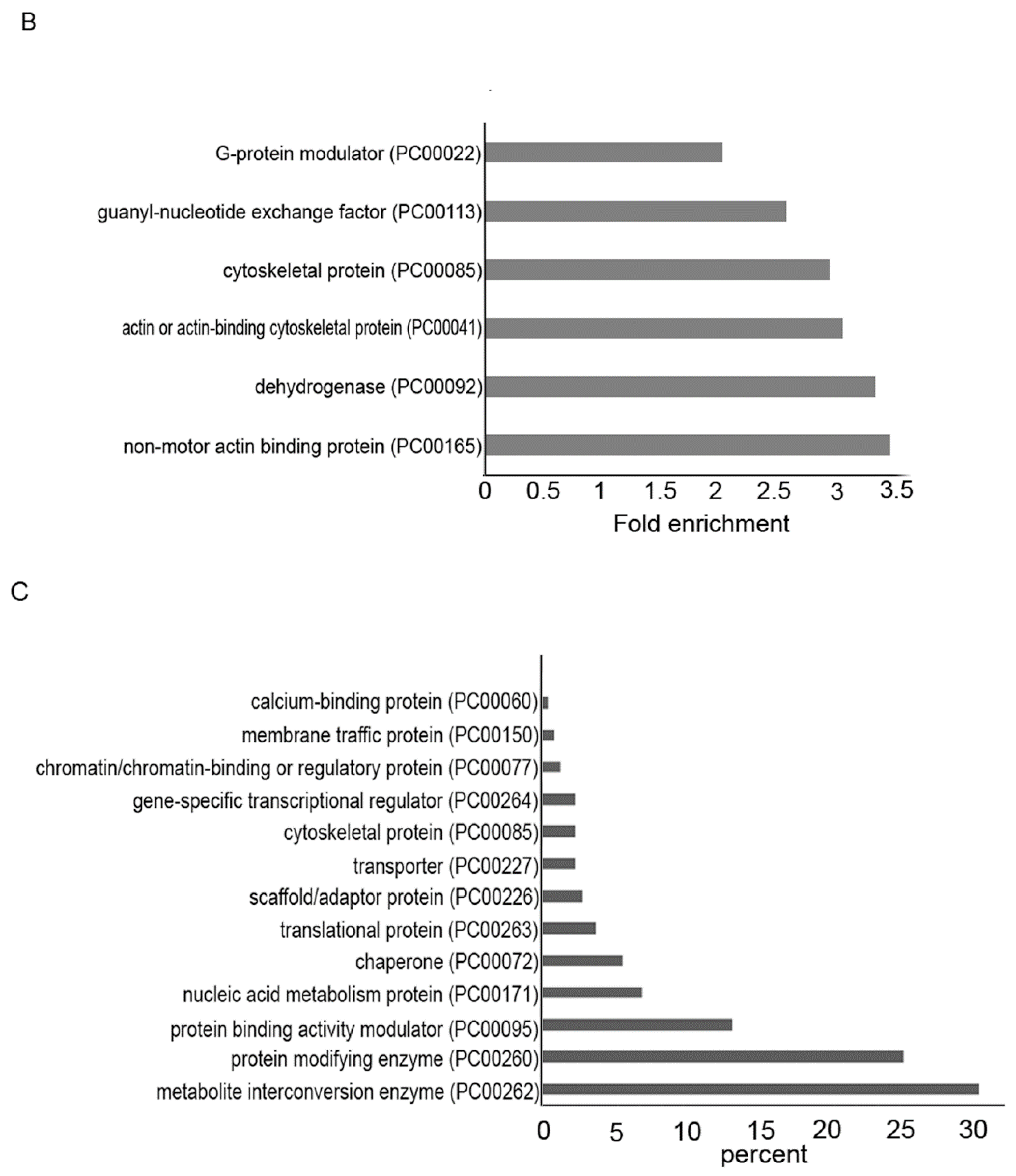
3.3. Transcriptomics of AFAT
3.4. Gene Categories Modulated in AFAT
3.5. Redoxomics of AFAT
3.6. Level of ROS in AFAT
3.7. Comparison between Transcriptomics and Redoxomics of AFAT
3.8. Comparison between Transcriptomics of AFAT and Redoxomics of Acute AF Trophozoites
3.9. Formation of F-Actin in AFAT
3.10. Overexpression of EhTrxR Does Not Protect E. histolytica Trophozoites against AF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turkeltaub, J.A.; McCarty, T.R., 3rd; Hotez, P.J. The intestinal protozoa: Emerging impact on global health and development. Curr. Opin. Gastroenterol. 2015, 31, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.J.; MacLeod, I.; Wilmot, A.J.; Elsdon-Dew, R. Metronidazole in amoebic dysentery and amoebic liver abscess. Lancet 1966, 2, 1329–1331. [Google Scholar] [CrossRef] [PubMed]
- Leitsch, D.; Kolarich, D.; Binder, M.; Wilson, I.B.H.; Altmann, F.; Duchene, M. Nitroimidazole action in Entamoeba histolytica: A central role for thioredoxin reductase. PLoS Biol. 2007, 5, e211. [Google Scholar] [CrossRef] [PubMed]
- Cowdrey, S.C. Letter: Hazards of metronidazole. N. Engl. J. Med. 1975, 293, 455. [Google Scholar]
- Andersson, K.E. Pharmacokinetics of Nitroimidazoles-Spectrum of Adverse Reactions. Scand. J. Infect. Dis. Suppl. 1981, 26, 60–67. [Google Scholar]
- Roe, F.J. Metronidazole: Review of uses and toxicity. J. Antimicrob. Chemother. 1977, 3, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Camacho, N.; Espinoza, C.; Rodriguez, C.; Rodriguez, E. Isolates of Clostridium perfringens recovered from Costa Rican patients with antibiotic-associated diarrhoea are mostly enterotoxin-negative and susceptible to first-choice antimicrobials. J. Med. Microbiol. 2008, 57, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.J.; Sheikh, A.F.; Goodarzi, H.; Yadyad, M.J.; Seyedian, S.S.; Aslani, S. Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders. Infect. Drug Resist. 2019, 12, 535–543. [Google Scholar] [CrossRef]
- Wassmann, C.; Bruchhaus, I. Superoxide dismutase reduces susceptibility to metronidazole of the pathogenic protozoan Entamoeba histolytica under microaerophilic but not under anaerobic conditions. Arch. Biochem. Biophys. 2000, 376, 236–238. [Google Scholar] [CrossRef]
- Wassmann, C.; Hellberg, A.; Tannich, E.; Bruchhaus, I. Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin 1 and flavin reductase. J. Biol. Chem. 1999, 274, 26051–26056. [Google Scholar] [CrossRef]
- Upcroft, J.A.; Upcroft, P. Drug susceptibility testing of anaerobic protozoa. Antimicrob. Agents Chemother. 2001, 45, 1810–1814. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, A.E.; Walz, D.T.; Batista, V.; Mizraji, M.; Roisman, F.; Misher, A. Auranofin. New oral gold compound for treatment of rheumatoid arthritis. Ann. Rheum. Dis. 1976, 35, 251–257. [Google Scholar] [CrossRef]
- Gromer, S.; Arscott, L.D.; Williams, C.H., Jr.; Schirmer, R.H.; Becker, K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J. Biol. Chem. 1998, 273, 20096–20101. [Google Scholar] [CrossRef] [PubMed]
- Onodera, T.; Momose, I.; Kawada, M. Potential Anticancer Activity of Auranofin. Chem. Pharm. Bull. 2019, 67, 186–191. [Google Scholar] [CrossRef]
- Ruth, M.M.; van Rossum, M.; Koeken, V.; Pennings, L.J.; Svensson, E.M.; Ruesen, C. Auranofin Activity Exposes Thioredoxin Reductase as a Viable Drug Target in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, e00449-19. [Google Scholar] [CrossRef]
- AbdelKhalek, A.; Abutaleb, N.S.; Mohammad, H.; Seleem, M.N. Antibacterial and antivirulence activities of auranofin against Clostridium difficile. Int. J. Antimicrob. Agents 2019, 53, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Jackson-Rosario, S.; Cowart, D.; Myers, A.; Tarrien, R.; Levine, R.L.; Scott, R.A. Auranofin disrupts selenium metabolism in Clostridium difficile by forming a stable Au-Se adduct. J. Biol. Inorg. Chem. 2009, 14, 507–519. [Google Scholar] [CrossRef]
- Abutaleb, N.S.; Seleem, M.N. Antivirulence activity of auranofin against vancomycin-resistant enterococci: In vitro and in vivo studies. Int. J. Antimicrob. Agents 2020, 55, 105828. [Google Scholar] [CrossRef]
- AbdelKhalek, A.; Abutaleb, N.S.; Elmagarmid, K.A.; Seleem, M.N. Repurposing auranofin as an intestinal decolonizing agent for vancomycin-resistant enterococci. Sci. Rep. 2018, 8, 8353. [Google Scholar] [CrossRef]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.; Sobreira, T.J.; Hedrick, V.E.; Paul, L.N. Antibacterial activity and mechanism of action of auranofin against multi-drug resistant bacterial pathogens. Sci. Rep. 2016, 6, 22571. [Google Scholar] [CrossRef]
- Angelucci, F.; Sayed, A.A.; Williams, D.L.; Boumis, G.; Brunori, M.; Dimastrogiovanni, D. Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: Structural and kinetic aspects. J. Biol. Chem. 2009, 284, 28977–28985. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, A.N.; Davioud-Charvet, E.; Sayed, A.A.; Califf, L.L.; Dessolin, J.; Arner, E.S. Thioredoxin glutathione reductase from Schistosoma mansoni: An essential parasite enzyme and a key drug target. PLoS Med. 2007, 4, e206. [Google Scholar] [CrossRef]
- Hopper, M.; Yun, J.F.; Zhou, B.; Le, C.; Kehoe, K.; Le, R. Auranofin inactivates Trichomonas vaginalis thioredoxin reductase and is effective against trichomonads in vitro and in vivo. Int. J. Antimicrob. Agents 2016, 48, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Tejman-Yarden, N.; Miyamoto, Y.; Leitsch, D.; Santini, J.; Debnath, A.; Gut, J. A reprofiled drug, auranofin, is effective against metronidazole-resistant Giardia lamblia. Antimicrob. Agents Chemother. 2013, 57, 2029–2035. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.; Parsonage, D.; Andrade, R.M.; He, C.; Cobo, E.R.; Hirata, K. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat. Med. 2012, 18, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Leitsch, D.; Muller, J.; Muller, N. Evaluation of Giardia lamblia thioredoxin reductase as drug activating enzyme and as drug target. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Mi-Ichi, F.; Ishikawa, T.; Tam, V.K.; Deloer, S.; Hamano, S.; Hamada, T.; Yoshida, H. Characterization of Entamoeba histolytica adenosine 5’-phosphosulfate (APS) kinase; validation as a target and provision of leads for the development of new drugs against amoebiasis. PLoS Negl. Trop. Dis. 2019, 13, e0007633. [Google Scholar] [CrossRef] [PubMed]
- Shaulov, Y.; Nagaraja, S.; Sarid, L.; Trebicz-Geffen, M.; Ankri, S. Formation of oxidised (OX) proteins in Entamoeba histolytica exposed to auranofin and consequences on the parasite virulence. Cell. Microbiol. 2020, 22, e13174. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.I.; Tirtorahardjo, J.A.; Jan, S.; Schweizer, S.S.; Rosario, S.A.C.; Du, Y. Auranofin Resistance in Toxoplasma gondii Decreases the Accumulation of Reactive Oxygen Species but Does Not Target Parasite Thioredoxin Reductase. Front. Cell. Infect. Microbiol. 2021, 11, 618994. [Google Scholar] [CrossRef]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Alterzon-Baumel, S.; Hertz, R.; Methling, K. Proteomic Identification of Oxidized Proteins in Entamoeba histolytica by Resin-Assisted Capture: Insights into the Role of Arginase in Resistance to Oxidative Stress. PLoS Negl. Trop. Dis. 2016, 10, e0004340. [Google Scholar] [CrossRef]
- Trebicz-Geffen, M.; Shahi, P.; Nagaraja, S.; Vanunu, S.; Manor, S.; Avrahami, A.; Ankri, S. Identification of S-Nitrosylated (SNO) Proteins in Entamoeba histolytica Adapted to Nitrosative Stress: Insights into the Role of SNO Actin and In vitro Virulence. Front. Cell. Infect. Microbiol. 2017, 7, 192. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Dastidar, P.G.; Majumder, S.; Lohia, A. Eh Klp5 is a divergent member of the kinesin 5 family that regulates genome content and microtubular assembly in Entamoeba histolytica. Cell. Microbiol. 2007, 9, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Lavi, T.; Isakov, E.; Harony, H.; Fisher, O.; Siman-Tov, R.; Ankri, S. Sensing DNA methylation in the protozoan parasite Entamoeba histolytica. Mol. Microbiol. 2006, 62, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Parsonage, D.; Sheng, F.; Hirata, K.; Debnath, A.; McKerrow, J.H.; Reed, S.L.; Abagyan, R.; Poole, L.B.; Podust, L.M. X-ray structures of thioredoxin and thioredoxin reductase from Entamoeba histolytica and prevailing hypothesis of the mechanism of Auranofin action. J. Struct. Biol. 2016, 194, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Hamann, L.; Nickel, R.; Tannich, E. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 1995, 92, 8975–8979. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, M.A.; Mi-ichi, F.; Nakada-Tsukui, K.; Nozaki, T. Localization and targeting of an unusual pyridine nucleotide transhydrogenase in Entamoeba histolytica. Eukaryot. Cell 2010, 9, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Ankri, S. Entamoeba histolytica-Gut Microbiota Interaction: More Than Meets the Eye. Microorganisms 2021, 9, 581. [Google Scholar] [CrossRef]
- Pineda, E.; Perdomo, D. Entamoeba histolytica under Oxidative Stress: What Countermeasure Mechanisms Are in Place? Cells 2017, 6, 44. [Google Scholar] [CrossRef]
- Chopra, I. Over-expression of target genes as a mechanism of antibiotic resistance in bacteria. J. Antimicrob. Chemother. 1998, 41, 584–588. [Google Scholar] [CrossRef]
- Capela, R.; Moreira, R.; Lopes, F. An Overview of Drug Resistance in Protozoal Diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef]
- Arias, D.G.; Regner, E.L.; Iglesias, A.A.; Guerrero, S.A. Entamoeba histolytica thioredoxin reductase: Molecular and functional characterization of its atypical properties. Biochim. Et Biophys. Acta 2012, 1820, 1859–1866. [Google Scholar] [CrossRef]
- Pineda, E.; Encalada, R.; Rodriguez-Zavala, J.S.; Olivos-Garcia, A.; Moreno-Sanchez, R.; Saavedra, E. Pyruvate:ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010, 277, 3382–3395. [Google Scholar] [CrossRef]
- Kumar, A.; Shen, P.S.; Descoteaux, S.; Pohl, J.; Bailey, G.; Samuelson, J. Cloning and expression of an NADP(+)-dependent alcohol dehydrogenase gene of Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 1992, 89, 10188–10192. [Google Scholar] [CrossRef]
- König, C.; Meyer, M.; Lender, C.; Nehls, S.; Wallaschkowski, T.; Holm, T.; Matthies, T.; Lercher, D.; Matthiesen, J.; Fehling, H.; et al. An Alcohol Dehydrogenase 3 (ADH3) from Entamoeba histolytica Is Involved in the Detoxification of Toxic Aldehydes. Microorganisms 2020, 8, 1608. [Google Scholar] [CrossRef]
- Klomsiri, C.; Karplus, P.A.; Poole, L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011, 14, 1065–1077. [Google Scholar] [CrossRef]
- Bosch, D.E.; Siderovski, D.P. G protein signaling in the parasite Entamoeba histolytica. Exp. Mol. Med. 2013, 45, e15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Accorsi, K.; Giglione, C.; Vanoni, M.; Parmeggiani, A. The Ras GDP/GTP cycle is regulated by oxidizing agents at the level of Ras regulators and effectors. FEBS Lett. 2001, 492, 139–145. [Google Scholar] [CrossRef]
- Ahmad, A.; Mishra, S.; Som, L.; Gourinath, S. Role of kinases in virulence and pathogenesis of protozoan parasite E. Histolytica. Front. Biosci. 2020, 25, 1617–1635. [Google Scholar]
- Corcoran, A.; Cotter, T.G. Redox regulation of protein kinases. FEBS J. 2013, 280, 1944–1965. [Google Scholar] [CrossRef]
- Froscio, M.; Murray, A.W.; Hurst, N.P. Inhibition of protein kinase C activity by the antirheumatic drug auranofin. Biochem. Pharmacol. 1989, 38, 2087–2089. [Google Scholar] [CrossRef]
- Penuliar, G.M.; Nakada-Tsukui, K.; Nozaki, T. Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance. Front. Microbiol. 2015, 6, 354. [Google Scholar] [CrossRef]
- Ehrenkaufer, G.M.; Suresh, S.; Solow-Cordero, D.; Singh, U. High-Throughput Screening of Entamoeba Identifies Compounds Which Target Both Life Cycle Stages and Which Are Effective Against Metronidazole Resistant Parasites. Front. Cell. Infect. Microbiol. 2018, 8, 276. [Google Scholar] [CrossRef]
- Baumel-Alterzon, S.; Weber, C.; Guillen, N.; Ankri, S. Identification of dihydropyrimidine dehydrogenase as a virulence factor essential for the survival of Entamoeba histolytica in glucose-poor environments. Cell. Microbiol. 2013, 15, 130–144. [Google Scholar] [CrossRef]
- Herencias, C.; Rodriguez-Beltran, J.; Leon-Sampedro, R.; Alonso-Del Valle, A.; Palkovicova, J.; Canton, R. Collateral sensitivity associated with antibiotic resistance plasmids. eLife 2021, 10, e65130. [Google Scholar] [CrossRef]
- Tekwani, B.L.; Mehlotra, R.K. Molecular basis of defence against oxidative stress in Entamoeba histolytica and Giardia lamblia. Microbes Infect 1999, 1, 385–394. [Google Scholar] [CrossRef]
- Bruchhaus, I.; Tannich, E. Induction of the iron-containing superoxide dismutase in Entamoeba histolytica by a superoxide anion-generating system or by iron chelation. Mol. Biochem. Parasitol. 1994, 67, 281–288. [Google Scholar] [CrossRef]
- Palmer, A.C.; Kishony, R. Opposing effects of target overexpression reveal drug mechanisms. Nat. Commun. 2014, 5, 4296. [Google Scholar] [CrossRef]
- Fiskus, W.; Rao, R.; Fernandez, P.; Herger, B.; Yang, Y.; Chen, J. Molecular and biologic characterization and drug sensitivity of pan-histone deacetylase inhibitor-resistant acute myeloid leukemia cells. Blood 2008, 112, 2896–2905. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaulov, Y.; Sarid, L.; Trebicz-Geffen, M.; Ankri, S. Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization. Antioxidants 2021, 10, 1240. https://doi.org/10.3390/antiox10081240
Shaulov Y, Sarid L, Trebicz-Geffen M, Ankri S. Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization. Antioxidants. 2021; 10(8):1240. https://doi.org/10.3390/antiox10081240
Chicago/Turabian StyleShaulov, Yana, Lotem Sarid, Meirav Trebicz-Geffen, and Serge Ankri. 2021. "Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization" Antioxidants 10, no. 8: 1240. https://doi.org/10.3390/antiox10081240
APA StyleShaulov, Y., Sarid, L., Trebicz-Geffen, M., & Ankri, S. (2021). Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization. Antioxidants, 10(8), 1240. https://doi.org/10.3390/antiox10081240