Mitochondria Content and Activity Are Crucial Parameters for Bull Sperm Quality Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material Collection and Preparation for Analyses
2.2. Flow Cytometry (Mitochondria Content and Activity)
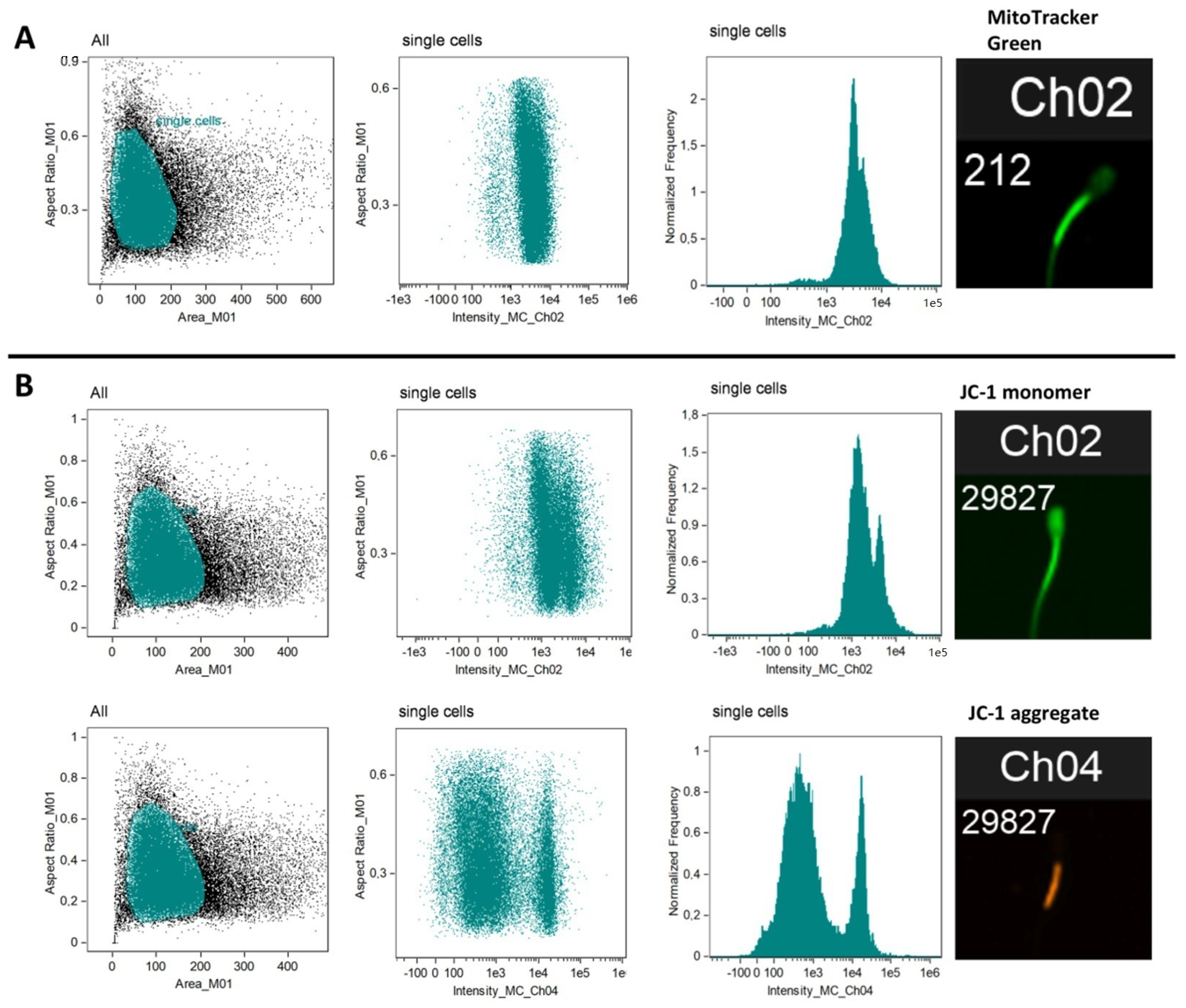
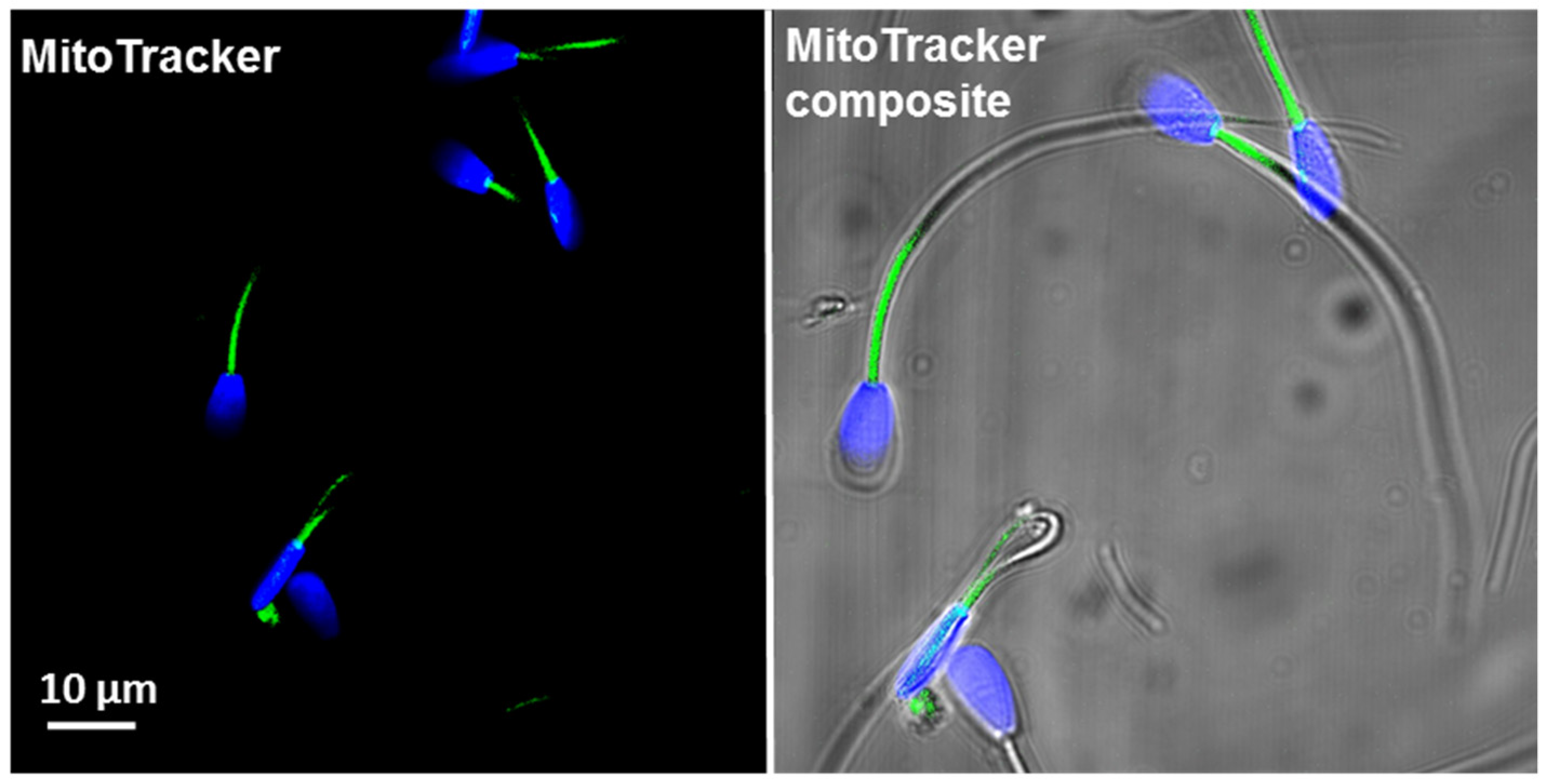
2.2.1. Mitochondria Content per Sperm Cell (MitoTrackerTM Green FM)
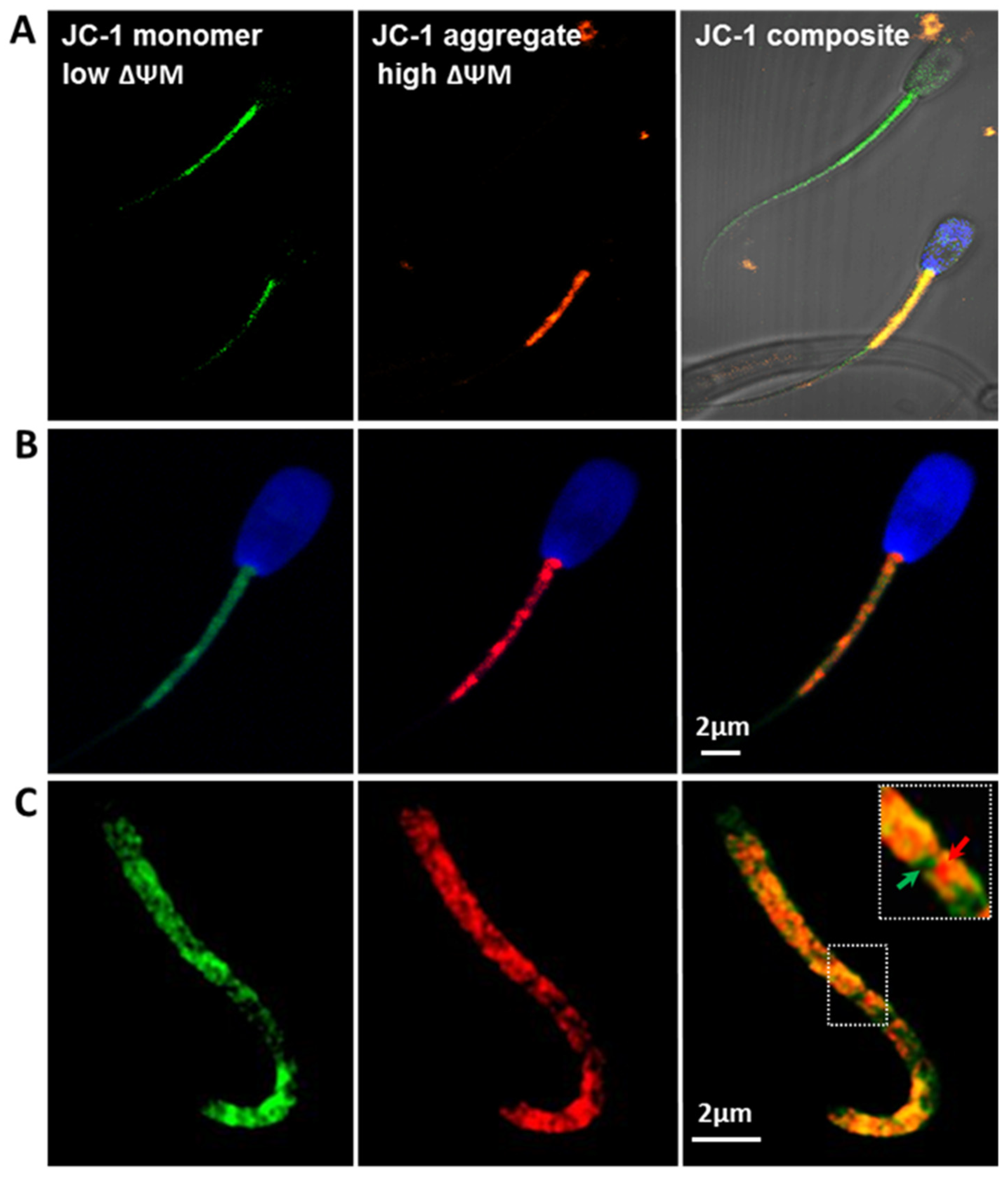
2.2.2. Mitochondria Activity (JC-1)
2.2.3. Flow Cytometry
2.3. Quantitative Gene Expression Analysis (Q-PCR)
2.3.1. RNA Extraction and cDNA Synthesis
2.3.2. Quantitative PCR (Q-PCR)
2.4. mtDNA Evaluation
2.5. Oxygen Consumption Rate Analysis (OCR)
2.5.1. Agilent XF Seahorse Analysis (Mitochondrial Respiration Analysis)
2.5.2. Preparation of Bovine Sperm Cells
2.5.3. Seahorse XF Cell Mito Stress Test Kit
2.6. Statistical Analysis
2.6.1. CASA Analysis
2.6.2. Flow Cytometry
2.6.3. Q-PCR and mtDNA Evaluation
3. Results
3.1. Sperm Motility and Morphology (CASA Analysis)
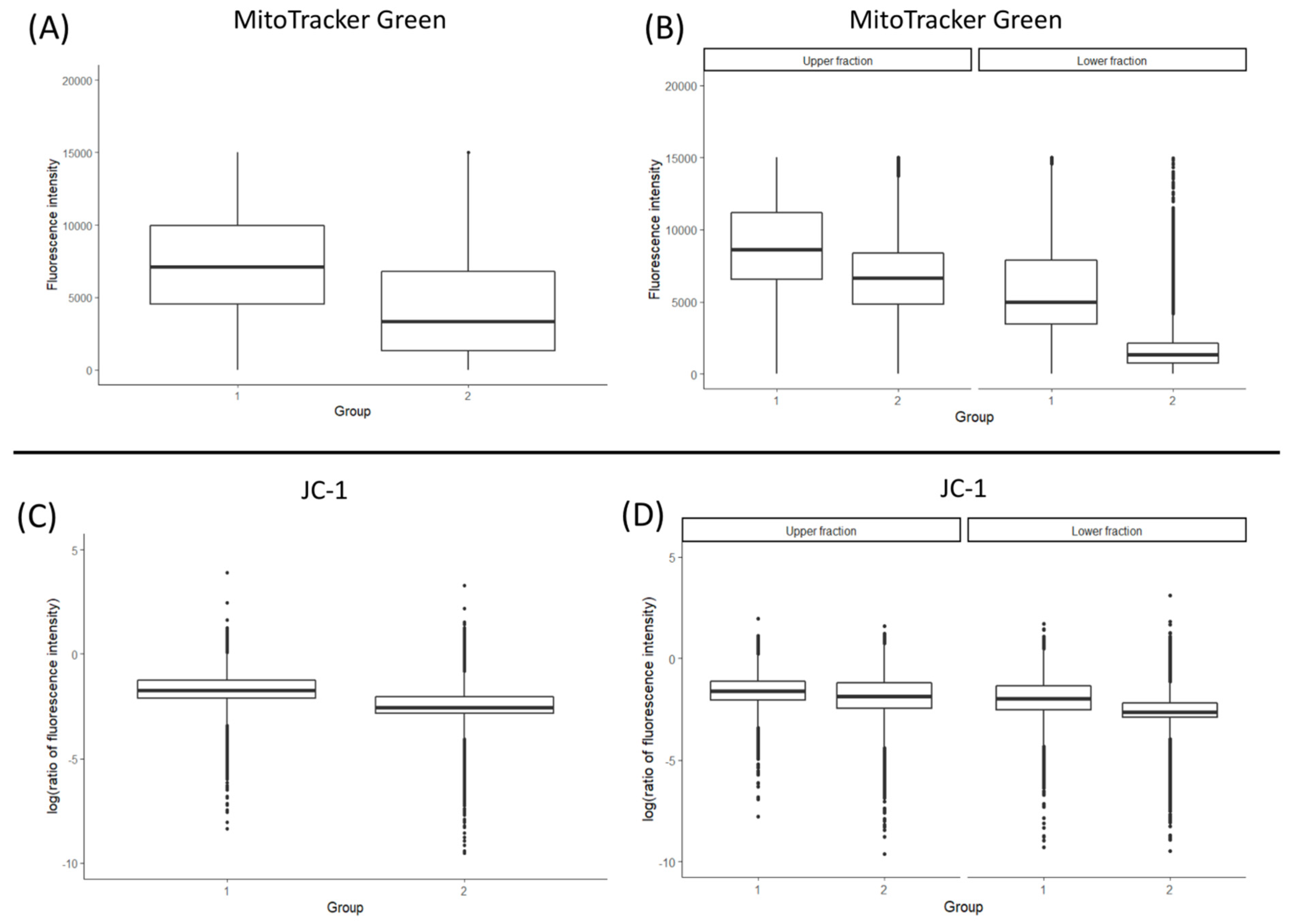
3.2. Total Mitochondrial Content (MitoTracker Green Staining and mtDNA Copy Number)
3.3. Mitochondrial Membrane Potential (ΔΨm)
3.4. Gene Expression
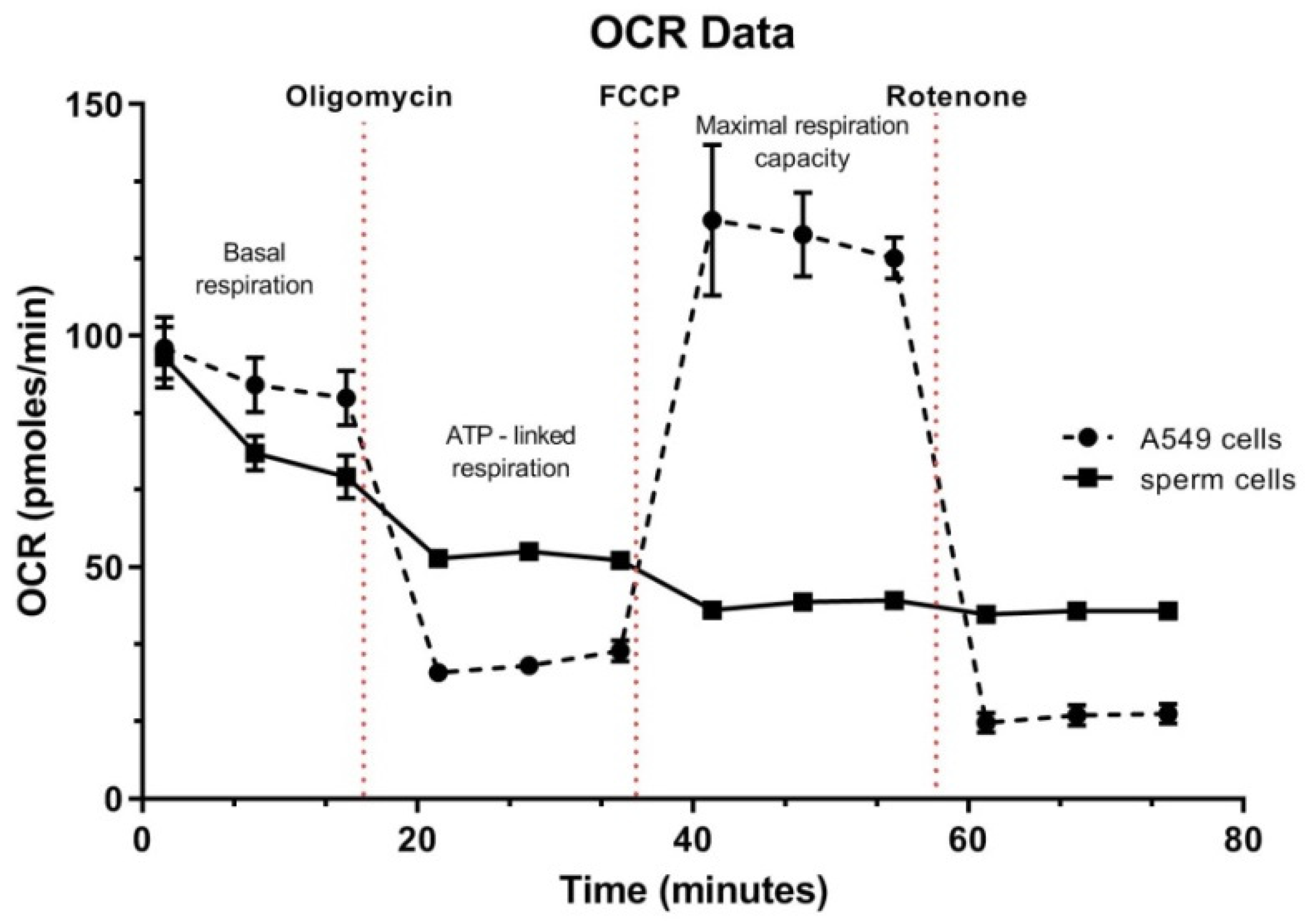
3.5. Spermatozoa OCR Analysis
4. Discussion
4.1. Mitochondrial Content Corresponds to mtDNA Copy Number in Bovine Spermatozoa
4.2. Mitochondrial Membrane Potential (ΔΨm) Is an Indicator of Bull Sperm Quality
4.3. Gene Expression
4.4. Oxygen Consumption
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, D.Y.; Baker, H.W. Defective sperm-zona pellucida interaction: A major cause of failure of fertilization in clinical in-vitro fertilization. Hum. Reprod. 2000, 15, 702–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandruvada, S.; Royfman, R.; Shah, T.A.; Sindhwani, P.; Dupree, J.M.; Schon, S.; Avidor-Reiss, T. Lack of trusted diagnostic tools for undetermined male infertility. J. Assist. Reprod. Genet. 2021, 38, 265–276. [Google Scholar] [CrossRef]
- Lessard, C.; Siqueira, L.G.; D’Amours, O.; Sullivan, R.; Leclerc, P.; Palmer, C. Infertility in a beef bull due to a failure in the capacitation process. Theriogenology 2011, 76, 891–899. [Google Scholar] [CrossRef]
- Losano, J.D.A.; Padín, J.F.; Méndez-López, I.; Angrimani, D.S.R.; García, A.G.; Barnabe, V.H.; Nichi, M. The stimulated glycolytic pathway is able to maintain atp levels and kinetic patterns of bovine epididymal sperm subjected to mitochondrial uncoupling. Oxidative Med. Cell. Longev. 2017, 2017, 1682393. [Google Scholar] [CrossRef]
- Espinoza, J.A.; Schulz, M.A.; Sánchez, R.; Villegas, J.V. Integrity of mitochondrial membrane potential reflects human sperm quality. Andrologia 2009, 41, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Zhuang, X.J.; Wei, Y.M.; Zhang, M.; Lu, S.S.; Lu, Y.Q.; Yang, X.G.; Lu, K.H. Comparison of mitochondrial function in boar and bull spermatozoa throughout cryopreservation based on jc-1 staining. CryoLetters 2017, 38, 75–79. [Google Scholar]
- Darr, C.R.; Cortopassi, G.A.; Datta, S.; Varner, D.D.; Meyers, S.A. Mitochondrial oxygen consumption is a unique indicator of stallion spermatozoal health and varies with cryopreservation media. Theriogenology 2016, 86, 1382–1392. [Google Scholar] [CrossRef]
- Cadenas, E. Mitochondrial free radical production and cell signaling. Mol. Asp. Med. 2004, 25, 17–26. [Google Scholar] [CrossRef]
- Alvarez, S.; Valdez, L.B.; Zaobornyj, T.; Boveris, A. Oxygen dependence of mitochondrial nitric oxide synthase activity. Biochem. Biophys. Res. Commun. 2003, 305, 771–775. [Google Scholar] [CrossRef]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef]
- Olsen, R.K.; Cornelius, N.; Gregersen, N. Redox signalling and mitochondrial stress responses; lessons from inborn errors of metabolism. J. Inherit. Metab. Dis. 2015, 38, 703–719. [Google Scholar] [CrossRef] [Green Version]
- Vertika, S.; Singh, K.K.; Rajender, S. Mitochondria, spermatogenesis, and male infertility—An update. Mitochondrion 2020, 54, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Baklouti-Gargouri, S.; Ghorbel, M.; Mahmoud, A.B.; Mkaouar-Rebai, E.; Cherif, M.; Chakroun, N.; Sellami, A.; Fakhfakh, F.; Ammar-Keskes, L. Mitochondrial DNA mutations and polymorphisms in asthenospermic infertile men. Mol. Biol. Rep. 2013, 40, 4705–4712. [Google Scholar] [CrossRef]
- La Vignera, S.; Condorelli, R.A.; Duca, Y.; Mongioi, L.M.; Cannarella, R.; Giacone, F.; Calogero, A.E. Fsh therapy for idiopathic male infertility: Four schemes are better than one. Aging Male Off. J. Int. Soc. Study Aging Male 2020, 5, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Nowicka-Bauer, K.; Nixon, B. Molecular changes induced by oxidative stress that impair human sperm motility. Antioxidants 2020, 9, 134. [Google Scholar] [CrossRef] [Green Version]
- Tvrda, E.; Straka, P.; Galbavy, D.; Ivanic, P. Epicatechin provides antioxidant protection to bovine spermatozoa subjected to induced oxidative stress. Molecules 2019, 24, 3226. [Google Scholar] [CrossRef] [Green Version]
- Bonanno, O.; Romeo, G.; Asero, P.; Pezzino, F.M.; Castiglione, R.; Burrello, N.; Sidoti, G.; Frajese, G.V.; Vicari, E.; D’Agata, R. Sperm of patients with severe asthenozoospermia show biochemical, molecular and genomic alterations. Reproduction 2016, 152, 695. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, C.; Jouy, N.; Leroy-Martin, B.; Defossez, A.; Formstecher, P.; Marchetti, P. Comparison of four fluorochromes for the detection of the inner mitochondrial membrane potential in human spermatozoa and their correlation with sperm motility. Hum. Reprod. 2004, 19, 2267–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Garrett, L.J.; Revell, S.G.; Leese, H.J. Adenosine triphosphate production by bovine spermatozoa and its relationship to semen fertilizing ability. J. Androl. 2008, 29, 449–458. [Google Scholar] [CrossRef] [Green Version]
- Petit, J.M.; Ratinaud, M.H.; Cordelli, E.; Spanò, M.; Julien, R. Mouse testis cell sorting according to DNA and mitochondrial changes during spermatogenesis. Cytometry 1995, 19, 304–312. [Google Scholar] [CrossRef]
- Hecht, N.B.; Bradley, F.M. Changes in mitochondrial protein composition during testicular differentiation in mouse and bull. Gamete Res. 1981, 4, 433–449. [Google Scholar] [CrossRef]
- Varuzhanyan, G.; Chan, D.C. Mitochondrial dynamics during spermatogenesis. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Varuzhanyan, G.; Rojansky, R.; Sweredoski, M.J.; Graham, R.L.; Hess, S.; Ladinsky, M.S.; Chan, D.C. Mitochondrial fusion is required for spermatogonial differentiation and meiosis. eLife 2019, 8, e51601. [Google Scholar] [CrossRef] [PubMed]
- Dietert, S.E. Fine structure of the formation and fate of the residual bodies of mouse spermatozoa with evidence for the participation of lysosomes. J. Morphol. 1966, 120, 317–346. [Google Scholar] [CrossRef]
- Otani, H.; Tanaka, O.; Kasai, K.; Yoshioka, T. Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: Regular dispositions and synchronized changes. Anat. Rec. 1988, 222, 26–33. [Google Scholar] [CrossRef]
- Gu, N.-H.; Zhao, W.-L.; Wang, G.-S.; Sun, F. Comparative analysis of mammalian sperm ultrastructure reveals relationships between sperm morphology, mitochondrial functions and motility. Reprod. Biol. Endocrinol. 2019, 17, 66. [Google Scholar] [CrossRef] [Green Version]
- Rocha, A.; Oliveira, E.; Vilhena, M.J.; Diaz, J.; Sousa, M. A novel apical midpiece defect in the spermatozoa of a bull without an apparent decrease in motility and fertility. A case study. Theriogenology 2006, 66, 913–922. [Google Scholar] [CrossRef] [PubMed]
- International Embryo Transfer Society. Available online: https://www.iets.org/Committees/Data-Retrieval-Committee (accessed on 17 June 2021).
- Baust, J.G.; Gao, D.; Baust, J.M. Cryopreservation: An emerging paradigm change. Organogenesis 2009, 5, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magdanz, V.; Boryshpolets, S.; Ridzewski, C.; Eckel, B.; Reinhardt, K. The motility-based swim-up technique separates bull sperm based on differences in metabolic rates and tail length. PLoS ONE 2019, 14, e0223576. [Google Scholar] [CrossRef]
- Bianchi, E.; Stermer, A.; Boekelheide, K.; Sigman, M.; Hall, S.J.; Reyes, G.; Dere, E.; Hwang, K. High-quality human and rat spermatozoal rna isolation for functional genomic studies. Andrology 2018, 6, 374–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, A.; Tang, C.; Xie, Y.; Ortogero, N.; Yuan, S.; Yan, W. Spermbase: A database for sperm-borne rna contents. Biol. Reprod. 2016, 95, 99. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, I.; Bissonnette, N.; Boissonneault, G.; Vallee, M.; Robert, C. A molecular analysis of the population of mrna in bovine spermatozoa. Reproduction 2007, 133, 1073–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsztynowicz, M.; Pawlak, P.; Podstawski, Z.; Nizanski, W.; Partyka, A.; Gotowiecka, M.; Kosiniak-Kamysz, K.; Lechniak, D. Mitochondrial DNA copy number in spermatozoa of fertile stallions. Reprod. Domest. Anim. 2016, 51, 378–385. [Google Scholar] [CrossRef] [PubMed]
- R Package Documentation. Available online: https://rdrr.io/cran/rstatix/ (accessed on 17 June 2021).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. 2020. Available online: https://www.R-project.org/ (accessed on 17 June 2021).
- Russell, V. Lenth. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.5.4. 2021. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 17 June 2021).
- Vincent, P.; Underwood, S.L.; Dolbec, C.; Bouchard, N.; Kroetsch, T.; Blondin, P. Bovine semen quality control in artificial insemination centers. Anim. Reprod. 2012, 9, 153–165. [Google Scholar]
- Zhang, G.; Wang, Z.; Ling, X.; Zou, P.; Yang, H.; Chen, Q.; Zhou, N.; Sun, L.; Gao, J.; Zhou, Z.; et al. Mitochondrial biomarkers reflect semen quality: Results from the marchs study in chongqing, China. PLoS ONE 2016, 11, e0168823. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.M.; Kelly, A.K.; O’Meara, C.; Eivers, B.; Lonergan, P.; Fair, S. Influence of bull age, ejaculate number, and season of collection on semen production and sperm motility parameters in holstein friesian bulls in a commercial artificial insemination centre. J. Anim. Sci. 2018, 96, 2408–2418. [Google Scholar] [CrossRef]
- Ogata, K.; Nagata, M.P.B.; Nishizono, H.; Yamanouchi, T.; Matsuda, H.; Ogata, Y.; Takeda, K.; Hashiyada, Y.; Yamashita, K. In vitro survival kinetics of microfluidic-sorted bovine spermatozoa. Andrology 2020, 9, 977–988. [Google Scholar] [CrossRef]
- Nagata, M.B.; Egashira, J.; Katafuchi, N.; Endo, K.; Ogata, K.; Yamanaka, K.; Yamanouchi, T.; Matsuda, H.; Hashiyada, Y.; Yamashita, K. Bovine sperm selection procedure prior to cryopreservation for improvement of post-thawed semen quality and fertility. J. Anim. Sci. Biotechnol. 2019, 10, 91. [Google Scholar] [CrossRef]
- Ugur, M.R.; Saber Abdelrahman, A.; Evans, H.C.; Gilmore, A.A.; Hitit, M.; Arifiantini, R.I.; Purwantara, B.; Kaya, A.; Memili, E. Advances in cryopreservation of bull sperm. Front. Vet. Sci. 2019, 6, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaveiro, A.; Liu, J.; Engel, B.; Critser, J.K.; Woelders, H. Significant variability among bulls in the sperm membrane permeability for water and glycerol: Possible implications for semen freezing protocols for individual males. Cryobiology 2006, 53, 349–359. [Google Scholar] [CrossRef]
- Loomis, P.R.; Graham, J.K. Commercial semen freezing: Individual male variation in cryosurvival and the response of stallion sperm to customized freezing protocols. Anim. Reprod. Sci. 2008, 105, 119–128. [Google Scholar] [CrossRef]
- Peris, S.I.; Morrier, A.; Dufour, M.; Bailey, J.L. Cryopreservation of ram semen facilitates sperm DNA damage: Relationship between sperm andrological parameters and the sperm chromatin structure assay. J. Androl. 2004, 25, 224–233. [Google Scholar] [CrossRef]
- O’Connell, M.; McClure, N.; Lewis, S.E. The effects of cryopreservation on sperm morphology, motility and mitochondrial function. Hum. Reprod. 2002, 17, 704–709. [Google Scholar] [CrossRef]
- Martínez, C.O.; Juárez-Mosqueda Mde, L.; Hernández, J.; Valencia, J. Cryopreservation of bull spermatozoa alters the perinuclear theca. Theriogenology 2006, 66, 1969–1975. [Google Scholar] [CrossRef] [PubMed]
- Cavelier, L.; Johannisson, A.; Gyllensten, U. Analysis of mtdna copy number and composition of single mitochondrial particles using flow cytometry and pcr. Exp. Cell Res. 2000, 259, 79–85. [Google Scholar] [CrossRef] [PubMed]
- May-Panloup, P.; Chre¬tien, M.-F.; Savagner, F.; Vasseur, C.; Jean, M.; Malthie¡ry, Y.; Reynier, P. Increased sperm mitochondrial DNA content in maleinfertility. Hum. Reprod. 2003, 18, 550–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wai, T.; Ao, A.; Zhang, X.; Cyr, D.; Dufort, D.; Shoubridge, E.A. The role of mitochondrial DNA copy number in mammalian fertility. Biol. Reprod. 2010, 83, 52–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hecht, N.B.; Liem, H.; Kleene, K.C.; Distel, R.J.; Ho, S.M. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev. Biol. 1984, 102, 452–461. [Google Scholar] [CrossRef]
- Diez-Sanchez, C.; Ruiz-Pesini, E.; Lapena, A.C.; Montoya, J.; Perez-Martos, A.; Enriquez, J.A.; Lopez-Perez, M.J. Mitochondrial DNA content of human spermatozoa. Biol. Reprod. 2003, 68, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Darr, C.R.; Moraes, L.E.; Connon, R.E.; Love, C.C.; Teague, S.; Varner, D.D.; Meyers, S.A. The relationship between mitochondrial DNA copy number and stallion sperm function. Theriogenology 2017, 94, 94–99. [Google Scholar] [CrossRef]
- Song, G.J.; Lewis, V. Mitochondrial DNA integrity and copy number in sperm from infertile men. Fertil. Steril. 2008, 90, 2238–2244. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.H.; Chao, H.T.; Liu, H.W.; Liao, T.L.; Wei, Y.H. Sperm mitochondrial DNA depletion in men with asthenospermia. Fertil. Steril. 2004, 82, 66–73. [Google Scholar] [CrossRef]
- Word Health Organization. Available online: https://www.who.int/publications/i/item/9789241547789 (accessed on 17 June 2021).
- Nakada, K.; Sato, A.; Yoshida, K.; Morita, T.; Tanaka, H.; Inoue, S.; Yonekawa, H.; Hayashi, J. Mitochondria-related male infertility. Proc. Natl. Acad. Sci. USA 2006, 103, 15148–15153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Pang, M.G. Mitochondrial functionality in male fertility: From spermatogenesis to fertilization. Antioxidants 2021, 10, 98. [Google Scholar] [CrossRef]
- Aitken, R.J. Impact of oxidative stress on male and female germ cells: Implications for fertility. Reproduction 2020, 159, R189–R201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.; Virk, G.; Ong, C.; du Plessis, S.S. Effect of oxidative stress on male reproduction. World J. Mens Health 2014, 32, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Said, T.M.; Gaglani, A.; Agarwal, A. Implication of apoptosis in sperm cryoinjury. Reprod. Biomed. Online 2010, 21, 456–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, K.E.; Gjeldnes, A.; Tverdal, A.; De Angelis, P.M.; Farstad, W.; Håård, M.; Kommisrud, E. Alterations of sperm DNA integrity during cryopreservation procedure and in vitro incubation of bull semen. Anim. Reprod. Sci. 2010, 117, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Hitit, M.; Ugur, M.R.; Dinh, T.T.N.; Sajeev, D.; Kaya, A.; Topper, E.; Tan, W.; Memili, E. Cellular and functional physiopathology of bull sperm with altered sperm freezability. Front. Vet. Sci. 2020, 7, 581137. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, R.; Kalthur, G.; Barbato, V.; Di Nardo, M.; Adiga, S.K.; Talevi, R. Mitochondrial dysfunction and oxidative stress caused by cryopreservation in reproductive cells. Antioxidants 2021, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.; Hoek, J.; Sheu, S.S. Mitochondrial Ca2+ and regulation of the permeability transition pore. J. Bioenerg. Biomembr. 2017, 49, 27–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; He, L.; Lemasters, J.J. Mitochondrial permeability transition: A common pathway to necrosis and apoptosis. Biochem. Biophys. Res. Commun. 2003, 304, 463–470. [Google Scholar] [CrossRef]
- Gallon, F.; Marchetti, C.; Jouy, N.; Marchetti, P. The functionality of mitochondria differentiates human spermatozoa with high and low fertilizing capability. Fertil. Steril. 2006, 86, 1526–1530. [Google Scholar] [CrossRef]
- Durairajanayagam, D.; Singh, D.; Agarwal, A.; Henkel, R. Causes and consequences of sperm mitochondrial dysfunction. Andrologia 2021, 53, e13666. [Google Scholar] [CrossRef]
- Sousa, A.P.; Amaral, A.; Baptista, M.; Tavares, R.; Caballero Campo, P.; Caballero Peregrin, P.; Freitas, A.; Paiva, A.; Almeida-Santos, T.; Ramalho-Santos, J. Not all sperm are equal: Functional mitochondria characterize a subpopulation of human sperm with better fertilization potential. PLoS ONE 2011, 6, e18112. [Google Scholar] [CrossRef] [Green Version]
- Garner, D.L.; Thomas, C.A. Organelle-specific probe jc-1 identifies membrane potential differences in the mitochondrial function of bovine sperm. Mol. Reprod. Dev. 1999, 53, 222–229. [Google Scholar] [CrossRef]
- Piasecka, M.; Kawiak, J. Sperm mitochondria of patients with normal sperm motility and with asthenozoospermia: Morphological and functional study. Folia Histochem. Cytobiol. 2003, 41, 125–139. [Google Scholar]
- Oguz, Y.; Guler, I.; Erdem, A.; Mutlu, M.F.; Gumuslu, S.; Oktem, M.; Bozkurt, N.; Erdem, M. The effect of swim-up and gradient sperm preparation techniques on deoxyribonucleic acid (DNA) fragmentation in subfertile patients. J. Assist. Reprod. Genet. 2018, 35, 1083–1089. [Google Scholar] [CrossRef]
- Adiga, S.K.; Kumar, P. Influence of swim-up method on the recovery of spermatozoa from different types of semen samples. J. Assist. Reprod. Genet. 2001, 18, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Amaral, S.; Oliveira, P.J.; Ramalho-Santos, J. Diabetes and the impairment of reproductive function: Possible role of mitochondria and reactive oxygen species. Curr. Diabetes Rev. 2008, 4, 46–54. [Google Scholar] [PubMed]
- Chandrashekar, K.N. Evidence of oxidative stress and mitochondrial dysfunctions in the testis of prepubertal diabetic rats. Int. J. Impot. Res. 2009, 21, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.G.; Farias, J.G.; Henriquez-Olavarrieta, S.; Madrid, E.; Parraga, M.; Zepeda, A.B.; Moreno, R.D. The hypoxic testicle: Physiology and pathophysiology. Oxidative Med. Cell. Longev. 2012, 2012, 929285. [Google Scholar] [CrossRef] [PubMed]
- Spiess, A.N.; Feig, C.; Schulze, W.; Chalmel, F.; Cappallo-Obermann, H.; Primig, M.; Kirchhoff, C. Cross-platform gene expression signature of human spermatogenic failure reveals inflammatory-like response. Hum. Reprod. 2007, 22, 2936–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, T.; Nixon, B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev. Cell 2020, 52, 399–411. [Google Scholar] [CrossRef]
- Mateus, I.; Feijo, M.; Espinola, L.M.; Vaz, C.V.; Correia, S.; Socorro, S. Glucose and glutamine handling in the sertoli cells of transgenic rats overexpressing regucalcin: Plasticity towards lactate production. Sci. Rep. 2018, 8, 10321. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, H.; Wu, H.; Chen, Y.; Han, D. Apoptotic spermatogenic cells can be energy sources for sertoli cells. Reproduction 2009, 137, 469–479. [Google Scholar] [CrossRef] [Green Version]
- Ge, R.; Chen, G.; Hardy, M.P. The role of the leydig cell in spermatogenic function. In Molecular Mechanisms in Spermatogenesis; Cheng, C.Y., Ed.; Springer: New York, NY, USA, 2008; pp. 255–269. [Google Scholar]
- Iwasaki, A.; Gagnon, C. Formation of reactive oxygen species in spermatozoa of infertile patients. Fertil. Steril. 1992, 57, 409–416. [Google Scholar] [CrossRef]
- Miller, D.; Ostermeier, G.C. Towards a better understanding of rna carriage by ejaculate spermatozoa. Hum. Reprod. Update 2006, 12, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Gur, Y.; Breitbart, H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006, 20, 411–416. [Google Scholar] [CrossRef] [Green Version]
- Selvaraju, S.; Parthipan, S.; Somashekar, L.; Kolte, A.P.; Binsila, B.K.; Arangasamy, A.; Ravindra, J.P. Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Sci. Rep. 2017, 7, 42392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Liu, J.; Zhu, P.; Wang, H.; Zhao, Z.; Sun, G.; Li, J. Expression of peroxiredoxins in the human testis, epididymis and spermatozoa and their role in preventing H2O2-induced damage to spermatozoa. Folia Histochem. Cytobiol. 2018, 56, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamatani, T. Human spermatozoal rnas. Fertil. Steril. 2012, 97, 275–281. [Google Scholar] [CrossRef]
- O’Flaherty, C. Peroxiredoxins: Hidden players in the antioxidant defence of human spermatozoa. Basic Clin. Androl. 2014, 24, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, D.Y.; Kim, K.U.; Kwon, W.S.; Rahman, M.S.; Khatun, A.; Pang, M.G. Peroxiredoxin activity is a major landmark of male fertility. Sci. Rep. 2017, 7, 17174. [Google Scholar] [CrossRef] [Green Version]
- Cassani, P.; Beconi, M.T.; O’Flaherty, C. Relationship between total superoxide dismutase activity with lipid peroxidation, dynamics and morphological parameters in canine semen. Anim. Reprod. Sci. 2005, 86, 163–173. [Google Scholar] [CrossRef]
- Grant, K.E.; de Oliveira, R.V.; Hennington, B.S.; Govindaraju, A.; Perkins, A.; Stokes, J.; Rowe, D.; Topper, E.; Kaya, A.; Moura, A.; et al. Sperm superoxide dismutase is associated with bull fertility. Reprod. Fertil. Dev. 2015. [Google Scholar] [CrossRef]
- Alvarez, J.G.; Storey, B.T. Lipid peroxidation and the reactions of superoxide and hydrogen peroxide in mouse spermatozoa. Biol. Reprod. 1984, 30, 833–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tramer, F.; Rocco, F.; Micali, F.; Sandri, G.; Panfili, E. Antioxidant systems in rat epididymal spermatozoa. Biol. Reprod. 1998, 59, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Jow, W.W.; Schlegel, P.N.; Cichon, Z.; Phillips, D.; Goldstein, M.; Bardin, C.W. Identification and localization of copper-zinc superoxide dismutase gene expression in rat testicular development. J. Androl. 1993, 14, 439–447. [Google Scholar] [PubMed]
- Faure, C.; Leveille, P.; Dupont, C.; Julia, C.; Chavatte-Palmer, P.; Sutton, A.; Levy, R. Are superoxide dismutase 2 and nitric oxide synthase polymorphisms associated with idiopathic infertility? Antioxid. Redox Signal. 2014, 21, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Garrido, N.; Meseguer, M.; Alvarez, J.; Simón, C.; Pellicer, A.; Remohí, J. Relationship among standard semen parameters, glutathione peroxidase/glutathione reductase activity, and mrna expression and reduced glutathione content in ejaculated spermatozoa from fertile and infertile men. Fertil. Steril. 2004, 82 (Suppl. 3), 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Nesci, S.; Spinaci, M.; Galeati, G.; Nerozzi, C.; Pagliarani, A.; Algieri, C.; Tamanini, C.; Bucci, D. Sperm function and mitochondrial activity: An insight on boar sperm metabolism. Theriogenology 2020, 144, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Gil, J.E.; Bonet, S. Current knowledge on boar sperm metabolism: Comparison with other mammalian species. Theriogenology 2016, 85, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Tourmente, M.; Varea-Sanchez, M.; Roldan, E.R.S. Faster and more efficient swimming: Energy consumption of murine spermatozoa under sperm competitiondagger. Biol. Reprod. 2019, 100, 420–428. [Google Scholar] [CrossRef] [PubMed]

| GENE | PRIMER SET | PROBE | ANNEALING (°C) | PRODUCT SIZE (bp) |
|---|---|---|---|---|
| ACTB (NCBI ref. sequence mRNA: NM_173979) | 5′CCTGGGCATGGAATCCTG3′ 5′GATGTCGACGTCACACTTCATGAT3′ | 5′6FAM- ATTGAAGGTAGTTTCGTGAATGCCGCAG-BBQ3′ | 59 | 69 |
| ACTB (DNA) (NCBI ref. sequence: NG_002441.3) | 5′GGCTCGTGTGACAAAAGC3′ 5′GCAGAAGAGTGCAAGGAACA3′ | 5′6FAM- CAGTAGGTGCACAGTACGTTCTGAAGTGAA-BBQ3′ | 60 | 137 |
| CYTB (NCBI ref. sequence: NC_006853) | 5′CCCGATTCTTCGCTTTCCAT3′ 5′CTACGTCTGAGGAAATTCCTGTTG3′ | 5′6FAM- CATCATAGCAATTGCCATAGTCCACC-BBQ3′ | 60 | 119 |
| GAPDH (NCBI ref. sequence mRNA: NC_037329.1) | 5′GGCTGGGGCTCACTTGAA3′ 5′CAGGAGGCATTGCTGACAATC3′ | 5′YAK- TCATCTCTGCACCTTCTGCCGATGC-BBQ3′ | 59 | 134 |
| GSS (NCBI ref. sequence mRNA: NM_001015630) | 5′CTCCGCAGCCCTGAAAC3′ 5′ACTGAGAACATGTCGATGCACA3′ | 5′6FAM- AGCTGGCAGAGACGGTGTTGATTTC-BBQ3′ | 59 | 96 |
| PRDX1 (NCBI ref. sequence mRNA: NM_174431) | 5′CCATAAACGACCTTCCTGTT3′ 5′GAGAAATATTCTTTGCTCTTCTGGAC3′ | 5′6FAM- CTGGAAGCCTGGCAGTGATACCATC-BBQ3′ | 59 | 159 |
| SOD1 (NCBI ref. sequence mRNA: NM_174156) | 5′TTCCCCCGAGTCATGGC3′ 5′AGCCTTGTGTATTGTCTCCAAAC3′ | 5′FAM- AGTCGTGGTAACTGGATCCATTACAGGA-BBQ3′ | 59 | 177 |
| BULLS | PM (%) | Motile (%) | Bent Tail (%) | Coiled Tail(%) | Dag Defect (%) | Distal Droplet % | Proximal Droplet % | Proper Morphology % | DMR % | ALH (μm) | BCF (Hz) | DAP (μm) | DCL (μm) | DSL (μm) | LIN (%) | VAP (μm/s) | VCL (μm/s) | VSL (μm/s) | WOB (%) | STR (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group A | 38.80 A (2.588) | 52.10 A (1.693) | 1.30 A (0.116) | 0.10 a (0.053) | 1.70 A (0.130) | 2.50 A (0.255) | 5.30 a (0.437) | 89.30 A (0.713) | 1.60 A (0.127) | 8.4 (0.297) | 36.80 (0.776) | 61.50 a (4.055) | 102.3 a (6.660) | 56.7 A (3.834) | 58.0 a (1.817) | 139.4 a (6.455) | 223.6 A (13.994) | 126.2 A (6.918) | 64.3 a (1.355) | 93.5 A (0.859) |
| Group B | 14.90 B (2.227) | 30.20 B (2.457) | 3.85 B (0.579) | 0.20 b (0.047) | 4.80 B (0.628) | 7.00 B (1.303) | 6.80 b (1.228) | 78.05 B (2.373) | 4.45 B (0.399) | 7.90 (0.7) | 33.40 (1.061) | 41.80 b (2.682) | 76.50 b (8.950) | 32.50 B (2.582) | 47.2 b (2.142) | 100.4 b (5.655) | 149.25 B (8.898) | 79.35 B (4.720) | 54.80 b (2.259) | 87.45 B (1.668) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madeja, Z.E.; Podralska, M.; Nadel, A.; Pszczola, M.; Pawlak, P.; Rozwadowska, N. Mitochondria Content and Activity Are Crucial Parameters for Bull Sperm Quality Evaluation. Antioxidants 2021, 10, 1204. https://doi.org/10.3390/antiox10081204
Madeja ZE, Podralska M, Nadel A, Pszczola M, Pawlak P, Rozwadowska N. Mitochondria Content and Activity Are Crucial Parameters for Bull Sperm Quality Evaluation. Antioxidants. 2021; 10(8):1204. https://doi.org/10.3390/antiox10081204
Chicago/Turabian StyleMadeja, Zofia E., Marta Podralska, Agnieszka Nadel, Marcin Pszczola, Piotr Pawlak, and Natalia Rozwadowska. 2021. "Mitochondria Content and Activity Are Crucial Parameters for Bull Sperm Quality Evaluation" Antioxidants 10, no. 8: 1204. https://doi.org/10.3390/antiox10081204
APA StyleMadeja, Z. E., Podralska, M., Nadel, A., Pszczola, M., Pawlak, P., & Rozwadowska, N. (2021). Mitochondria Content and Activity Are Crucial Parameters for Bull Sperm Quality Evaluation. Antioxidants, 10(8), 1204. https://doi.org/10.3390/antiox10081204






