LC-ESI-LTQ-Orbitrap-MS for Profiling the Distribution of Oleacein and Its Metabolites in Rat Tissues
Abstract
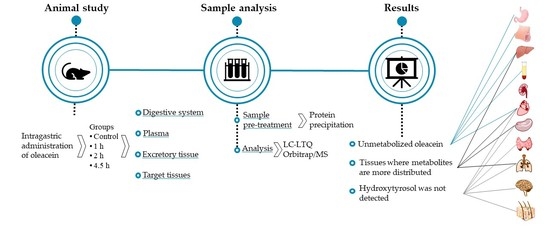
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animals
2.3. Refined Olive Oil
2.4. Dosage Information
2.5. Sample Pre-Treatment for OLEA Metabolite Analysis
2.5.1. Plasma
2.5.2. Tissues
2.6. Instruments and Analytical Conditions
2.7. Data Analysis
3. Results and Discussion
3.1. Pre-Systemic Metabolism
3.2. Plasma
3.3. Accumulation and Distribution of OLEA Derivatives in Rat Tissues
3.3.1. Heart
3.3.2. Spleen, Lungs and Thyroids
3.3.3. Brain
3.3.4. Kidneys
3.3.5. Skin
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andrewes, P.; Busch, J.L.H.C.; De Joode, T.; Groenewegen, A.; Alexandre, H. Sensory Properties of Virgin Olive Oil Polyphenols: Identification of Deacetoxy-ligstroside Aglycon as a Key Contributor to Pungency. J. Agric. Food Chem. 2003, 51, 1415–1420. [Google Scholar] [CrossRef]
- López-Yerena, A.; Ninot, A.; Lozano-Castellón, J.; Escribano-Ferrer, E.; Romero-Aroca, A.J.; Belaj, A.; Vallverdú-Queralt, A.; Lamuela-Raventós, R.M. Conservation of Native Wild Ivory-White Olives from the MEDES Islands Natural Reserve to Maintain Virgin Olive Oil Diversity. Antioxidants 2020, 9, 1009. [Google Scholar] [CrossRef]
- Miho, H.; Díez, C.; Mena-Bravo, A.; de Medina, V.S.; Moral, J.; Melliou, E.; Magiatis, P.; Rallo, L.; Barranco, D.; Priego-Capote, F. Cultivar influence on variability in olive oil phenolic profiles determined through an extensive germplasm survey. Food Chem. 2018, 266, 192–199. [Google Scholar] [CrossRef]
- Miho, H.; Moral, J.; López-González, M.A.; Díez, C.M.; Priego-Capote, F. The phenolic profile of virgin olive oil is influenced by malaxation conditions and determines the oxidative stability. Food Chem. 2020, 314, 126183. [Google Scholar] [CrossRef]
- Czerwińska, M.; Kiss, A.K.; Naruszewicz, M. A comparison of antioxidant activities of oleuropein and its dialdehydic derivative from olive oil, oleacein. Food Chem. 2012, 131, 940–947. [Google Scholar] [CrossRef]
- Czerwińska, M.; Kiss, A.K.; Naruszewicz, M. Inhibition of human neutrophils NEP activity, CD11b/CD18 expression and elastase release by 3,4-dihydroxyphenylethanol-elenolic acid dialdehyde, oleacein. Food Chem. 2014, 153, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Filipek, A.; Czerwińska, M.; Kiss, A.K.; Wrzosek, M.; Naruszewicz, M. Oleacein enhances anti-inflammatory activity of human macrophages by increasing CD163 receptor expression. Phytomedicine 2015, 22, 1255–1261. [Google Scholar] [CrossRef] [PubMed]
- Naruszewicz, M.; Czerwińska, M.; Kiss, A. Oleacein. Translation from Mediterranean Diet to Potential Antiatherosclerotic Drug. Curr. Pharm. Des. 2015, 21, 1205–1212. [Google Scholar] [CrossRef]
- Filipek, A.; Czerwińska, M.; Kiss, A.K.; Polański, J.A.; Naruszewicz, M. Oleacein may inhibit destabilization of carotid plaques from hypertensive patients. Impact on high mobility group protein-1. Phytomedicine 2017, 32, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.; Adsersen, A.; Christensen, S.B.; Jensen, S.R.; Nyman, U.; Smitt, U.W. Isolation of an angiotensin converting enzyme (ACE) inhibitor from Olea europaea and Olea lancea. Phytomedicine 1996, 2, 319–325. [Google Scholar] [CrossRef]
- Fabiani, R.; Rosignoli, P.; De Bartolomeo, A.; Fuccelli, R.; Servili, M.; Montedoro, G.F.; Morozzi, G. Oxidative DNA Damage Is Prevented by Extracts of Olive Oil, Hydroxytyrosol, and Other Olive Phenolic Compounds in Human Blood Mononuclear Cells and HL60 Cells. J. Nutr. 2008, 138, 1411–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polini, B.; Digiacomo, M.; Carpi, S.; Bertini, S.; Gado, F.; Saccomanni, G.; Macchia, M.; Nieri, P.; Manera, C.; Fogli, S. Oleocanthal and oleacein contribute to the in vitro therapeutic potential of extra virgin oil-derived extracts in non-melanoma skin cancer. Toxicol. In Vitro 2018, 52, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Cantafio, M.E.G.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A.; et al. Anti-tumor Activity and Epigenetic Impact of the Polyphenol Oleacein in Multiple Myeloma. Cancers 2019, 11, 990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikou, T.; Liaki, V.; Stathopoulos, P.; Sklirou, A.D.; Tsakiri, E.; Jakschitz, T.; Bonn, G.; Trougakos, I.P.; Halabalaki, M.; Skaltsounis, L.A. Comparison survey of EVOO polyphenols and exploration of healthy aging-promoting properties of oleocanthal and oleacein. Food Chem. Toxicol. 2019, 125, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Carpi, S.; Scoditti, E.; Massaro, M.; Polini, B.; Manera, C.; Digiacomo, M.; Salsano, J.E.; Poli, G.; Tuccinardi, T.; Doccini, S.; et al. The Extra-Virgin Olive Oil Polyphenols Oleocanthal and Oleacein Counteract Inflammation-Related Gene and miRNA Expression in Adipocytes by Attenuating NF-κB Activation. Nutrients 2019, 11, 2855. [Google Scholar] [CrossRef] [Green Version]
- Lepore, S.M.; Maggisano, V.; Bulotta, S.; Mignogna, C.; Arcidiacono, B.; Procopio, A.; Brunetti, A.; Russo, D.; Celano, M. Oleacein Prevents High Fat Diet-Induced A Diposity and Ameliorates Some Biochemical Parameters of Insulin Sensitivity in Mice. Nutrients 2019, 11, 1829. [Google Scholar] [CrossRef] [Green Version]
- López-Yerena, A.; Pérez, M.; Vallverdú-Queralt, A.; Miliarakis, E.; Lamuela-Raventós, R.; Escribano-Ferrer, E. Oleacein Intestinal Permeation and Metabolism in Rats Using an In Situ Perfusion Technique. Pharmaceutics 2021, 13, 719. [Google Scholar] [CrossRef]
- Pinto, J.; Paiva-Martins, F.; Corona, G.; Debnam, E.S.; Concha, M.J.O.; Vauzour, D.; Gordon, M.H.; Spencer, J.P.E. Absorption and metabolism of olive oil secoiridoids in the small intestine. Br. J. Nutr. 2011, 105, 1607–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López de las Hazas, M.C.; Piñol, C.; Macià, A.; Romero, M.-P.; Pedret, A.; Solà, R.; Rubió, L.; Motilva, M.-J. Differential absorption and metabolism of hydroxytyrosol and its precursors oleuropein and secoiridoids. J. Funct. Foods 2016, 22, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Serra, A.; Rubió, L.; Borràs, X.; Macià, A.; Romero, M.-P.; Motilva, M.-J. Distribution of olive oil phenolic compounds in rat tissues after administration of a phenolic extract from olive cake. Mol. Nutr. Food Res. 2011, 56, 486–496. [Google Scholar] [CrossRef]
- López-Yerena, A.; Vallverdú-Queralt, A.; Jáuregui, O.; Garcia-Sala, X.; Lamuela-Raventós, R.; Escribano-Ferrer, E. Tissue Distribution of Oleocanthal and Its Metabolites after Oral Ingestion in Rats. Antioxidants 2021, 10, 688. [Google Scholar] [CrossRef]
- Lozano-Castellón, J.; López-Yerena, A.; Olmo-Cunillera, A.; Jáuregui, O.; Pérez, M.; Lamuela-Raventós, R.M.; Vallverdú-Queralt, A. Total Analysis of the Major Secoiridoids in Extra Virgin Olive Oil: Validation of an UHPLC-ESI-MS/MS Method. Antioxidants 2021, 10, 540. [Google Scholar] [CrossRef]
- Panel, E.; Nda, A. Scientific Opinion on the substantiation of health claims related to polyphenols in olive and protection of LDL particles from oxidative damage (ID 1333, 1638, 1639, 1696, 2865), maintenance of normal blood HDL cholesterol concentrations (ID 1639), mainte. EFSA J. 2011, 9, 2033. [Google Scholar] [CrossRef]
- Diehl, K.-H.; Hull, R.; Morton, D.; Pfister, R.; Rabemampianina, Y.; Smith, D.; Vidal, J.-M.; Van De Vorstenbosch, C. A good practice guide to the administration of substances and removal of blood, including routes and volumes. J. Appl. Toxicol. 2001, 21, 15–23. [Google Scholar] [CrossRef]
- Brown, A.P.; Dinger, N.; Levine, B.S. Stress produced by gavage administration in the rat. Contemp. Top. Lab. Anim. Sci. 2000, 39, 17–21. [Google Scholar] [PubMed]
- Serreli, G.; Deiana, M. Biological Relevance of Extra Virgin Olive Oil Polyphenols Metabolites. Antioxidants 2018, 7, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manna, C.; Galletti, P.; Maisto, G.; Cucciolla, V.; D’Angelo, S.; Zappia, V. Transport mechanism and metabolism of olive oil hydroxytyrosol in Caco-2 cells. FEBS Lett. 2000, 470, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Wani, T.A.; Masoodi, F.A.; Gani, A.; Baba, W.N.; Rahmanian, N.; Akhter, R.; Wani, I.A.; Ahmad, M. Olive oil and its principal bioactive compound: Hydroxytyrosol–A review of the recent literature. Trends Food Sci. Technol. 2018, 77, 77–90. [Google Scholar] [CrossRef]
- European Union. Commission Regulation (EU) 432/2012 of 16 May 2012 Establishing a List of Permitted Health Claims Made on Foods, Other than Those Referring to the Reduction of Disease Risk and to Children’s Development and Health. Off. J. Eur. Union 2012, L136, 1–40. [Google Scholar]
- López de las Hazas, M.-C.; Rubio, L.; Kotronoulas, A.; De La Torre, R.; Solà, R.; Motilva, M.-J. Dose effect on the uptake and accumulation of hydroxytyrosol and its metabolites in target tissues in rats. Mol. Nutr. Food Res. 2015, 59, 1395–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Villalba, R.; Carrasco-Pancorbo, A.; Nevedomskaya, E.; Mayboroda, O.A.; Deelder, A.M.; Carretero, A.S.; Fernández-Gutiérrez, A. Exploratory analysis of human urine by LC–ESI-TOF MS after high intake of olive oil: Understanding the metabolism of polyphenols. Anal. Bioanal. Chem. 2010, 398, 463–475. [Google Scholar] [CrossRef]
- Jin, Y.; Penning, T. Aldo-Keto Reductases and Bioactivation/Detoxication. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 263–292. [Google Scholar] [CrossRef] [PubMed]
- Iyanagi, T. Molecular Mechanism of Phase I and Phase II Drug-Metabolizing Enzymes: Implications for Detoxification. Adv. Virus Res. 2007, 260, 35–112. [Google Scholar] [CrossRef]
- Silva, S.; Garcia-Aloy, M.; Figueira, M.E.; Combet, E.; Mullen, W.; Bronze, M. High Resolution Mass Spectrometric Analysis of Secoiridoids and Metabolites as Biomarkers of Acute Olive Oil Intake—An Approach to Study Interindividual Variability in Humans. Mol. Nutr. Food Res. 2017, 62, 1700065. [Google Scholar] [CrossRef] [Green Version]
- Karhunen, T.; Tilgmann, C.; Ulmanen, I.; Julkunen, I.; Panula, P. Distribution of catechol-O-methyltransferase enzyme in rat tissues. J. Histochem. Cytochem. 1994, 42, 1079–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef]
- de Sousa, I.P.; Bernkop-Schnürch, A. Pre-systemic metabolism of orally administered drugs and strategies to overcome it. J. Control. Release 2014, 192, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Serra, A.; Macià, A.; Piñol, C.; Romero, M.-P.; Motilva, M.-J.; Romero-Fabregat, M.-P. In vivo distribution and deconjugation of hydroxytyrosol phase II metabolites in red blood cells: A potential new target for hydroxytyrosol. J. Funct. Foods 2014, 10, 139–143. [Google Scholar] [CrossRef]
- López de las Hazas, M.C.; Motilva, M.J.; Piñol, C.; Macià, A. Application of dried blood spot cards to determine olive oil phenols (hydroxytyrosol metabolites) in human blood. Talanta 2016, 159, 189–193. [Google Scholar] [CrossRef]
- Visioli, F.; Galli, C.; Grande, S.; Colonnelli, K.; Patelli, C.; Galli, G.; Caruso, D. Hydroxytyrosol Excretion Differs between Rats and Humans and Depends on the Vehicle of Administration. J. Nutr. 2003, 133, 2612–2615. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Castellón, J.; López-Yerena, A.; De Alvarenga, J.F.R.; Del Castillo-Alba, J.R.; Vallverdú-Queralt, A.; Escribano-Ferrer, E.; Lamuela-Raventós, R.M. Health-promoting properties of oleocanthal and oleacein: Two secoiridoids from extra-virgin olive oil. Crit. Rev. Food Sci. Nutr. 2020, 60, 2532–2548. [Google Scholar] [CrossRef] [PubMed]
- López-Yerena, A.; Domínguez-López, I.; Vallverdú-Queralt, A.; Pérez, M.; Jáuregui, O.; Escribano-Ferrer, E.; Lamuela-Raventós, R. Metabolomics Technologies for the Identification and Quantification of Dietary Phenolic Compound Metabolites: An Overview. Antioxidants 2021, 10, 846. [Google Scholar] [CrossRef]
- Chan, J.M. Drug Metabolism and Pharmacogenetics. In Pharmacology and Physiology for Anesthesia; Elsevier: Amsterdam, The Netherlands, 2019; pp. 70–90. ISBN 978-0-323-48110-6. [Google Scholar]
- Yilmaz, Y.; Williams, G.; Walles, M.; Manevski, N.; Krähenbühl, S.; Camenisch, G. Comparison of Rat and Human Pulmonary Metabolism Using Precision-cut Lung Slices (PCLS). Drug Metab. Lett. 2019, 13, 53–63. [Google Scholar] [CrossRef] [PubMed]
- López de las Hazas, M.-C.; Godinho-Pereira, J.; Macià, A.; Almeida, A.F.; Ventura, M.R.; Motilva, M.-J.; Santos, C.N. Brain Uptake of Hydroxytyrosol and Its Main Circulating Metabolites: Protective Potential in Neuronal Cells. J. Funct. Foods 2018, 46, 110–117. [Google Scholar] [CrossRef]
- Gutiérrez-Miranda, B.; Gallardo, I.; Melliou, E.; Cabero, I.; Álvarez, Y.; Magiatis, P.; Hernández, M.; Nieto, M.L. Oleacein Attenuates the Pathogenesis of Experimental Autoimmune Encephalomyelitis through Both Antioxidant and Anti-Inflammatory Effects. Antioxidants 2020, 9, 1161. [Google Scholar] [CrossRef]
- Miners, J.; Yang, X.; Knights, K.M.; Zhang, L. The Role of the Kidney in Drug Elimination: Transport, Metabolism, and the Impact of Kidney Disease on Drug Clearance. Clin. Pharmacol. Ther. 2017, 102, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Rowland, M.; Tozer, T.N. Clinical Pharmacokinetics and Pharmacodynamics: Concepts and Applications, 4th ed.; Wolters Kluwer Health/Lippincott William & Wilkins: Philadelphia, PA, USA, 2019; ISBN 978-0-7817-5009-7. [Google Scholar]
| Metabolite (nmol OLEA Equivalents/g Tissue) | Time (h) | Stomach | Small Intestine | Liver |
|---|---|---|---|---|
| OLEA | 1 | 8.73 ± 0.29 a | 2.24 ± 0.14 a | 0.28 ± 0.02 |
| 2 | 7.39 ± 0.25 b | 1.69 ± 0.25 b | n.d. | |
| 4.5 | 5.77 ± 0.54 c | 0.51 ± 0.10 c | n.d. | |
| OH-TY | 1 | 13.37 ± 1.27 a | 4.71 ± 0.32 a | 3.34 ± 0.08 b |
| 2 | 7.81 ± 0.69 b | 3.79 ± 0.29 b | 3.83 ± 0.21 a | |
| 4.5 | 5.23 ± 0.37 c | 2.70 ± 0.21 c | 2.51 ± 0.07 c | |
| EA | 1 | 7.53 ± 0.56 a | 1.81 ± 0.05 a | 1.07 ± 0.06 b |
| 2 | 4.07 ± 0.36 b | 0.67 ± 0.08 b | 1.34 ± 0.08 a | |
| 4.5 | 1.81 ± 0.13 c | 0.16 ± 0.02 c | 0.76 ± 0.02 c | |
| OLEA + H2 | 1 | 1.34 ± 0.01 a | 5.31 ± 0.48 a | n.d. |
| 2 | 0.82 ± 0.01 b | 3.01 ± 0.35 b | n.d. | |
| 4.5 | 0.81 ± 0.01 b | 2.16 ± 0.10 c | n.d. | |
| OLEA + H2O | 1 | 1.90 ± 0.18 a | 1.29 ± 0.14 a | 0.33 ± 0.02 b |
| 2 | 1.66 ± 0.12 a | 0.96 ± 0.06 b | 0.77 ± 0.01 a | |
| 4.5 | 0.80 ± 0.03 b | 0.53 ± 0.01 c | 0.26 ± 0.04 c | |
| OLEA + CH3 | 1 | 0.46 ± 0.05 a | 3.69 ± 0.19 a | 3.88 ± 0.45 b |
| 2 | 0.41 ± 0.01 a | 2.99 ± 0.46 b | 5.79 ± 0.36 a | |
| 4.5 | 0.26 ± 0.00 b | 2.26 ± 0.22 c | 1.90 ± 0.24 c | |
| OLEA + H2O + CH3 | 1 | n.d. | n.d. | 1.37 ± 0.12 b |
| 2 | n.d. | n.d. | 1.78 ± 0.07 a | |
| 4.5 | n.d. | n.d. | 1.10 ± 0.07 c | |
| OLEA + H2 + glucu | 1 | n.d. | n.d. | 0.77 ± 0.05 b |
| 2 | n.d. | n.d. | 1.18 ± 0.06 a | |
| 4.5 | n.d. | n.d. | 0.46 ± 0.05 c | |
| OLEA+ H2O + CH3 + glucu | 1 | n.d. | n.d. | 0.38 ± 0.03 b |
| 2 | n.d. | n.d. | 0.69 ± 0.04 a | |
| 4.5 | n.d. | n.d. | 0.26 ± 0.01 c |
| Metabolites (nmol OLEA Equivalents/mL) | Time (h) | ||
|---|---|---|---|
| 1 | 2 | 4.5 | |
| OLEA | 1.37 ± 0.20 a | 0.65 ± 0.07 b | 0.67 ± 0.10 b |
| OH-TY | 2.11 ± 0.10 b | 3.02 ± 0.16 a | 1.62 ± 0.14 c |
| EA | 0.43 ± 0.03 b | 0.76 ± 0.04 a | 0.16 ± 0.02 c |
| OLEA + H2 | 0.87 ± 0.02 b | 1.08 ± 0.02 a | 0.87 ± 0.02 b |
| OLEA + H2O | 0.91 ± 0.03 b | 1.14 ± 0.02 a | 0.84 ± 0.02 c |
| OLEA + CH3 | 2.52 ± 0.24 b | 3.50 ± 0.18 a | 2.80 ± 0.22 b |
| OLEA + H2O + CH3 | 1.04 ± 0.01b | 1.35 ± 0.05 a | 0.91 ± 0.02 c |
| OLEA + H2 + glucu | 0.88 ± 0.04 b | 1.10 ± 0.04 a | 0.71 ± 0.02 c |
| OLEA + H2O + glucu | 1.26 ± 0.03 b | 1.44 ± 0.04 a | 1.03 ± 0.03 c |
| OLEA + H2O + CH3 + glucu | 0.57 ± 0.01 b | 0.65 ± 0.01 a | 0.50 ± 0.01 b |
| Metabolites (nmol OLEA Equivalents/g Tissue) | Time (h) | Heart | Spleen | Thyroids | Lungs | Brain | Kidney |
|---|---|---|---|---|---|---|---|
| OLEA | 1 | 0.44 ± 0.07 a | n.d. | n.d. | n.d. | n.d. | n.d. |
| 2 | 0.45 ± 0.04 a | n.d. | n.d. | n.d. | n.d. | n.d. | |
| 4.5 | 0.44 ± 0.06 a | n.d. | n.d. | n.d. | n.d. | n.d. | |
| OLEA + H2 | 1 | 0.17 ± 0.02 b | 0.30 ± 0.05 c | 0.10 ± 0.01 c | 0.49 ± 0.04 c | n.d. | n.d. |
| 2 | 0.30 ± 0.04 a | 0.54 ± 0.03 a | 0.24 ± 0.01 a | 0.70 ± 0.05 a | n.d. | n.d. | |
| 4.5 | 0.18 ± 0.01 b | 0.34 ± 0.02 b | 0.12 ± 0.01 b | 0.52 ± 0.01 b | n.d. | n.d. | |
| OLEA + H2O | 1 | 0.07 ± 0.01 b | 0.42 ± 0.03 b | 0.12 ± 0.02 b | 0.49 ± 0.02 b | 0.13 ± 0.03 b | n.d. |
| 2 | 0.17 ± 0.02 a | 0.61 ± 0.07 a | 0.24 ± 0.02 a | 1.02 ± 0.12 a | 0.22 ± 0.02 a | n.d. | |
| 4.5 | 0.07 ± 0.01 b | 0.42 ± 0.02 b | 0.13 ± 0.03 b | 0.42 ± 0.05 b | 0.10 ± 0.01 b | n.d. | |
| OLEA + CH3 | 1 | 0.20 ± 0.02 b | 0.31 ± 0.03 b | 0.21 ± 0.02 b | 0.35 ± 0.03 b | 0.32 ± 0.03 b | 0.17 ± 0.01 b |
| 2 | 0.36 ± 0.03 a | 0.39 ± 0.04 a | 0.31 ± 0.02 a | 0.45 ± 0.04 a | 0.52 ± 0.04 a | 0.20 ± 0.02 b | |
| 4.5 | 0.25 ± 0.03 b | 0.27 ± 0.03 b | 0.24 ± 0.01 b | 0.31 ± 0.02 b | 0.30 ± 0.03 b | 0.34 ± 0.04 a | |
| OLEA + H2O + CH3 | 1 | 0.11 ± 0.01 b | 0.18 ± 0.03 c | 0.14 ± 0.01 b | 0.62 ± 0.06 b | 0.17 ± 0.02 b | 0.13 ± 0.02 c |
| 2 | 0.18 ± 0.03 a | 0.68 ± 0.03 a | 0.22 ± 0.03 a | 0.84 ± 0.04 a | 0.35 ± 0.02 a | 0.23 ± 0.03 b | |
| 4.5 | 0.08 ± 0.01 c | 0.44 ± 0.04 b | 0.09 ± 0.09 c | 0.48 ± 0.04 c | 0.15 ± 0.02 b | 0.43 ± 0.06 a | |
| OLEA + H2 + glucu | 1 | 0.13 ± 0.02 b | 0.08 ± 0.01 b | 0.17 ± 0.02 b | 0.24 ± 0.01 b | 0.28 ± 0.03 b | 0.10 ± 0.02 c |
| 2 | 0.20 ± 0.01 a | 0.18 ± 0.01 a | 0.28 ± 0.01 a | 0.37 ± 0.01 a | 0.40 ± 0.03 a | 0.35 ± 0.13 b | |
| 4.5 | 0.11 ± 0.01 b | 0.08 ± 0.01 b | 0.07 ± 0.01 c | 0.11 ± 0.02 c | 0.09 ± 0.02 c | 0.64 ± 0.03 a | |
| OLEA + H2O + glucu | 1 | 0.42 ± 0.07 a | 0.30 ± 0.03 b | 0.21 ± 0.02 b | 0.32 ± 0.02 b | 0.28 ± 0.03 b | 0.20 ± 0.03 b |
| 2 | 0.43 ± 0.06 a | 0.43 ± 0.03 a | 0.35 ± 0.05 a | 0.45 ± 0.03 a | 0.41 ± 0.03 a | 0.25 ± 0.04 b | |
| 4.5 | 0.17 ± 0.02 b | 0.13 ± 0.02 c | 0.01 ± 0.02 c | 0.15 ± 0.02 c | 0.10 ± 0.02 c | 0.91 ± 0.05 a | |
| OLEA + H2O + CH3 + glucu | 1 | 0.05 ± 0.00 c | 0.11 ± 0.02 b | n.d. | 0.06 ± 0.01 b | 0.04 ± 0.00 b | 0.05 ± 0.00 c |
| 2 | 0.20 ± 0.01 a | 0.42 ± 0.04 a | n.d. | 0.10 ± 0.02 a | 0.08 ± 0.01 a | 0.14 ± 0.02 b | |
| 4.5 | 0.13 ± 0.01 b | 0.05 ± 0.00 c | n.d. | 0.04 ± 0.01 b | 0.05 ± 0.01 b | 0.40 ± 0.02 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Yerena, A.; Vallverdú-Queralt, A.; Lamuela-Raventós, R.M.; Escribano-Ferrer, E. LC-ESI-LTQ-Orbitrap-MS for Profiling the Distribution of Oleacein and Its Metabolites in Rat Tissues. Antioxidants 2021, 10, 1083. https://doi.org/10.3390/antiox10071083
López-Yerena A, Vallverdú-Queralt A, Lamuela-Raventós RM, Escribano-Ferrer E. LC-ESI-LTQ-Orbitrap-MS for Profiling the Distribution of Oleacein and Its Metabolites in Rat Tissues. Antioxidants. 2021; 10(7):1083. https://doi.org/10.3390/antiox10071083
Chicago/Turabian StyleLópez-Yerena, Anallely, Anna Vallverdú-Queralt, Rosa M. Lamuela-Raventós, and Elvira Escribano-Ferrer. 2021. "LC-ESI-LTQ-Orbitrap-MS for Profiling the Distribution of Oleacein and Its Metabolites in Rat Tissues" Antioxidants 10, no. 7: 1083. https://doi.org/10.3390/antiox10071083
APA StyleLópez-Yerena, A., Vallverdú-Queralt, A., Lamuela-Raventós, R. M., & Escribano-Ferrer, E. (2021). LC-ESI-LTQ-Orbitrap-MS for Profiling the Distribution of Oleacein and Its Metabolites in Rat Tissues. Antioxidants, 10(7), 1083. https://doi.org/10.3390/antiox10071083