Genome-Wide Screening of Oxidizing Agent Resistance Genes in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Bacterial Strains
2.2. ASKA Library Screening and Minimal Inhibitory Concentration (MIC) Determination
2.3. Bioinformatical Analyses
3. Results
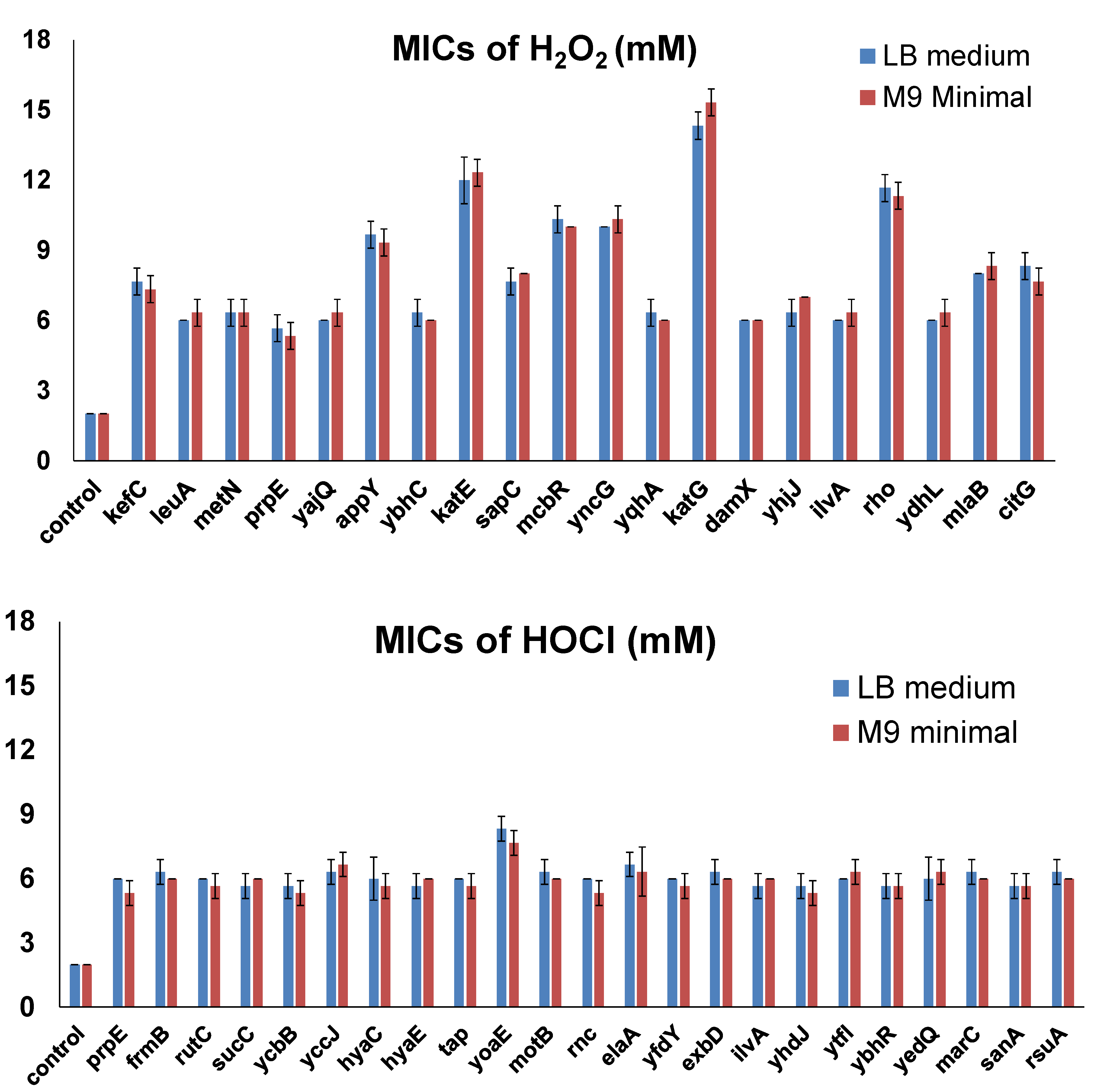
3.1. Genome-Wide Screening of Resistance Genes against H2O2
3.2. Genome-Wide Screening of Resistance Genes against HOCl
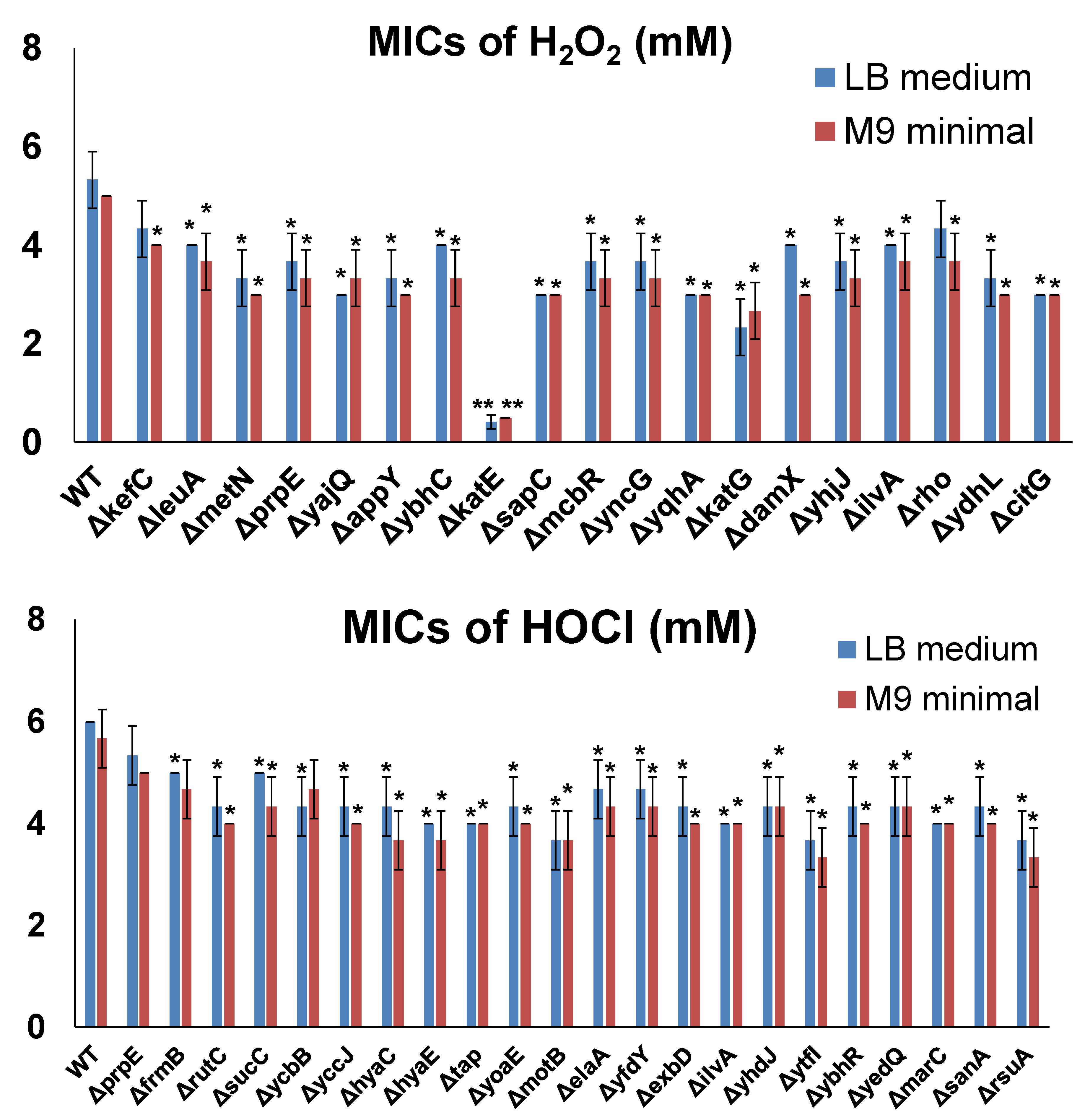
3.3. Effects of Overexpressing Selected Resistance Genes in the MG1655 Strain
3.4. Effects of Inactivating Selected Resistance Genes in the E. coli Genome
4. Discussion
4.1. Summary of the Study
4.2. H2O2-Resistance Genes
4.3. HOCl-Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Stockwell, B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014, 10, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Et Biophys. Acta Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 2019, 21, 521–530. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Biological production, detection, and fate of hydrogen peroxide. Antioxid. Redox Signal. 2018, 29, 541–551. [Google Scholar] [CrossRef]
- Cornelis, P.; Wei, Q.; Andrews, S.C.; Vinckx, T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 2011, 3, 540–549. [Google Scholar] [CrossRef]
- Jang, S.; Imlay, J.A. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 2010, 78, 1448–1467. [Google Scholar] [CrossRef]
- Brudzynski, K.; Abubaker, K.; Miotto, D. Unraveling a mechanism of honey antibacterial action: Polyphenol/H2O2-induced oxidative effect on bacterial cell growth and on DNA degradation. Food Chem. 2012, 133, 329–336. [Google Scholar] [CrossRef]
- Dahl, J.-U.; Gray, M.J.; Jakob, U. Protein quality control under oxidative stress conditions. J. Mol. Biol. 2015, 427, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, R.R.; Lehka, L.V.; Terenzi, A.; Matselyukh, B.P.; Rohr, J.; Jha, A.K.; Downey, T.; Kril, I.J.; Herbacek, I.; van Schoonhoven, S. Rapid generation of hydrogen peroxide contributes to the complex cell death induction by the angucycline antibiotic landomycin E. Free Radic. Biol. Med. 2017, 106, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef]
- Imlay, J.A. Diagnosing oxidative stress in bacteria: Not as easy as you might think. Curr. Opin. Microbiol. 2015, 24, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Jo, I.; Chung, I.-Y.; Bae, H.-W.; Kim, J.-S.; Song, S.; Cho, Y.-H.; Ha, N.-C. Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. USA 2015, 112, 6443–6448. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.W.; Kim, D.; Szubin, R.; Palsson, B.O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 2015, 12, 1289–1299. [Google Scholar] [CrossRef]
- Niu, W.; Zhang, Y.; Liu, J.; Wen, T.; Miao, T.; Basit, A.; Jiang, W. OxyR controls magnetosome formation by regulating magnetosome island (MAI) genes, iron metabolism, and redox state. Free Radic. Biol. Med. 2020, 161, 272–282. [Google Scholar] [CrossRef]
- Sultana, S.; Foti, A.; Dahl, J.-U. Bacterial Defense Systems against the Neutrophilic Oxidant Hypochlorous Acid. Infect. Immun. 2020, 88, e00964-19. [Google Scholar] [CrossRef]
- Goemans, C.V.; Collet, J.F. Stress-induced chaperones: A first line of defense against the powerful oxidant hypochlorous acid. F1000Res 2019, 8, 1678. [Google Scholar] [CrossRef] [PubMed]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving reactive chlorine stress: Responses of gram-negative bacteria to hypochlorous acid. Microorganisms 2020, 8, 1220. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Miura, H.; Peschek, J.; Le, Y.; Bach, N.C.; Kriehuber, T.; Winter, J. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 9493–9498. [Google Scholar] [CrossRef]
- Soo, V.W.; Hanson-Manful, P.; Patrick, W.M. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 1484–1489. [Google Scholar] [CrossRef] [PubMed]
- Qimron, U.; Marintcheva, B.; Tabor, S.; Richardson, C.C. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. USA 2006, 103, 19039–19044. [Google Scholar] [CrossRef]
- Chen, H.; Venkat, S.; Wilson, J.; McGuire, P.; Chang, A.L.; Gan, Q.; Fan, C. Genome-wide quantification of the effect of gene overexpression on Escherichia coli growth. Genes 2018, 9, 414. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martinez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017, 45, D543–D550. [Google Scholar] [CrossRef]
- Huntley, R.P.; Sawford, T.; Mutowo-Meullenet, P.; Shypitsyna, A.; Bonilla, C.; Martin, M.J.; O’Donovan, C. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015, 43, D1057–D1063. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. String: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Ferguson, G.P.; Nikolaev, Y.; McLaggan, D.; Maclean, M.; Booth, I.R. Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli: Role of KefB and KefC potassium channels. J. Bacteriol. 1997, 179, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Stieglitz, B.I.; Calvo, J.M. Distribution of the isopropylmalate pathway to leucine among diverse bacteria. J. Bacteriol. 1974, 118, 935–941. [Google Scholar] [CrossRef]
- Kadaba, N.S.; Kaiser, J.T.; Johnson, E.; Lee, A.; Rees, D.C. The high-affinity E. coli methionine ABC transporter: Structure and allosteric regulation. Science 2008, 321, 250–253. [Google Scholar] [CrossRef]
- Brock, M.; Maerker, C.; Schütz, A.; Völker, U.; Buckel, W. Oxidation of propionate to pyruvate in Escherichia coli: Involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 2002, 269, 6184–6194. [Google Scholar] [CrossRef] [PubMed]
- Saveanu, C.; Miron, S.; Borza, T.; Craescu, C.T.; Labesse, G.; Gagyi, C.; Popescu, A.; Schaeffer, F.; Namane, A.; Laurent-Winter, C. Structural and nucleotide-binding properties of YajQ and YnaF, two Escherichia coli proteins of unknown function. Protein Sci. 2002, 11, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Atlung, T.; Brøndsted, L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J. Bacteriol. 1994, 176, 5414–5422. [Google Scholar] [CrossRef]
- Molloy, M.P.; Herbert, B.R.; Slade, M.B.; Rabilloud, T.; Nouwens, A.S.; Williams, K.L.; Gooley, A.A. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 2000, 267, 2871–2881. [Google Scholar] [CrossRef]
- Loewen, P.C.; Switala, J. Purification and characterization of catalase HPII from Escherichia coli K12. Biochem. Cell Biol. 1986, 64, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, Y.; Nakamura, A.; Matsumoto, M.; Kanbe, A.; Sakanaka, M.; Higashi, K.; Igarashi, K.; Katayama, T.; Suzuki, H.; Kurihara, S. A novel putrescine exporter SapBCDF of Escherichia coli. J. Biol. Chem. 2016, 291, 26343–26351. [Google Scholar] [CrossRef] [PubMed]
- Lord, D.M.; Uzgoren Baran, A.; Soo, V.W.C.; Wood, T.K.; Peti, W.; Page, R. McbR/YncC: Implications for the mechanism of ligand and DNA binding by a bacterial GntR transcriptional regulator involved in biofilm formation. Biochemistry 2014, 53, 7223–7231. [Google Scholar] [CrossRef]
- Kanai, T.; Takahashi, K.; Inoue, H. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J. Biochem. 2006, 140, 703–711. [Google Scholar] [CrossRef]
- Loewen, P.C.; Triggs, B.L.; George, C.S.; Hrabarchuk, B.E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J. Bacteriol. 1985, 162, 661–667. [Google Scholar] [CrossRef]
- Yahashiri, A.; Jorgenson, M.A.; Weiss, D.S. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. USA 2015, 112, 11347–11352. [Google Scholar] [CrossRef]
- Grimminger, H.; Rahimi-Laridjani, I.; Koerner, K.; Lingens, F. Purification of threonine deaminase from Escherichia coli. Febs Lett. 1973, 35, 273–275. [Google Scholar] [CrossRef]
- Mitra, P.; Ghosh, G.; Hafeezunnisa, M.; Sen, R. Rho protein: Roles and mechanisms. Annu. Rev. Microbiol. 2017, 71, 687–709. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.W.; Hall, S.C.L.; Laxton, C.S.; Sridhar, P.; Mahadi, A.H.; Hatton, C.; Piggot, T.J.; Wotherspoon, P.J.; Leney, A.C.; Ward, D.G. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 2019, 4, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Dimroth, P.; Bott, M. Identification of triphosphoribosyl-dephospho-CoA as precursor of the citrate lyase prosthetic group. Febs Lett. 2000, 483, 165–168. [Google Scholar] [CrossRef]
- Gonzalez, C.F.; Proudfoot, M.; Brown, G.; Korniyenko, Y.; Mori, H.; Savchenko, A.V.; Yakunin, A.F. Molecular basis of formaldehyde detoxification: Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 2006, 281, 14514–14522. [Google Scholar] [CrossRef]
- Knapik, A.A.; Petkowski, J.J.; Otwinowski, Z.; Cymborowski, M.T.; Cooper, D.R.; Chruszcz, M.; Krajewska, W.M.; Minor, W. Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1294–1299. [Google Scholar] [CrossRef]
- Buck, D.; Guest, J.R. Overexpression and site-directed mutagenesis of the succinyl-CoA synthetase of Escherichia coli and nucleotide sequence of a gene (g30) that is adjacent to the suc operon. Biochem. J. 1989, 260, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Morè, N.; Martorana, A.M.; Biboy, J.; Otten, C.; Winkle, M.; Serrano, C.K.G.; Silva, A.M.; Atkinson, L.; Yau, H.; Breukink, E. Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio 2019, 10, e02729-18. [Google Scholar] [CrossRef] [PubMed]
- Volbeda, A.; Darnault, C.; Parkin, A.; Sargent, F.; Armstrong, F.A.; Fontecilla-Camps, J.C. Crystal structure of the O2-tolerant membrane-bound hydrogenase 1 from Escherichia coli in complex with its cognate cytochrome b. Structure 2013, 21, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Dubini, A.; Sargent, F. Assembly of Tat-dependent [NiFe] hydrogenases: Identification of precursor-binding accessory proteins. Febs Lett. 2003, 549, 141–146. [Google Scholar] [CrossRef]
- Manson, M.D.; Blank, V.; Brade, G.; Higgins, C.F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 1986, 321, 253–256. [Google Scholar] [CrossRef]
- Daley, D.O.; Rapp, M.; Granseth, E.; Melén, K.; Drew, D.; Von Heijne, G. Global topology analysis of the Escherichia coli inner membrane proteome. Science 2005, 308, 1321–1323. [Google Scholar] [CrossRef]
- Ridgway, H.G.; Silverman, M.; Simon, M.I. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J. Bacteriol. 1977, 132, 657–665. [Google Scholar] [CrossRef]
- Robertson, H.D.; Dunn, J.J. Ribonucleic acid processing activity of Escherichia coli ribonuclease III. J. Biol. Chem. 1975, 250, 3050–3056. [Google Scholar] [CrossRef]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef]
- Ollis, A.A.; Manning, M.; Held, K.G.; Postle, K. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 2009, 73, 466–481. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, S.E.; Balbontin, R.; Casadesus, J.; Marinus, M.G.; van der Woude, M. YhdJ, a nonessential CcrM-like DNA methyltransferase of Escherichia coli and Salmonella enterica. J. Bacteriol. 2007, 189, 4325–4327. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Shimada, T.; Yamamoto, K.; Ishihama, A. Transcription factor CecR (YbiH) regulates a set of genes affecting the sensitivity of Escherichia coli against cefoperazone and chloramphenicol. Microbiology 2016, 162, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R.; Galperin, M.Y.; Ghigo, J.-M.; Gomelsky, M.; Green, J.; Hughes, K.T.; Jenal, U.; Landini, P. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J. Bacteriol. 2016, 198, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.M.; Wang, W.; Silhavy, T.J. Novel RpoS-dependent mechanisms strengthen the envelope permeability barrier during stationary phase. J. Bacteriol. 2017, 199, e00708-16. [Google Scholar] [CrossRef]
- Conrad, J.; Niu, L.; Rudd, K.; Lane, B.G.; Ofengand, J. 16S ribosomal RNA pseudouridine synthase RsuA of Escherichia coli: Deletion, mutation of the conserved Asp102 residue, and sequence comparison among all other pseudouridine synthases. RNA 1999, 5, 751–763. [Google Scholar] [CrossRef]
- Zheng, M.; Aslund, F.; Storz, G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 1998, 279, 1718–1721. [Google Scholar] [CrossRef] [PubMed]
- Volbeda, A.; Amara, P.; Darnault, C.; Mouesca, J.-M.; Parkin, A.; Roessler, M.M.; Armstrong, F.A.; Fontecilla-Camps, J.C. X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, 5305–5310. [Google Scholar] [CrossRef] [PubMed]
- Gegner, J.A.; Dahlquist, F.W. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc. Natl. Acad. Sci. USA 1991, 88, 750–754. [Google Scholar] [CrossRef]
- Hantke, K. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 2005, 8, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Kutsukake, K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. MGG 1994, 243, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Clugston, S.L.; Barnard, J.F.J.; Kinach, R.; Miedema, D.; Ruman, R.; Daub, E.; Honek, J.F. Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: Evidence for optimal activation by nickel ions. Biochemistry 1998, 37, 8754–8763. [Google Scholar] [CrossRef]
- Masuda, H.; Awano, N.; Inouye, M. ydfD encodes a novel lytic protein in Escherichia coli. Fems Microbiol. Lett. 2016, 363, fnw039. [Google Scholar] [CrossRef][Green Version]
- Maddalo, G.; Stenberg-Bruzell, F.; Götzke, H.R.; Toddo, S.; Björkholm, P.; Eriksson, H.; Chovanec, P.; Genevaux, P.; Lehtiö, J.; Ilag, L.L. Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 2011, 10, 1848–1859. [Google Scholar] [CrossRef]
- Panina, E.M.; Mironov, A.A.; Gelfand, M.S. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 2001, 29, 5195–5206. [Google Scholar] [CrossRef]
- Chatterji, M.; Nagaraja, V. GyrI: A counter-defensive strategy against proteinaceous inhibitors of DNA gyrase. Embo Rep. 2002, 3, 261–267. [Google Scholar] [CrossRef]
- Andersen, P.S.; Smith, J.M.; Mygind, B. Characterization of the upp gene encoding uracil phosphoribosyltransferase of Escherichia coli K12. Eur. J. Biochem. 1992, 204, 51–56. [Google Scholar] [CrossRef]
- Palevsky, N.; Shemer, B.; Connolly, J.P.R.; Belkin, S. The highly conserved Escherichia coli transcription factor yhaJ regulates aromatic compound degradation. Front. Microbiol. 2016, 7, 1490. [Google Scholar] [CrossRef]
- Ray, W.K.; Larson, T.J. Application of AgaR repressor and dominant repressor variants for verification of a gene cluster involved in N-acetylgalactosamine metabolism in Escherichia coli K-12. Mol. Microbiol. 2004, 51, 813–826. [Google Scholar] [CrossRef]
- Leppik, R.A.; Young, I.G.; Gibson, F. Membrane-associated reactions in ubiquinone biosynthesis in Escherichia coli. 3-Octaprenyl-4-hydroxybenzoate carboxy-lyase. Biochim. Et Biophys. Acta Biomembr. 1976, 436, 800–810. [Google Scholar] [CrossRef]
- Hui, D.; Khaiat, S.; Uy, T.; Xu, H. Partial confirmation of single katG and katE knockouts and double katG/katE knockouts created from isogenic background of Escherichia coli K-12 strains. J. Exp. Microbiol. Immunol. 2014, 18, 139–145. [Google Scholar]
- Ivanova, A.; Miller, C.; Glinsky, G.; Eisenstark, A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol. Microbiol. 1994, 12, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Schellhorn, H.E.; Hassan, H.M. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 1988, 170, 4286–4292. [Google Scholar] [CrossRef]
- Wang, M.; Herrmann, C.J.; Simonovic, M.; Szklarczyk, D.; von Mering, C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 2015, 15, 3163–3168. [Google Scholar] [CrossRef]
- Bidnenko, E.; Bidnenko, V. Transcription termination factor Rho and microbial phenotypic heterogeneity. Curr. Genet. 2018, 64, 541–546. [Google Scholar] [CrossRef]
- Lindqvist, A.; Membrillo-Hernández, J.; Poole, R.K.; Cook, G.M. Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek 2000, 78, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, W.; Qi, K.; Wang, S.; Chen, X.; Ni, J.; Deng, R.; Shang, F.; Xue, T. McbR is involved in biofilm formation and H2O2 stress response in avian pathogenic Escherichia coli X40. Poult. Sci. 2019, 98, 4094–4103. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Favaloro, B.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Proteus mirabilis glutathione S-transferase B1-1 is involved in protective mechanisms against oxidative and chemical stresses. Biochem. J. 2003, 373, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hu, M.; Zhou, X.; Liu, Y.; Shi, C.; Shi, X. Role of yoaE Gene Regulated by CpxR in the Survival of Salmonella enterica Serovar Enteritidis in Antibacterial Egg White. Msphere 2020, 5, e00638-19. [Google Scholar] [CrossRef]
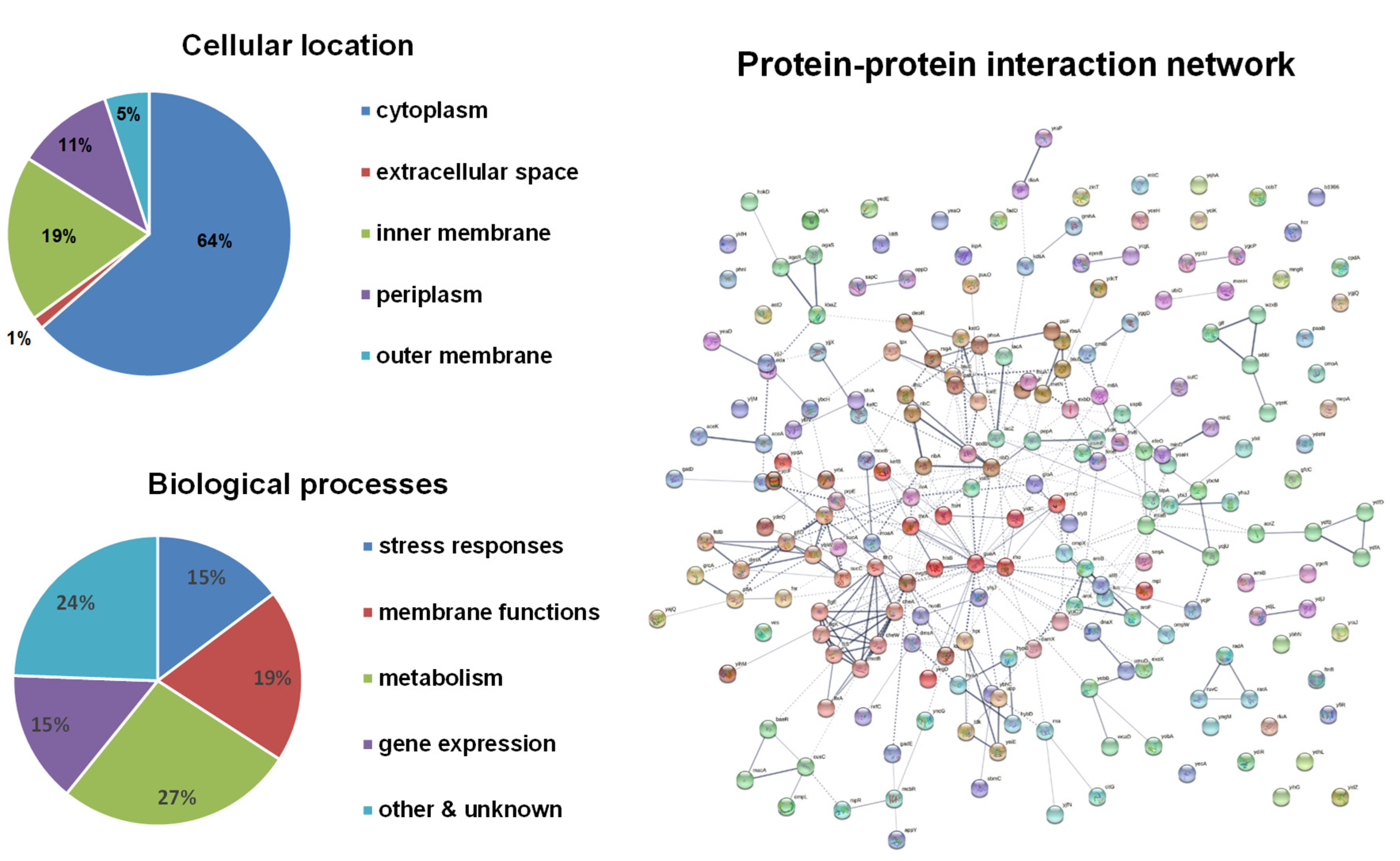

| Biological Functions 1 | Genes 2 |
|---|---|
| Stress responses | btuE, cheA, cheW, evgS, exoX, katE, katG, mepA, osmE, phoA, psiF, radA, rarA, ruvC, sbmC, sodB, sufC, tpx, ttdB, yajQ, ybcM, ybhT (acrZ), ybiJ, ycdO (efeO), ycjU (pgmB), ydjR (ves), yeaD, yegD, yfiD (grcA), yhiO (uspB), yjjX, yodA (zinT), ypdA (pyrS), yraJ |
| Membrane components and transporters | amiB, btuD, cmtB, cusC, damX, dmsA, exbD, feoB, fhuA, frvB, ftsH, gatC, hyaA, kefB, kefC, macA, metN, mltC, mtlA, nrfC, nuoB, ompL, ompW, ompX, oppD, rbsA, rfbX, sapC, shiA, slyB, wcaD, ybhC, ycjP, ydcT, ygjQ, yidC, yobA, yrbB (mlaB) |
| Metabolism | aceA, aceK, agaS, allB, aroB, aroF, aroL, astD, citG, cobT, eda, fadD, galU, gatD, gloA, guaA, hcr, hisB, hpt, hybD, hypB, ilvA, ispA, kbaZ, kdsA, lacA, lacZ, leuA, lpcA (gmhA), moaA, moeB, paaB, pflA, pflD, phnI, prpD, prpE, puuD, ribA, ribC, ribD, sucA, sucC, tdk, thiL, thrA, ubiD, upp, wbbI, yaiE (ppnP), ybdK, ybiS (ldtB), ycfS (idtC), ydeN, ydiR, ydjA, yeaU (dmlA), yfbB (menH) |
| DNA replication, gene expression and regulation | agaR, appY, baeR, cpdA, deoR, dnaX, flhD, fnr, gadE, mngR, pepA, rho, rluA, rna, rpmG, rsgA, stpA, tus, umuD, valS, ydfH, yecO (cmoA), yggD (fumE), yhaJ, yidZ, yjeK (epmB), yjfN, yncC (mcbR), yraO (diaA) |
| Cell division and mobility | flgE, flgK, fliS, flxA, minD, minE, motB, ybgF, ydeQ, yfiR, yihG, yraP |
| Other and unknown | glf, hokD, mpl, yabQ, yagM, yaiX, ybcH, ybfH, ybhN, ybiI, ybiV, ybiW, yceH, ycgL, yciC, yciK, yddK, ydfA, ydfB, ydfD, ydhL, ydjJ, ydjL, yeaO, yecA, yecI, yedE, yedM, yfdM, yfjM, ygcP, ygcU, ygeR, yhjJ, yihM, yjcF, yjjJ, ykfH, ymcB, yncG, yoaH, yobB, yoeE, yqeK, yqhA, yrbL |
| Gene | Known or Projected Functions |
|---|---|
| kefC | K+: H+ antiporter; plays a role in protecting the cell from electrophile toxicity [28]. |
| leuA | 2-isopropylmalate synthase; involved in the first step of leucine biosynthesis [29]. |
| metN | L-methionine/D-methionine ABC transporter ATP-binding subunit [30]. |
| prpE | Propionyl-CoA synthetase; catalyzes formation of propionyl-CoA, the first reaction in propionate catabolism via the methylcitrate cycle [31]. |
| yajQ | A nucleotide binding protein [32]. |
| appY | DNA-binding transcriptional activator; induces the expression of energy metabolism genes under anaerobiosis, stationary phase, and phosphate starvation [33]. |
| ybhC | An outer membrane lipoprotein [34]. |
| katE | Catalase HPII; the primary scavenger at high H2O2 concentrations [35]. |
| sapC | Putrescine ABC exporter membrane protein; putrescine efflux [36]. |
| mcbR | A member of the FadR C-terminal domain (FCD) family in the GntR superfamily of transcriptional regulators [37]. |
| yncG | Putative glutathione S-transferase [38]. |
| yqhA | Uncharacterized protein; predicted to be an integral membrane protein. |
| katG | Catalase/hydroperoxidase; bifunctional with both catalase and peroxidase activity [39]. |
| damX | Non-essential cell division protein [40]. |
| yhjJ | Peptidase M16 family protein. |
| ilvA | Threonine deaminase; carries out the first step in the synthesis of isoleucine [41]. |
| rho | Transcription termination factor; required for one of the two major types of termination of RNA transcription [42]. |
| ydhL | DUF1289 domain-containing protein. |
| mlaB | Intermembrane phospholipid transport system protein; forms a stable complex with MlaF, MlaE, and MlaD and is required for the stability of this complex [43]. |
| citG | Triphosphoribosyl-dephospho-CoA synthase [44]. |
| Biological Functions | Genes |
|---|---|
| Stress responses | cheW, dacB, frmB, gloA, groL, hyaE, hyaF, hybG, inaA, otsA, rfbC, sanA, sbmC, solA, ssuD, ycbB (ldtD), yghW, yhaK, yodA (ZinT), yqjA |
| Membrane components and transporters | appC, chbB, exbD, gspK, hyaA, hyaC, hydN, marC, mpaA, nmpC, nrfC, rfe, slyB, tap, ybhC, ybhR, ybiM, ybiO, yceJ, ydgK, yfdY, yggR, ygjE (ttdT), yhiP (dtpB), yoaE, yoeE, ypfJ, yrbB |
| Metabolism | acnA, agaS, aspC, bioA, cobS, cynT, dadA, hisA, ilvA, leuA, mtlD, nagD, paaG, prpE, purB, sucC, ubiD, upp, ycdK (rutC), ygbL |
| DNA replication, gene expression and regulation | agar, gyrA, lhr, rnc, rpoS, rsuA, trmU, uvrY, yccK (tusE), ydaV, ydcP (rlhA), ydiP, ydjF, yhaJ, yhdJ, ykgM, ymfG (xisE), yneJ |
| Cell division & mobility | flip, fliS, motB, ydeQ, yedQ (dgcQ), ygcF (queE), yneF (dgcF) |
| Other and unknown | dsrB, elaA, smf, sprT, yagM, ybbV, yccJ, ycgY, yddJ, ydfB, ydfD, ydhL, yeeX, yfiH (pgeF), yhcG, ykfC, ymfE, yncH, yncM, ynfN, ytfI |
| Gene | Known or Projected Functions |
|---|---|
| prpE | Propionate-CoA ligase; catalyzes the synthesis of propionyl-CoA from propionate and CoA [31]. |
| frmB | S-formylglutathione hydrolase; hydrolyzes S-formylglutathione to glutathione and formate [45]. |
| rutC | Putative aminoacrylate peracid reductase [46]. |
| sucC | Succinate-CoA ligase (ADP-forming) subunit beta; functions in the citric acid cycle [47]. |
| ycbB | Periplasmic L,D-transpeptidase; plays a role in the protective remodeling of peptidoglycan during cell envelope stress [48]. |
| yccJ | PF13993 family protein. |
| hyaC | Probable Ni/Fe-hydrogenase 1 b-type cytochrome subunit; functions in anchoring hydrogenase to the membrane [49]. |
| hyaE | Hydrogenase-1 operon protein [50]. |
| tap | Methyl-accepting chemotaxis protein IV [51]. |
| yoaE | UPF0053 inner membrane protein; putative transport protein [52]. |
| motB | Motility protein B; comprises the stator element of the flagellar motor complex with MotA [53]. |
| rnc | Ribonuclease 3 for rRNA processing [54]. |
| elaA | Putative N-acetyltransferase. |
| yfdY | DUF2545 domain-containing protein [55]. |
| exbD | A component of the energy-transducing Ton system [56]. |
| ilvA | Threonine deaminase; carries out the first step in the synthesis of isoleucine [41]. |
| yhdJ | Overexpression of YdhJ leads to methylation of genomic DNA at the NsiI recognition sequence (5′-ATGCAT-3′) [57]. |
| ytfI | Uncharacterized gene. |
| ybhR | One of two integral membrane subunits of a putative ABC exporter [58]. |
| yedQ | A probable inner membrane protein whose expression is dependent on σS under a number of stress conditions [59]. |
| marC | An inner membrane protein with six predicted transmembrane domains [52]. |
| sanA | Multi-copy expression of sanA complements the vancomycin sensitivity of an E. coli K-12 mutant with outer membrane permeability defects [60]. |
| rsuA | Pseudo-uridine synthase that is responsible for pseudouridylation of 16S rRNA at position 516 [61]. |
| Gene | Known or Projected Functions |
|---|---|
| leuA | 2-isopropylmalate synthase; involved in the first committed step in leucine biosynthesis [29]. |
| prpE | Propionyl-CoA synthetase; catalyzes formation of propionyl-CoA via the methylcitrate cycle [31]. |
| sucC | β subunit of succinyl-CoA synthetase [47]. |
| ybhC | An outer membrane lipoprotein [34]. |
| hyaA | Small subunit of hydrogenase-1; contains a unique proximal [4Fe-3S] cluster that is essential for oxygen tolerance [63]. |
| ydeQ | Uncharacterized gene. |
| cheW | Chemotaxis protein; in the ternary receptor complexes of two-component signaling pathways [64]. |
| motB | Motility protein B; comprises the stator element of the flagellar motor complex with MotA [53]. |
| yodA | Metal-binding protein; may function as a periplasmic zinc chaperone delivering zinc to apo-enzymes in this compartment [65]. |
| ydfB | Uncharacterized gene. |
| fliS | Flagellar biosynthesis protein; substrate-specific chaperones of the flagellar export system [66]. |
| gloA | Glyoxalase I; catalyzes the first of two sequential steps in the conversion of methylglyoxal to D-lactate [67]. |
| ydfD | A lysis protein encoded by the Qin prophage [68]. |
| slyB | Outer membrane lipoprotein [69]. |
| yoeE | TonB-dependent receptor plug domain-containing protein; may be regulated by Fur regulon [70]. |
| sbmC | DNA gyrase inhibitor; protects cell from DNA damage cause by DNA-bound gyrase [71]. |
| upp | Uracil phosphoribosyltransferase; a pyrimidine salvage enzyme that catalyzes the synthesis of uridine 5′-monophosphate from uracil and 5-phospho-α-D-ribose 1-diphosphate [72]. |
| yhaJ | DNA-binding transcriptional activator; a member of the LysR protein family [73]. |
| agaR | DNA-binding transcriptional repressor [74]. |
| exbD | A component of the energy-transducing Ton system [56]. |
| agaS | Putative galactosamine-6-phosphate deaminase/isomerase. |
| ilvA | Threonine deaminase; carries out the first step in the synthesis of isoleucine [41]. |
| ubiD | 3-octaprenyl-4-hydroxybenzoate decarboxylase; an enzyme of the ubiquinol-8 biosynthesis pathway that catalyzes the decarboxylation of 3-octaprenyl-4-hydroxybenzoate [75]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Wilson, J.; Ercanbrack, C.; Smith, H.; Gan, Q.; Fan, C. Genome-Wide Screening of Oxidizing Agent Resistance Genes in Escherichia coli. Antioxidants 2021, 10, 861. https://doi.org/10.3390/antiox10060861
Chen H, Wilson J, Ercanbrack C, Smith H, Gan Q, Fan C. Genome-Wide Screening of Oxidizing Agent Resistance Genes in Escherichia coli. Antioxidants. 2021; 10(6):861. https://doi.org/10.3390/antiox10060861
Chicago/Turabian StyleChen, Hao, Jessica Wilson, Carson Ercanbrack, Hannah Smith, Qinglei Gan, and Chenguang Fan. 2021. "Genome-Wide Screening of Oxidizing Agent Resistance Genes in Escherichia coli" Antioxidants 10, no. 6: 861. https://doi.org/10.3390/antiox10060861
APA StyleChen, H., Wilson, J., Ercanbrack, C., Smith, H., Gan, Q., & Fan, C. (2021). Genome-Wide Screening of Oxidizing Agent Resistance Genes in Escherichia coli. Antioxidants, 10(6), 861. https://doi.org/10.3390/antiox10060861







