Abstract
The purpose of the current study was to investigate antioxidant and anti-inflammatory effects of spray dry powder containing 40% curcumin (CM-SD) in C2C12 myoblast cells. CM-SD increased DPPH radical scavenging activity in a dose-dependent manner, and up to 30 μg/mL of CM-SD did not express cytotoxicity in C2C12 cells. Exposure to hydrogen peroxide (H2O2) drastically decreased the viability of C2C12 cells, but pre-treatment of CM-SD significantly increased the cell viability (p < 0.01). CM-SD significantly transactivated the nuclear factor erythroid-2-related factor 2 (Nrf2)-dependent luciferase activity in a dose-dependent manner and enhanced the levels of heme oxygenase (HO)-1, glutamate cysteine ligase catalytic subunit (GCLC), and NAD(P)H-dependent quinone oxidoreductase (NQO)-1. CM-SD also significantly reduced reactive oxygen species (ROS) production and lipid peroxidation and restored glutathione (GSH) depletion in H2O2-treated C2C12 cells. Moreover, CM-SD significantly reduced lipopolysaccharides (LPS)-mediated interleukin (IL)-6 production in the conditioned medium. Results from the current study suggest that CM-SD could be a useful candidate against oxidative stress and inflammation-related muscle disorders.
1. Introduction
Sarcopenia is the degenerative loss of skeletal muscle mass (0.5–1% loss per year after the age of 50), quality, and strength associated with aging [1]. Nutritional imbalance, accumulation of aged proteins, depletion of growth hormones, proinflammatory cytokines, oxidative stress-mediated apoptosis of skeletal muscles, and inactivation of satellite cells have been known to cause sarcopenia [2,3]. Although the detailed molecular mechanisms which contribute to sarcopenia are largely unknown and need to be further elucidated, recent evidence indicates that apoptosis is associated with the aging-related loss of skeletal muscle since increases in Bax/Bcl-2 ratio, cytoplasmic release of cytochrome c, activation of p53 and caspases, and fragmentation of chromosomal DNA are observed in aged muscles [4,5,6]. The current estimates suggest that about 200 million people worldwide will experience sarcopenia to a degree that could affect their health over the next four decades [7]. Although there are no approved mediations for the treatment of sarcopenia, exercise and nutrition for stimulating muscle protein synthesis have been suggested to delay the progression of sarcopenia. Thus, the development of strategies to counteract the negative impact of sarcopenia is warranted. Medicinal herbs are gaining space and importance in the pharmaceutical industry as well as inspiring the search for new potential sources of bioactive molecules, and they are considered to be potential sources to combat various diseases including muscle weakness [8,9,10,11,12]. As regulation of oxidative stress seems to be essential for sarcopenia [3,9], traditional herbs possessing potent antioxidant activities are attractive resources for managing sarcopenia with fewer side effects.
Curcuma longa L., turmeric or curcuma, an herbaceous perennial of the Zingiberaceae, is distributed in Africa, Latin America, and Asia, including India, China, and Japan [13,14]. Curcumin, a representative substance from the root of turmeric, includes about 2–8% turmeric and is consumed daily throughout Asian countries reported without toxicity [15,16]. Curcumin has also been reported to possess diverse pharmacological effects including antioxidant and anti-inflammatory activities [15,17]. Recently, it has been suggested that curcumin alleviated chronic kidney disease-induced muscle atrophy by inhibiting glycogen synthase kinase (GSK)-3β [18]. Nevertheless, the effects of curcumin on wasting muscle have not been fully established yet. Therefore, the objective of the present study was to evaluate in vitro antioxidant and anti-inflammatory effects of curcumin in a murine myoblast C2C12 cell lines, as a process to develop potent myoprotective medicinal foods.
2. Materials and Methods
2.1. Preparation of Test Articles
Spray dry powders of turmeric containing 40% curcumin (CM-SD) were supplied by Aribio Co., Ltd. (Seongnam-si, Gyeonggi-do, Republic of Korea) on 17 October 2018. In brief, commercial ethanol extract of turmeric (Curcumin C3 Complex®, 95% curcuminoids; Sabinsa Korea Corporation, Seoul, Republic of Korea) was blended with purified water, phosphoric acid, and Ghatti gum, and then the mixture was suspended for more than 20 h for particle sizes less than 1000 nm. After filtering, 6.76% of dextrin was added to the suspension and spray dried to produce Ari C4 Complex as CM-SD with 400 mg/g of curcumin. Some specimens of turmeric were deposited in the herbarium of the Department of Anatomy and Histology, College of Korean Medicine, Daegu Haany University. One hundred milligrams of CM-SD were dissolved in 1 mL of dimethyl sulfoxide (DMSO) and then filter-sterilized by using a 0.2 μm syringe filter (Nalgene, NY, USA). One hundred millimolar of 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox; Merck Millipore, Burlington, MA, USA) and 30 μM of isorhamnetin (IsoR), a reference drug as a positive control, were also dissolved in DMSO. All test articles were stored at –20 °C to protect light and humidity until use.
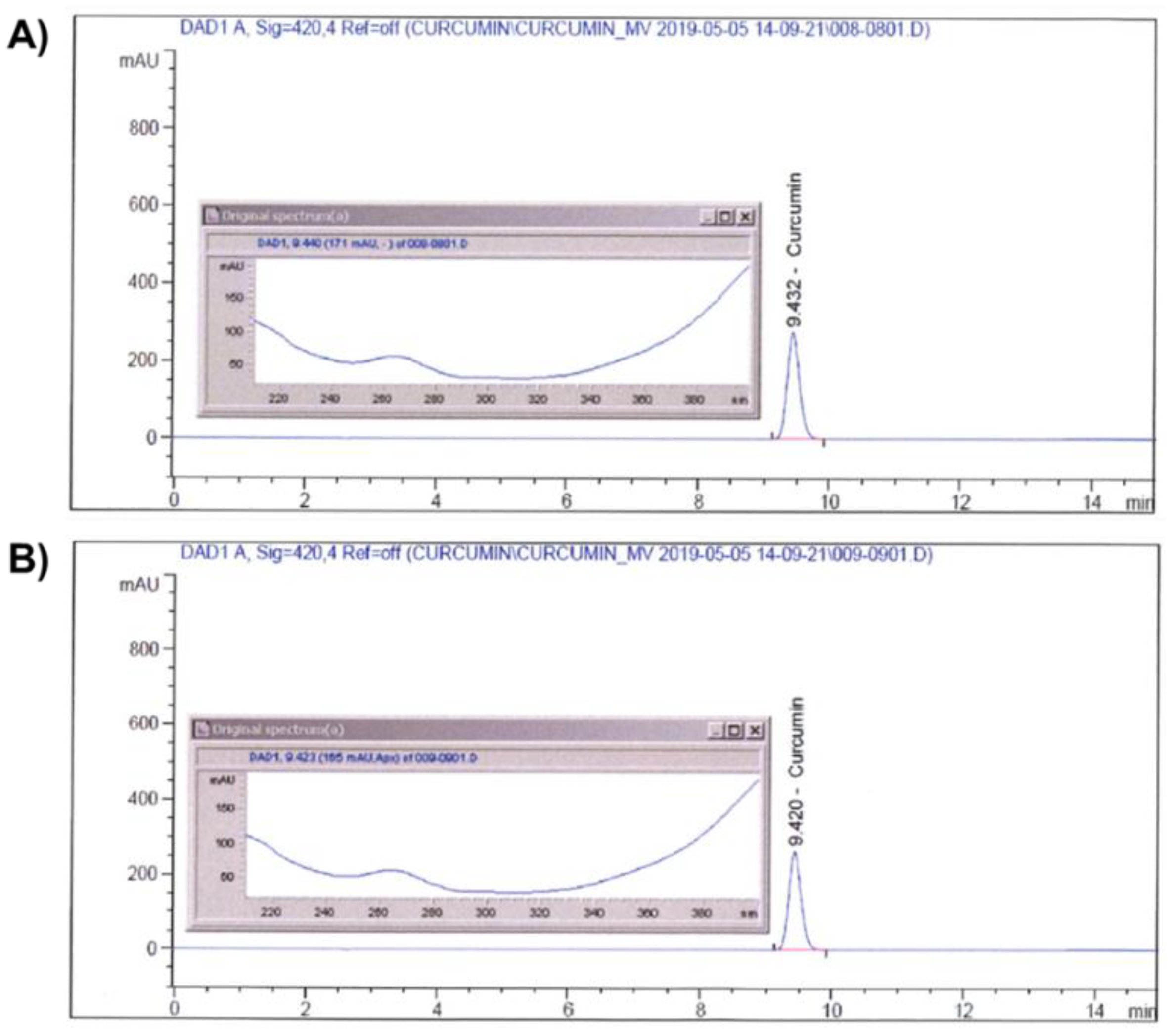
Identification of curcumin in CM-SD was performed by an Agilent 1100 Series high-performance liquid chromatography (HPLC) System (Agilent Technologies, Inc., Santa Clara, CA, USA) coupled with diode array detector. Curcumin was analyzed by a Capcell Pak® C18 column (4.6 × 250 mm, 5 μm; Shiseido Co., Ltd., Kyoto, Japan) at 40 °C. A mobile phase containing 0.1% (v/v) phosphoric acid (Samchun Chemicals Co., Ltd., Pyeongtaek-si, Gyeonggi-do, Republic of Korea) (A) and acetonitrile (HPLC grade; Burdick & Jackson, Charlotte, NC, USA) (B) was used with the following gradient: 0–15 min, 50% B. The sample injection volume was 10 μL with a flow rate of 1 mL/min. The UV wavelength was set at 420 nm. Curcumin standard (Sigma-Aldrich, St. Louis, MO, USA) and CM-SD were both dissolved in HPLC grade methanol (Daejung Chemicals & Materials, Siheung-si, Gyeonggi-do, Republic of Korea) for the analyses. The chromatograms of curcumin in standard chemical and CM-SD are shown in Figure 1A,B, respectively.
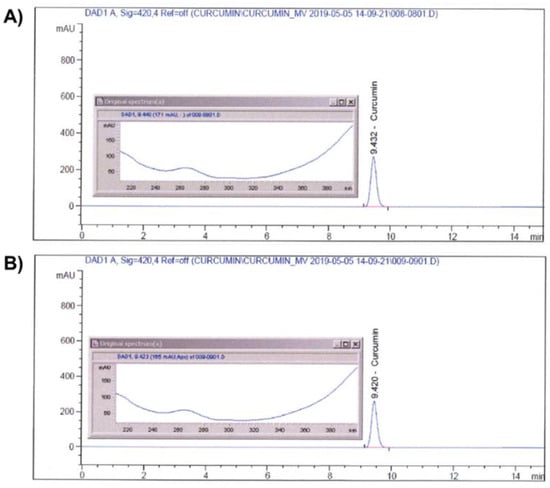
Figure 1.
Identification of curcumin on standard (A) and CM-SD (B) by HPLC.
2.2. Determination of Radical Scavenging Activity
Radical scavenging activity was determined using the 2,2-diphenyl-1-picrylhydrazyl (DPPH) method [19]. DPPH (Sigma-Aldrich) in ethanol solution (150 μM) was reacted with CM-SD (1–100 μg/mL) or Trolox (100 μM), a water-soluble analog of vitamin E. Samples were incubated at room temperature for 30 min under light protection. The absorbance was measured at 517 nm using a Synergy HTX microplate reader (BioTek Instruments, Inc., Winooski, VT, USA). Radical scavenging activity was calculated according to the following equation:
where S and S0 indicate absorbance of DPPH solution with or without samples, and C and C0 indicate absorbance of solvent with or without sample.
2.3. C2C12 Myoblast Cell Culture
C2C12 cells, a murine myoblast cell line, were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained under standard conditions, as previously described [20]. Briefly, C2C12 myoblast cells were cultured in growth medium comprising 90% Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum, 50 units/mL penicillin, and 50 μg/mL streptomycin at 37 °C in a humidified atmosphere with 5% CO2. When C2C12 cells were grown to about 70% of confluency, the cells were sub-cultured on appropriated new culture dishes to maintain phenotype of the myoblast cells. Recombinant HepG2 cells expressing antioxidant response element (ARE)-driven luciferase were generated, as described previously [21]. Briefly, pGL4.37[luc2P/ARE/Hygro] Vector, ARE-driven reporter gene construct, was obtained from Promega Corporation (Madison, WI, USA). HepG2 cells were stably transfected with pGL4.37 plasmid using FuGENE® HD Transfection Reagent (Promega) according to manufacturer’s instruction, and 80 μg/mL of hygromycin was added to select the resistant colonies. The selected resistant colonies were pooled and used for reporter gene analysis.
2.4. Cell Viability Assay
C2C12 cells were seeded at a density of 2 × 104 cells/well in a 96-well plate to determine the cytotoxicity activity of CM-SD. To examine the cytotoxicity of CM-SD, C2C12 cells were treated with 1–30 μg/mL of CM-SD for 24 h. To investigate the cytoprotective effects of CM-SD in C2C12 cells, 1–30 μg/mL of CM-SD were pre-incubated 1 h prior to the addition of 100 μM of hydrogen peroxide (H2O2; Sigma-Aldrich) for 6 h. Thirty micromolar of IsoR was used as a reference drug [22]. After incubation of the cells, viable cells were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 0.1 μg/mL; Sigma-Aldrich) for 2 h, according to a previous study with some modifications [23]. The media were removed, and then the produced formazan crystals in each well were dissolved by 200 μL of DMSO. Absorbance was measured at 570 nm using an automatic microplate reader (BioTek Instruments, Inc.). Relative cell viabilities were calculated according to the following equation:
2.5. Nuclear Factor Erythroid-2-Related Factor 2 (Nrf2)-Dependent Reporter Gene Assay
To determine the luciferase activity, recombinant HepG2 cells which express pGL4.37 were replated in 12-well plates (5 × 105 cells/well) overnight and exposed to 1–30 μg/mL of CM-SD for 18 h, as previously described [21]. The relative luciferase activity was calculated as the relative change to protein content measured by the bicinchoninic acid method. To investigate Nrf2 transactivation in C2C12 cells, reporter gene assay was carried out as previously established [24]. Briefly, C2C12 cells were plated at a density of 5 × 104 cells per well in a 12-well plate and allowed to grow overnight. The cells were co-transfected with 500 ng of pGL4.37 and 100 ng of pRL-TK (Renilla luciferase expression plasmid under an HSV-thymidine kinase promoter) for 6 h in the presence of FuGENE® HD transfection reagent (Promega), and then exposed to 3–30 μg/mL of CM-SD for 18 h. Thirty micromolar of IsoR was used as a reference drug [22]. The luminescence intensity of treated cell lysates was measured using Dual-Luciferase Reporter Assay System and GloMax® 20/20 Luminometer (Promega). The luminescence intensity of firefly luciferase was divided by that of Renilla luciferase to calculate the relative luciferase activity.
2.6. Total RNA Isolation and Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
After C2C12 cells were treated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 12 h, total RNA was isolated from treated cells using Tri-solution (Bioscience Technology, Daegu, Republic of Korea). To obtain cDNA, the RNA (2 μg each) and oligo-d(T)16 primer were added to AccuPower® RT PreMix (Bioneer Corporation, Daejeon, Korea), and resulting mixtures were reverse-transcribed using a Mastercycler® Nexus Gradient Thermal Cycler (Eppendorf, Hamburg, Germany). Real-time PCR was performed using a CFX-96 (Bio-Rad, Hercules, CA, USA) and a SYBR® Green Premix (Takara Bio Inc., Shiga, Japan). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene and melting curve analysis was performed to verify amplicon accuracy. Relative expression of each gene was calculated as 2−ΔΔCT, as previously reported [25]. Primers for the amplification of specific genes were synthesized from Bioneer Corporation, and nucleotide sequences are listed in Table 1.

Table 1.
Oligonucleotide sequences used in the present study.
2.7. Determination of Superoxide Dismutase (SOD) Activity
After C2C12 cells were treated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 24 h, cells in phosphate buffered saline were homogenized by sonication and clarified by centrifugation at 1500× g for 10 min. SOD activity in cell homogenates was determined using a commercial kit (Cayman Chemical, Ann Arbor, MI, USA) which utilizes a tetrazolium salt for detection of superoxide radicals generated by xanthine oxidase and hypoxanthine at 450 nm. SOD activity was determined from a standard curve, and specific activity was obtained by dividing SOD activity by protein concentration.
2.8. Determination of Glutathione (GSH) Contents
Glutathione contents were determined using GSH/GSSG-Glo™ Assay Kit (Promega), according to manufacturer’s instructions. Briefly, C2C12 cells were pretreated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 1 h, and then exposed to 100 μM of H2O2 for 6 h. Cells were lysed with glutathione lysis reagent and then incubated with luciferin generating reagent for 30 min and luciferin detection reagent for 15 min. Luminescence intensities were measured using an automated microplate reader (Infinite 200 PRO; Tecan Group Ltd., Männedorf, Switzerland), and the concentration of GSH was determined from a standard curve.
2.9. Determination of Reactive Oxygen Species (ROS) Production
ROS production was analyzed as previously described [26] with some modifications. Briefly, C2C12 cells were replated in 96-well black plate (2 × 104 cells/well) overnight, pretreated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 1 h, and exposed to 100 μM of H2O2 and 1 μM of 2′,7′-dichlorofluorescein diacetate (DCFH-DA; Sigma-Aldrich) for 3 h. Thirty micromolar of IsoR was used as reference drug. The fluorescence emitted by dichlorofluorescein was measured at excitation/emission wavelengths of 485/530 nm using an automated microplate reader (Tecan Group Ltd.).
2.10. Determination of Malondialdehyde (MDA) Contents
C2C12 cells were pretreated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 1 h, and subsequently exposed to 100 μM of H2O2 for 12 h. After treatment, the cells were lysed by a radioimmunoprecipitation assay buffer containing 1 mM sodium fluoride, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, and protease inhibitor cocktail (GenDEPOT, Barker, TX, USA) for 1 h on ice and centrifuged at 15,000× g for 10 min. MDA levels in the resulting lysates (approximately 250 μg of protein lysates) were assayed according to the manufacturer’s instruction (TBARS Assay Kit; Cayman Chemical, Ann Arbor, MI, USA). The absorbance of the sample was monitored at 530 nm using a microplate reader (BioTek Instruments, Inc.), and the concentration of MDA was determined from a standard curve.
2.11. Quantification of Tumor Necrosis Factor (TNF)-α, Interleukin (IL)-1β, and Interleukin (IL)-6 Levels
To quantify proinflammatory cytokines in the conditioned medium, C2C12 cells were pretreated with 3–30 μg/mL of CM-SD or 30 μM of IsoR for 1 h and subsequently exposed to 1 μg/mL of lipopolysaccharide (LPS) for 12 h, as previously described [23]. Levels of murine TNF-α, IL-1β, and IL-6 in conditioned media were measured by BD OptEIA™ ELISA sets (BD Biosciences, San Diego, CA, USA), according to manufacturer instructions. Briefly, Nunc MaxiSorp™ 96-well microplate (Thermo Fisher Scientific, Waltham, MA, USA) was coated with each diluted capture antibodies, blocked with phosphate buffered saline containing 10% fetal bovine serum, bound with diluted conditioned media, and incubated with streptavidin-conjugated horseradish peroxidase and detection antibodies. After intense washing with phosphate buffered saline containing 0.05% Tween 20, the wells were developed with TMB Substrate Reagent Set (BD Biosciences) and added 2 N of H2SO4 to stop color development. Absorbance was measured at 420 nm with an automated microplate reader (BioTek Instruments). Absorbance at 570 nm was used as reference absorbance. Levels of IL-6 were calculated by interpolation of standard curve.
2.12. Statistical Analyses
All numerical data were expressed as mean ± standard deviation (SD) from at least three separated experiments. One-way analysis of variance test (ANOVA) was used to determine the significances of differences among experimental groups, and Levene’s test was used to examine variance homogeneity. Fisher’s least significant difference (LSD) test for variance homogeneity or Dunnett’s T3 test for variance non-homogeneity was used as post hoc analysis to identify significant differences between pairs of groups. Differences were considered significant at p < 0.05. Statistical analyses were conducted using SPSS for Windows (Release 14.0K; SPSS Inc., Chicago, IL, USA).
3. Results and Discussion
3.1. Radical Scavenging Activity of CM-SD
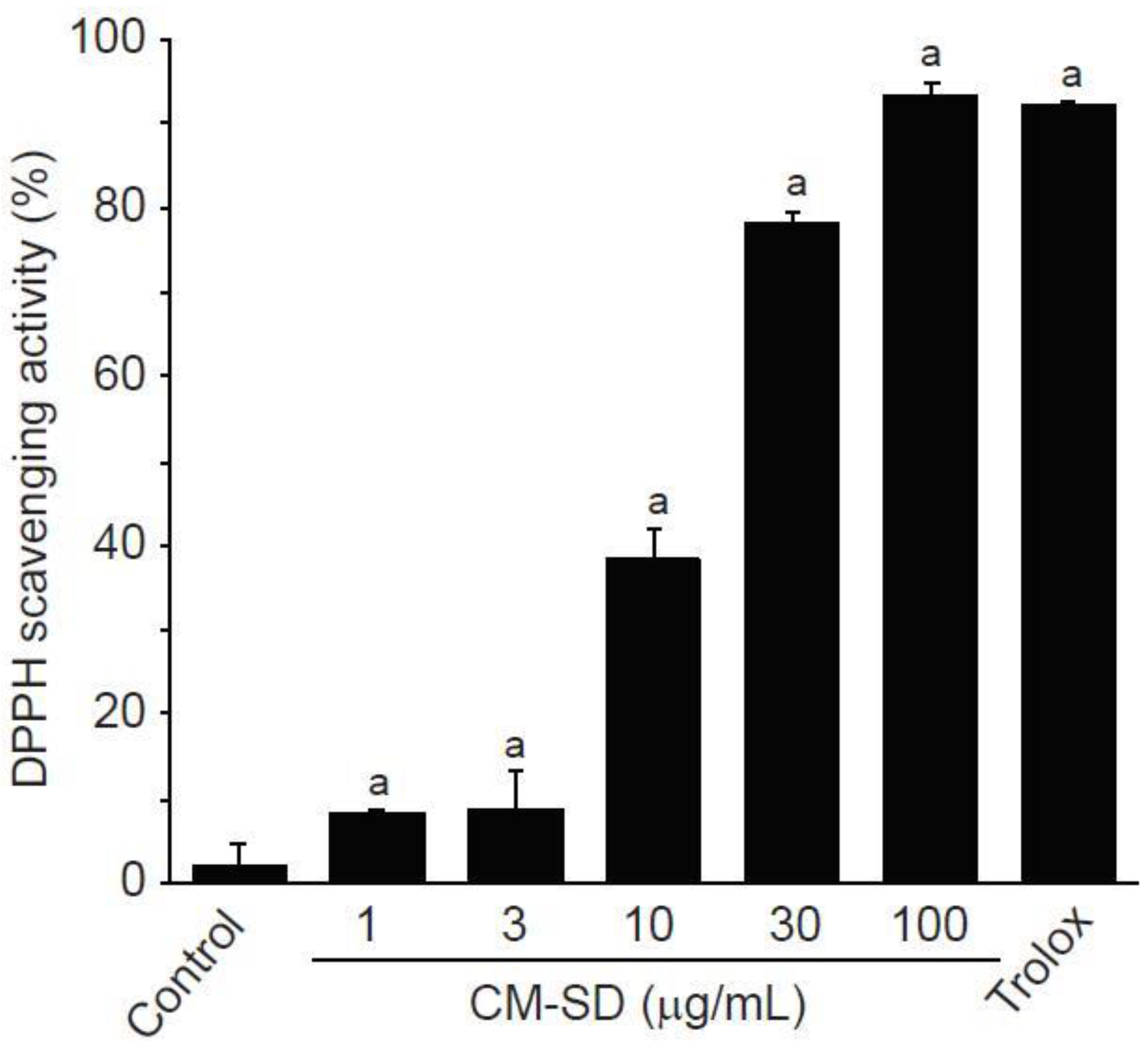
Antioxidant activity is regarded as one of the most important mechanisms for the regulation of degenerative loss of skeletal muscle. To investigate in vitro antioxidant capacity of CM-SD, the current study examined radical scavenging activity by DPPH assay. CM-SD increased radical scavenging activity in a dose-dependent manner, as shown in Figure 2. Relative radical scavenging activities by 1, 3, 10, 30, and 100 μg/mL of CM-SD were 8.19 ± 0.47%, 8.85 ± 4.49%, 35.25 ± 3.67%, 78.42 ± 1.02%, and 93.53 ± 1.38%, respectively. Radical scavenging activity by 100 μM of Trolox was 92.30 ± 0.17%. Significant differences in radical scavenging activity were observed in 1–100 μg/mL of CM-SD, and the level for 100 μg/mL of CM-SD was comparable to that by 100 μM of Trolox (p = 0.562 in LSD test).
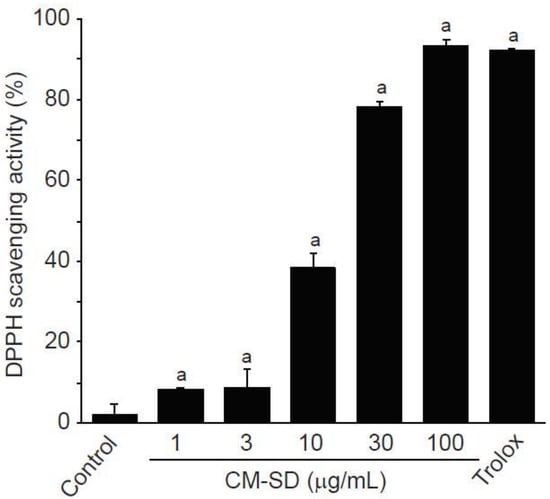
Figure 2.
DPPH scavenging activity of curcumin 40% spray dry powders (CM-SD). Values are expressed as mean ± SD of three separate experiments. a p < 0.01, significant as compared with control by LSD test.
Enhancing antioxidant capacity plays an essential role in protecting cells from oxidative stress and reducing inflammatory responses in skeletal muscle. Therefore, we firstly investigated the antioxidant potential of CM-SD using in vitro DPPH radical, a well-known radical generator. Results from the present study exhibited that CM-SD increased radical scavenging activity in a dose-dependent manner. Similarly, a previous study using in vitro models found that curcumin proved antioxidant and free radical scavenging abilities [27]. Thus, CM-SD seems to have potent radical scavenging activities without any cytotoxicity at least in our experimental system, while further studies would be required to evaluate in vitro and in vivo toxicity studies for CM-SD.
3.2. Effects of CM-SD on C2C12 Cell Viability and H2O2-Induced Cytotoxicity
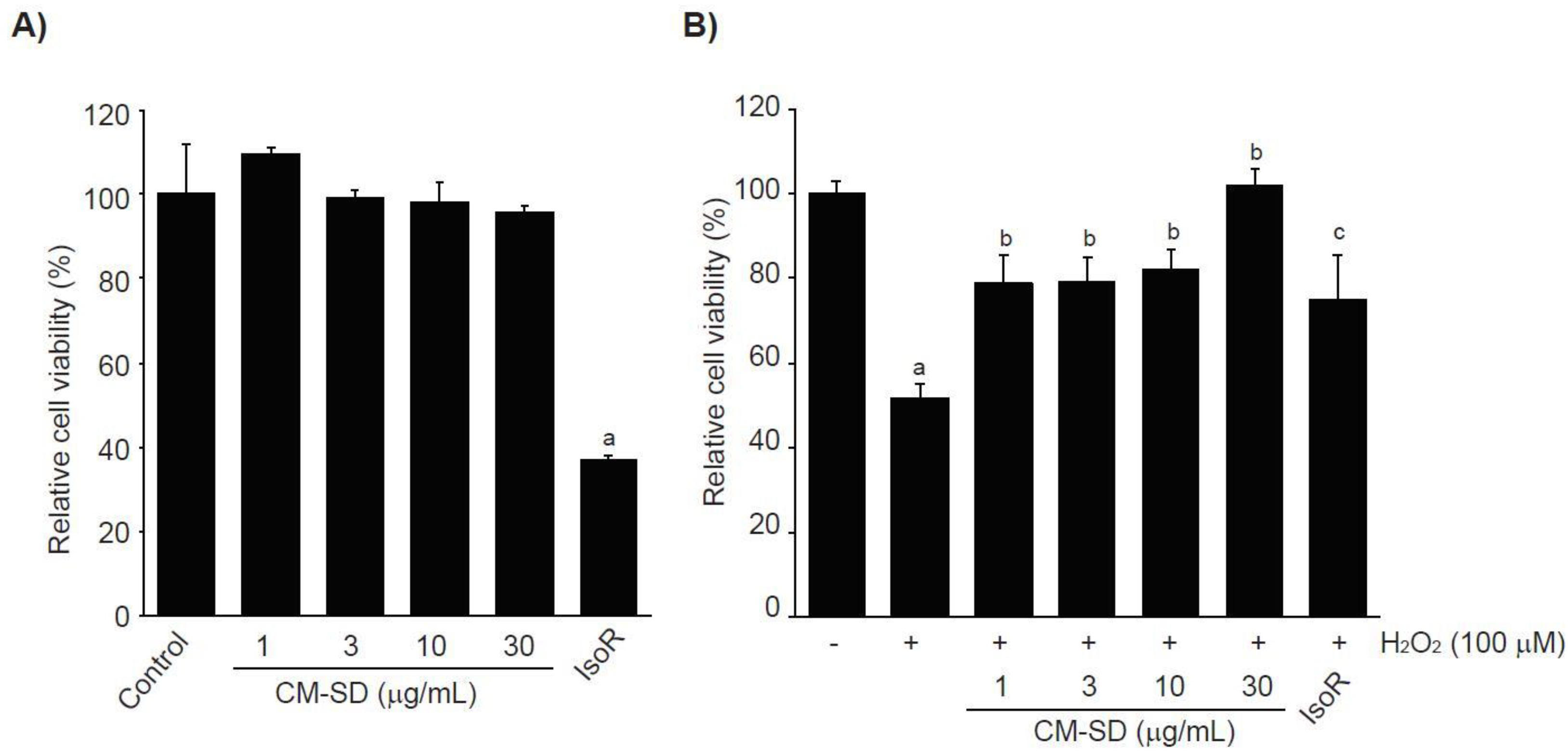
Prior to evaluating the antioxidant potentials, the effects of CM-SD on cell viabilities were investigated after treating C2C12 cells with 1–30 μg/mL of CM-SD for 24 h. The results from MTT assay indicate that treatment with CM-SD did not show any cytotoxicity. However, treatment of IsoR (30 μM, 24 h), a reference drug, significantly decreased the viability of C2C12 cells (Figure 3A). Relative cell viability by treatment with 1, 3, 10, and 30 μg/mL of CM-SD was 109.60 ± 1.55%, 99.37 ± 1.89%, 98.38 ± 4.69%, and 95.74 ± 1.66% of control cells, respectively. Cell viability by 30 μM of IsoR was 37.19 ± 0.55% of control cells. Since 1–30 μg/mL of CM-SD did not show any cytotoxicity, the current study chose up to 30 μg/mL of CM-SD as a maximum treatment concentration in subsequent experiments in C2C12 cells.
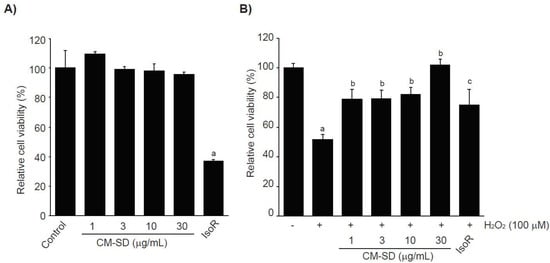
Figure 3.
Effect of CM-SD on cell viability (A) and H2O2-mediated cytotoxicity (B) of C2C12 cells. Values are expressed as mean ± SD of six separate experiments. a p < 0.01, significant as compared with control by Dunnett’s T3 test; b p < 0.01, c p < 0.05, significant as compared with H2O2-treated cells by Dunnett’s T3 test.
It was well established that H2O2, lactate, and LPS are representative agents to provoke cytotoxicity of myoblasts by inducing oxidative stress, metabolic stress, and inflammation [28,29,30,31], The current study preliminary tested whether H2O2, sodium L-lactate (20 mM), or LPS (1 μg/mL) reduced the viability of C2C12 cells or not. Only treatment of H2O2 (1 mM, 24 h) significantly decreased the viability of C2C12 cells (data not shown). To examine the effect of CM-SD on oxidative stress-mediated cytotoxicity, C2C12 cells were pretreated with 1–30 μg/mL of CM-SD for 1 h, subsequently exposed to 100 μM of H2O2 for 6 h, and then determined the cell viability by MTT assay. Exposure of H2O2 for 6 h significantly decreased the cell viability (51.80 ± 3.36% of control). However, pretreatment of CM-SD increased cell viability, and the cell viability by pretreatment with 30 μg/mL of CM-SD was comparable to control cells (p = 0.998). Statistical significances of differences were observed in all four different concentrations of CM-SD-pretreated cells as compared with H2O2 alone-treated cells (Figure 3B). When comparing with 30 μM of IsoR, pretreatment of CM-SD (30 μg/mL) showed a more potent cytoprotective activity against H2O2-induced cytotoxicity (p = 0.013). Relative cell viability by pretreatment with 1, 3, 10, and 30 μg/mL of CM-SD were 78.91 ± 6.47%, 79.39 ± 5.55%, 81.97 ± 4.97%, and 101.87 ± 3.78% of control cells, respectively. Relative cell viability by pretreatment with 30 μM of IsoR was 74.97 ± 10.67% of control cells.
Hydrogen peroxide (H2O2), generated in oxidative stress, easily penetrates inside and outside of the cells and acts as a second messenger for maintaining redox homeostasis [32]. However, H2O2, beyond adaptation capacity, produces more aggressive oxygen radicals, leads to dysfunction of biological macromolecules, and thereby induces DNA damage, apoptosis, and necrosis [33]. In this regard, H2O2 has been widely used as a pro-oxidant in in vitro cultured cell lines including C2C12 cells [22]. In parallel with the previous report, the present results showed that the treatment of 100 μM of hydrogen peroxide for 6 h significantly reduced the viability of C2C12 cells. However, pretreatment of CM-SD concentration-dependently inhibited the H2O2-mediated cytotoxicity.
3.3. Effects of CM-SD on Transcription of Antioxidant Genes
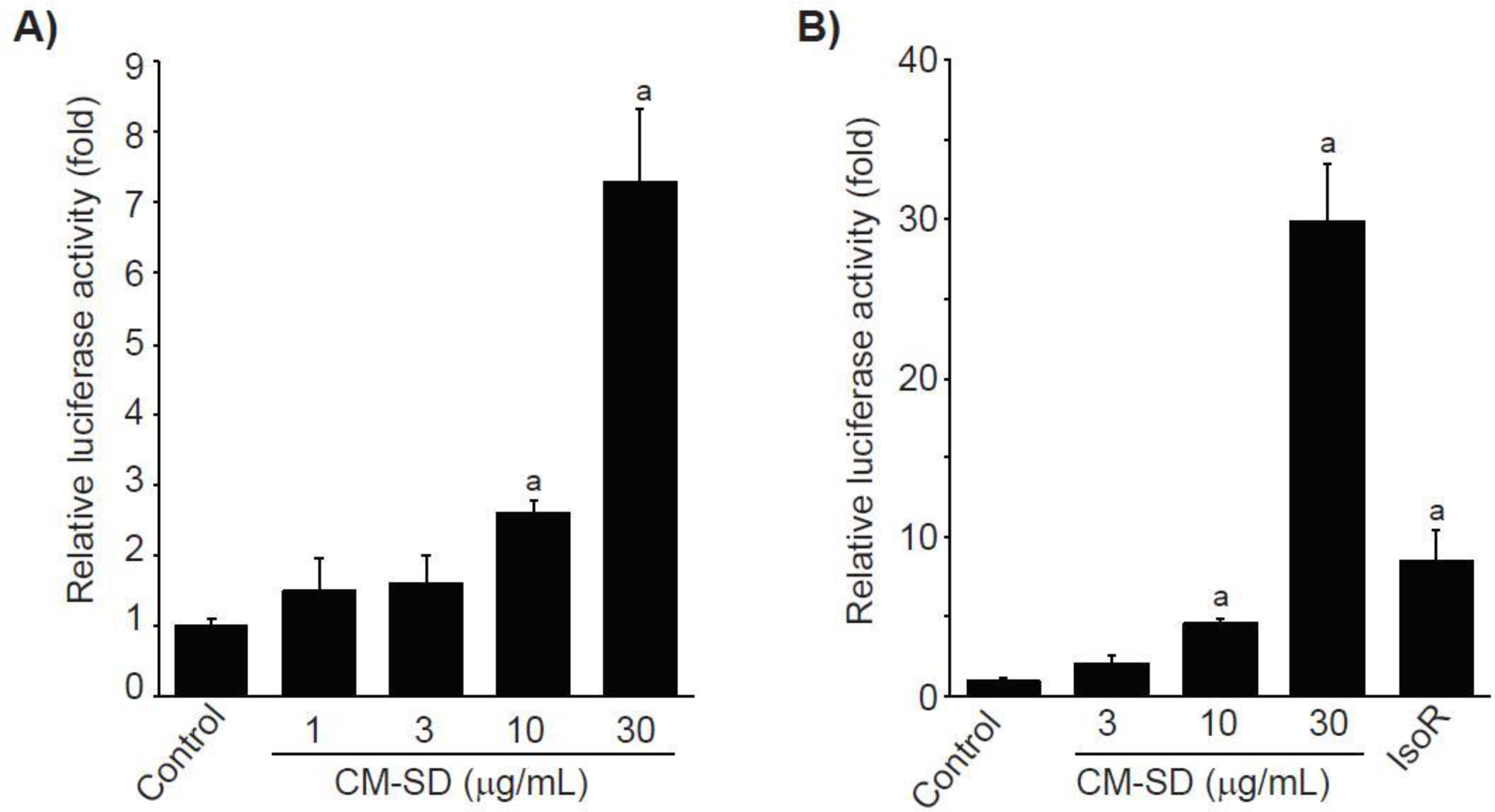
To examine whether Nrf2 is involved in CM-SD-mediated cytoprotectant, recombinant HepG2 cells, which harbor the ARE-driven reporter gene, were treated with 1–30 μg/mL of CM-SD for 18 h, and then the CM-SD-mediated luciferase activities were monitored. Results from reporter gene assay indicate that treatment with CM-SD significantly increased ARE-driven luciferase activities in a concentration-dependent manner. Statistical significances of differences were observed in 10 and 30 μg/mL of CM-SD-treated cells as compared with control cells (Figure 4A). Relative luciferase activities by treatment with 1, 3, 10, and 30 μg/mL of CM-SD were 1.48 ± 0.46, 1.60 ± 0.39, 2.60 ± 0.17, and 7.30 ± 1.02 folds of control cells, respectively. To further confirm whether CM-SD transactivates Nrf2 in C2C12 cells, C2C12 cells were transiently co-transfected with pGL4.37 and pRL-TK reporter plasmids and treated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 18 h. In parallel with the results obtained from recombinant HepG2 cells, treatment with 10 and 30 μg/mL of CM-SD significantly increased Nrf2-mediated luciferase activity. When comparing with 30 μM of IsoR (8.53 ± 1.87-fold the control cells), 30 μg/mL of CM-SD showed a more potent Nrf2 transactivation (p = 0.002 in Dunnett’s T3 test) (Figure 4B). Relative luciferase activities treated with 3, 10, and 30 μg/mL of CM-SD were 2.04 ± 0.44, 4.52 ± 0.34, and 29.88 ± 3.64 folds of control cells, respectively.
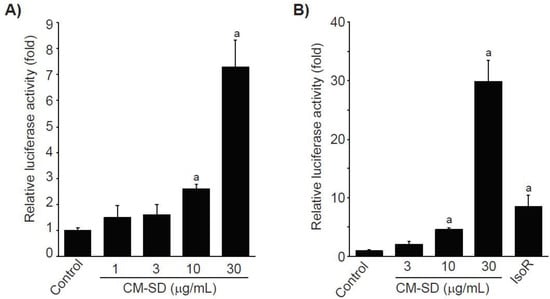
Figure 4.
Effect of CM-SD on Nrf2 transactivation in recombinant HepG2 cells (which harbor ARE-driven reporter gene) (A) and C2C12 cells (B). Values are expressed as mean ± SD at least three separate experiments. Experimental data from HepG2 and C2C12 cells were analyzed by LSD and Dunnett’s T3 test, respectively. a p < 0.01, significant as compared with control.
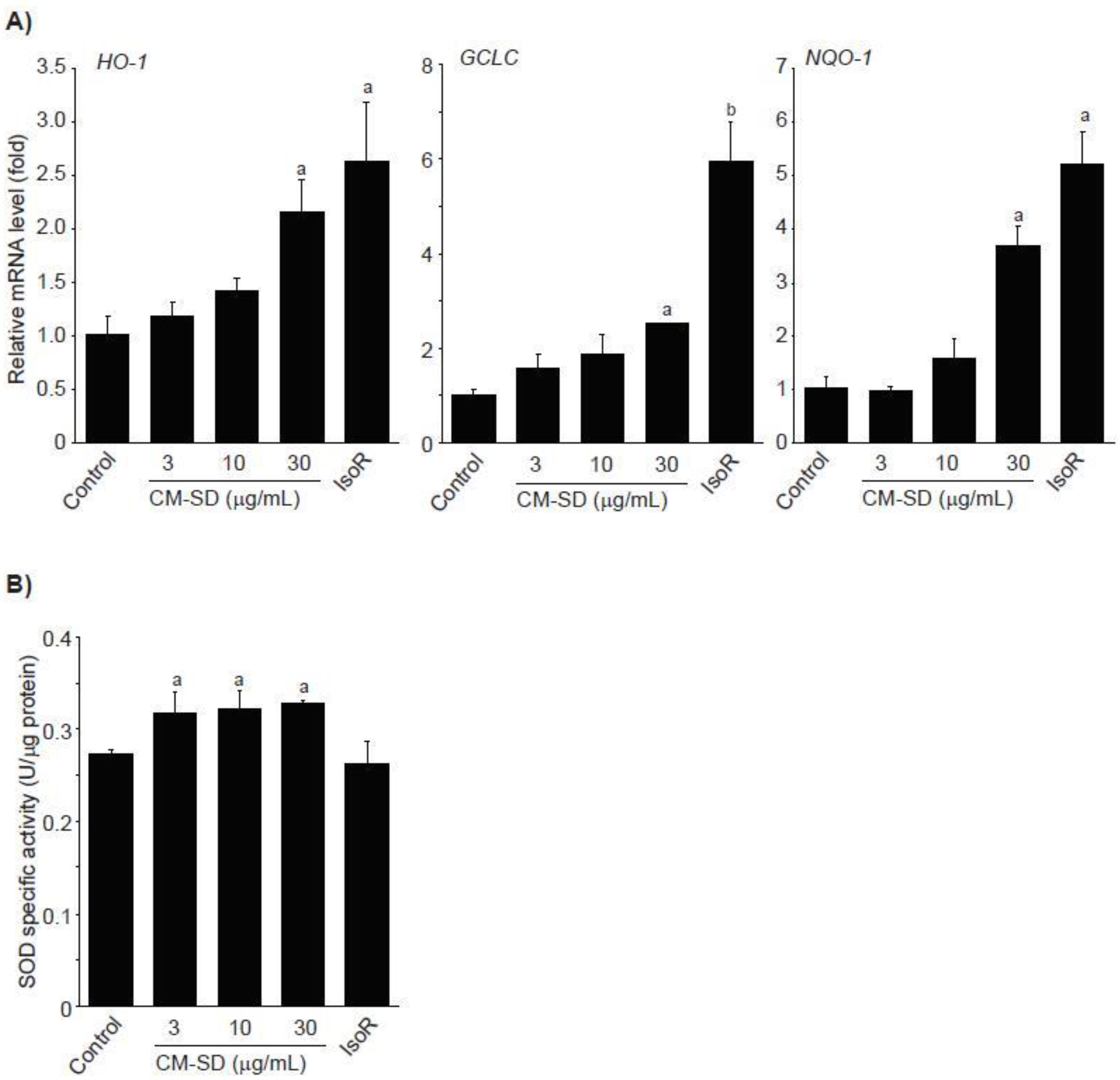
Activated Nrf2 binds to ARE and enhances transcription of antioxidant genes such as heme oxygenase (HO)-1, glutamate cysteine ligase catalytic subunit (GCLC), NADH-dependent quinone oxidoreductase (NQO)-1, and SOD [34,35]. To examine whether Nrf2 activation by CM-SD increases activities of antioxidant genes, C2C12 cells were treated with 3–30 μg/mL of CM-SD for 12 h, and then monitored mRNA levels of antioxidant genes by real-time PCR analysis. Results from real-time PCR analysis indicate that treatment with CM-SD tended to increase mRNA levels of HO-1, GCLC, and NQO-1, while induction of antioxidant genes treated by 30 μg/mL of CM-SD was only statistically significant as compared with control cells. As expected, IsoR (30 μM) significantly increased the mRNA levels of HO-1, GCLC, and NQO-1 (Figure 5A). Relative HO-1 mRNA levels treated with 3 μg/mL CM-SD, 10 μg/mL CM-SD, 30 μg/mL CM-SD, and 30 μM IsoR were 1.19 ± 0.12, 1.41 ± 0.12, 2.16 ± 0.31, and 2.64 ± 0.56 folds of control cells, respectively. Relative GCLC mRNA levels treated with 3 μg/mL CM-SD, 10 μg/mL CM-SD, 30 μg/mL CM-SD, and 30 μM IsoR were 1.57 ± 0.31, 1.88 ± 0.38, 2.51 ± 0.02, and 5.94 ± 0.88 folds of control cells, respectively. Relative NQO-1 mRNA levels treated with 3 μg/mL CM-SD, 10 μg/mL CM-SD, 30 μg/mL CM-SD, and 30 μM IsoR were 0.96 ± 0.09, 1.58 ± 0.36, 3.69 ± 0.38, and 5.22 ± 0.60 folds of control cells, respectively. When C2C12 cells were treated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 24 h, SOD activities in cell homogenates were marginally, but significantly increased by three different dosages of CM-SD. However, IsoR had no effect on SOD activity (Figure 5B). Specific activities of SOD in control, 3 μg/mL CM-SD-, 10 μg/mL CM-SD-, 30 μg/mL CM-SD-, and 30 μM IsoR-treated cells were 0.27 ± 0.005, 0.32 ± 0.02, 0.32 ± 0.02, 0.33 ± 0.003, and 0.26 ± 0.24 U/μg of protein, respectively.
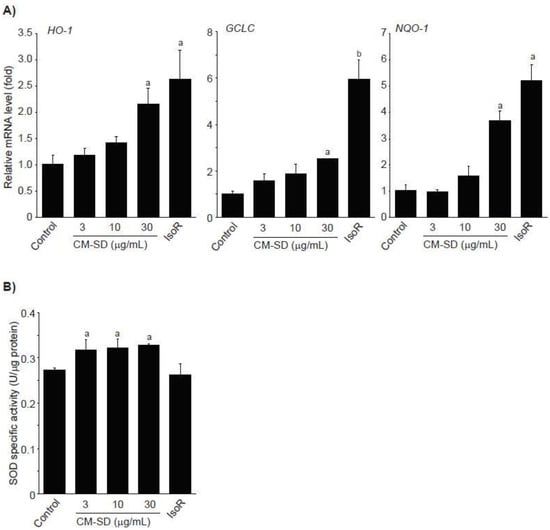
Figure 5.
Effect of CM-SD on HO-1, GCLC, and NQO-1 activation (A) and SOD activity (B). Values are expressed mean ± SD of three separate experiments. Experimental data for HO-1, NQO-1, and SOD were analyzed by LSD test, and the data for GCLC were for Dunnett’s T3 test. a p < 0.01 and b p < 0.05, significant as compared with control.
Nrf2 is an essential master regulator that protects cells against oxidative stress and enhances the cellular defense system through the induction of antioxidant genes [36,37]. Activated Nrf2 is released from its cytosolic repressor Keap1, translocates into the nucleus, binds to ARE in the promoter regions, and then induces the expression of antioxidant genes [38,39]. Therefore, Nrf2 binding to the ARE is crucial for enhancing the antioxidant capacity of cells. In addition, it has been reported that Nrf2 knockout reduces cannabinoid type 2 receptor-mediated myoblast differentiation, which suggests Nrf2 contributes to the differentiation of myoblasts to myotubes [40]. Results from the ARE-driven reporter gene assay using recombinant HepG2 cells and C2C12 cells suggest treatment with CM-SD significantly increased ARE-driven luciferase activities in a concentration-dependent manner. As curcumin and polyphenolic compounds are well-known Nrf2 inducers [22,41], these compounds existing in CM-SD could contribute to activate Nrf2 and to protect cells from excess oxidative stress. Further studies are needed to elucidate essential compounds in CM-SD to activate Nrf2. In addition, it has been reported that a variety of cellular signaling pathways such as p38 mitogen-activated protein kinase, extracellular-regulated protein kinase, phosphatidylinositol 3-kinase, protein kinase C, or casein kinase are involved in activation of Nrf2 [34,42,43,44]. Therefore, upstream signaling molecules contributed to Nrf2 activation by CM-SD should also be elucidated in the future.
Expression of Nrf2-mediated target gene promotes cell survival in oxidizing environments via enhancement of free radical metabolism, regulation of proteasome function, maintaining of glutathione homeostasis, inhibition of cytokine-mediated inflammation, and recognition of damaged DNA [34]. It has been well-known that several antioxidant genes such as HO-1, GCLC, and NQO-1 contain AREs in their promoter regions, and they are induced by Nrf2 activation [34,36]. HO-1 is an inducible rate-limiting enzyme initially identified as facilitating degradation of heme to biliverdin, ferrous iron, and carbon monoxide. Because the final products (e.g., carbon monoxide) of heme degradation exert antioxidant effects by neutralizing oxidative stress, HO-1 induction represents a potential therapeutic target in the management of oxidative stress-related disorders [45]. GCLC, the first rate-limiting enzyme of glutathione biosynthesis, is also involved in the protection of cells from oxidative stress by accelerating GSH biosynthesis [46]. NQO-1 is a member of NAD(P)H dehydrogenase family and involves two-electron reduction of quinones to hydroquinones. Therefore, NQO-1 prevents the one electron reduction of quinonoid compounds that results in the production of reactive oxygen species [47]. In the present study, treatment with CM-SD tended to increase mRNA levels of HO-1, GCLC, and NQO-1. Another antioxidant enzyme such as SOD is also essential in both scavenging ROS and maintaining cellular integrity [48]. SOD diminishes ROS by conversion of superoxide radical to H2O2. The present results indicate that CM-SD marginally, but significantly, increased SOD activity. Taken together, CM-SD seems to enhance antioxidant capacity via activating Nrf2-dependent antioxidant gene induction.
3.4. Effects of CM-SD on H2O2-Mediated Oxidative Stress
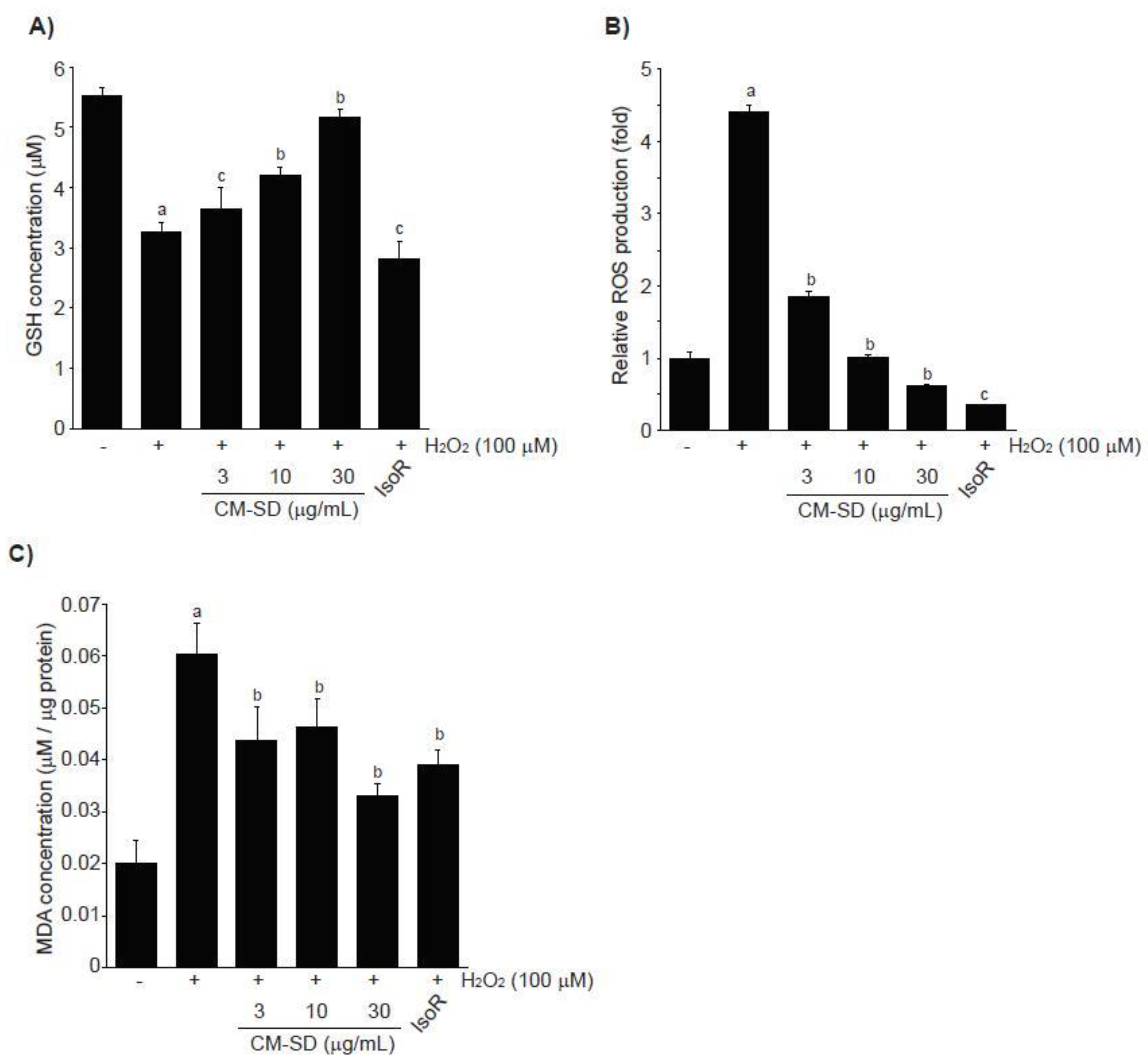
Depletion of endogenous antioxidants (e.g., GSH) and acceleration of ROS production and lipid peroxidation are associated with H2O2-mediated cytotoxicity. To examine whether CM-SD-dependent activation of antioxidant defense system contributes to protect cells from H2O2-mediated oxidative stress, we first measured GSH levels in H2O2-exposed C2C12 cells. H2O2 (100 μM, 6 h) significantly depleted endogenous GSH in C2C12 cells. However, pretreatment with 3–30 μg/mL of CM-SD for 1 h concentration-dependently attenuated the depletion of intracellular GSH by H2O2. GSH levels with 30 μg/mL of CM-SD pretreatment were comparable to those of control cells (p = 0.077 in LSD test). IsoR (30 μM) did not alter the GSH reduction by H2O2 treatment (Figure 6A). GSH levels in control, H2O2, H2O2 + 3 μg/mL CM-SD-, H2O2 + 10 μg/mL CM-SD-, H2O2 + 30 μg/mL CM-SD-, and H2O2 + 30 μM IsoR-treated cells were 5.51 ± 0.14, 3.25 ± 0.19, 3.66 ± 0.33, 4.20 ± 0.12, 5.16 ± 0.14, and 2.81 ± 0.30 μM, respectively.
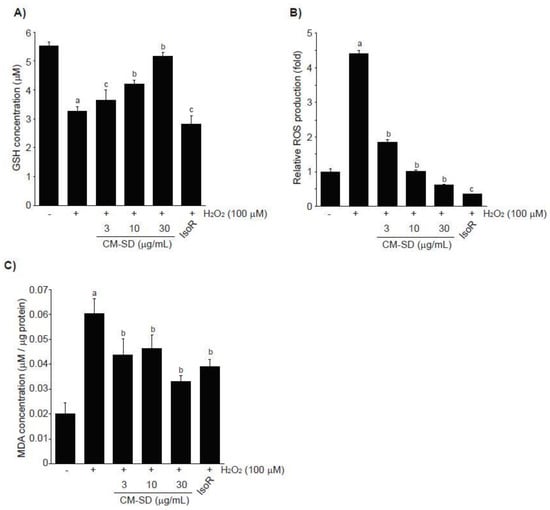
Figure 6.
Effect of CM-SD on GSH (A), ROS (B), and MDA (C) levels followed by H2O2-induced oxidative stress in C2C12 cells. Values are expressed mean ± SD of three separate experiments. Experimental data for GSH and MDA were analyzed by LSD test, and the data for ROS were for Dunnett’s T3 test. a p < 0.01, significant as compared with control; b p < 0.01 and c p < 0.05, significant as compared with H2O2-treated cells.
In addition, exposure of H2O2 for 3 h increased ROS production (4.42 ± 0.08 folds of control cells), and pretreatment with either 3–30 μg/mL of CM-SD or with 30 μM of IsoR significantly inhibited the ROS production by H2O2. Relative ROS levels in 3 μg/mL CM-SD-, 10 μg/mL CM-SD-, 30 μg/mL CM-SD-, and 30 μM IsoR-pretreated cells were 1.85 ± 0.07, 1.02 ± 0.02, 0.61 ± 0.02, and 0.37 ± 0.01 folds, respectively (Figure 6B). Moreover, pretreatment with 3–30 μg/mL of CM-SD significantly reduced MDA levels by H2O2 in a concentration-dependent manner, as compared with H2O2-treated cells (100 μM, 12 h). Inhibition of MDA with 30 μg/mL of CM-SD was comparable to that by 30 μM of IsoR (Figure 6C). MDA levels in control, H2O2, H2O2 + 3 μg/mL CM-SD-, H2O2 + 10 μg/mL CM-SD-, H2O2 + 30 μg/mL CM-SD-, and H2O2 + 30 μM IsoR-treated cells were 0.0200 ± 0.0047, 0.0603 ± 0.0057, 0.0437 ± 0.0067, 0.0463 ± 0.0056, 0.0331 ± 0.0024, and 0.039 ± 0.0031 μM/μg protein, respectively.
3.5. Effects of CM-SD on LPS-Induced Proinflammatory Cytokines Production
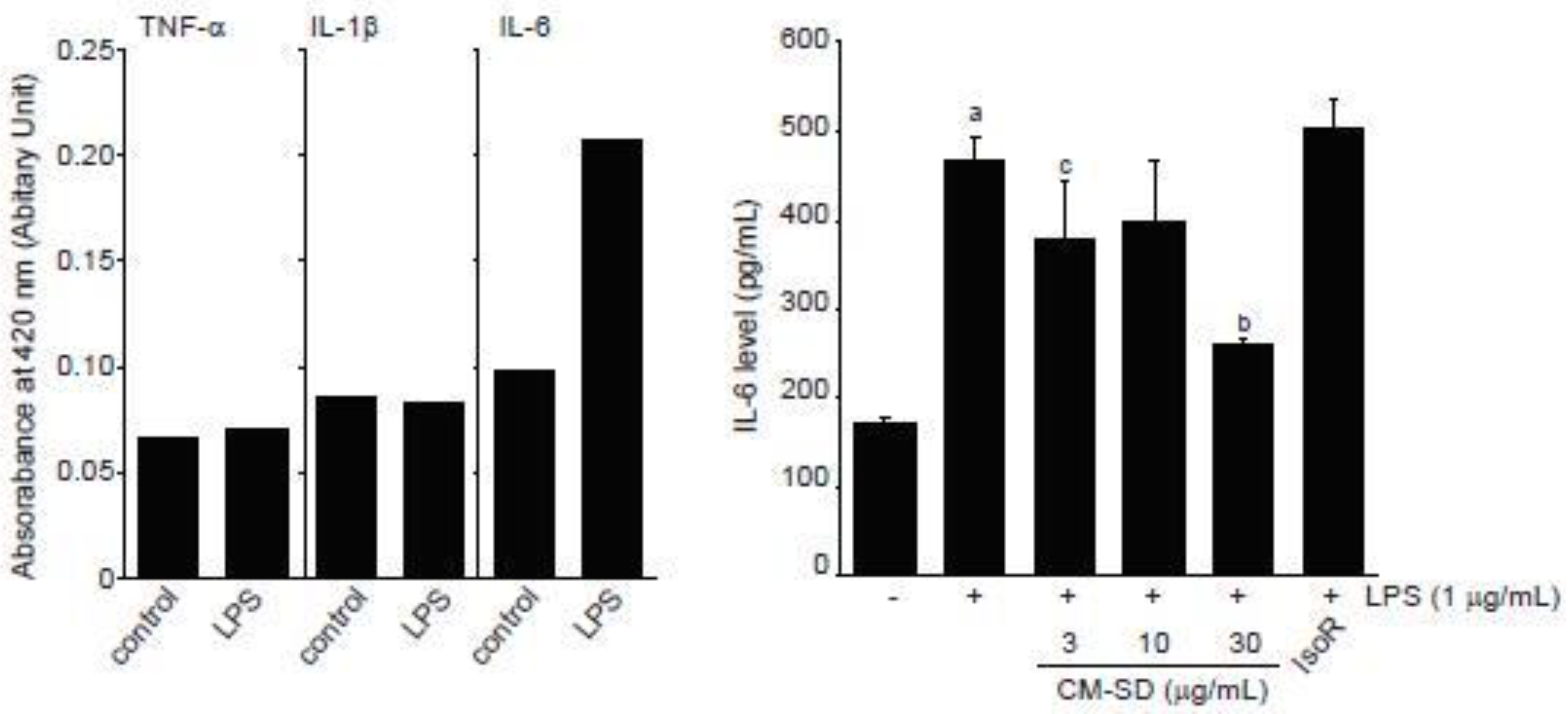
LPS is endotoxin of gram-negative bacteria that binds to toll-like receptor (TLR) 4 and activates an acute inflammatory response in myoblast as well as immune cells [31,49]. When C2C12 cells were exposed to 1 μg/mL of LPS for 12 h, our preliminary experiments by ELISA showed that IL-6, but not TNF-α and IL-1β, increased in conditioned medium obtained from LPS-treated C2C12 cells (Figure 7, left). To examine the effect of CM-SD on LPS-mediated proinflammatory cytokines production, C2C12 cells were pretreated with either 3–30 μg/mL of CM-SD or 30 μM of IsoR for 1 h, subsequently exposed to 1 μg/mL of LPS for 12 h, and then the IL-6 production in culture medium was determined. When compared to the control, treatment with LPS (1 μg/mL) significantly increased the production of IL-6. Pretreatment of CM-SD significantly decreased LPS-inducible IL-6 production. However, IsoR pretreatment did not inhibit IL-6 production by LPS (Figure 7, right). IL-6 levels in control, LPS alone, LPS + 3 μg/mL CM-SD-, LPS + 10 μg/mL CM-SD-, LPS + 30 μg/mL CM-SD-, and LPS + 30 μM IsoR-treated cells were 171.43 ± 6.23, 468.10 ± 27.86, 380.95 ± 62.14, 400.48 ± 64.81, 259.05 ± 7.87, and 502.38 ± 33.57 pg/mL, respectively.
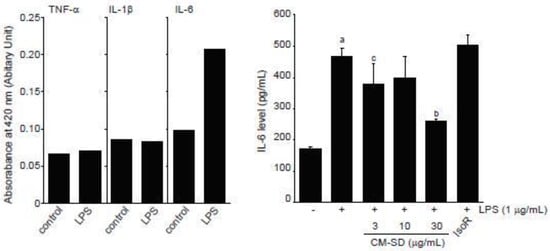
Figure 7.
Effect of CM-SD on proinflammatory cytokines production. Values are expressed mean ± SD of three separate experiments. a p < 0.01, significant as compared with control by Dunnett’s T3 test; b p < 0.01, c p < 0.05, significant as compared with LPS-treated cells by Dunnett’s T3 test.
Since myoblast cells have been known to constitutively express TLR4, an LPS receptor [31], LPS stimulates expression of proinflammatory cytokines in myoblast and myotube cells [50,51] and inhibits myogenic differentiation via activating TLR4/nuclear factor-kappa B (NF-κB) and secreting TNF-α [31]. Among numerous proinflammatory cytokines, IL-6 is a pleiotropic cytokine that is elevated by inflammatory response, exercise, muscle contraction, and denervation in skeletal muscle [50,52,53,54]. In the present study, we showed that CM-SD tended to decrease IL-6 secretion in the medium of LPS-stimulated C2C12 cells, and IL-6 reductions by 3 and 30 mg/mL of CM-SD treatment were statistically significant as compared to LPS alone treated cells. Therefore, these results raise the possibility that CM-SD has anti-inflammatory activity in myoblast cells. Unfortunately, our preliminary ELISA results could not detect secretion of TNF-α and IL-1β in the medium of LPS (1 μg/mL, 12 h)-stimulated C2C12 cells. Since TNF-α and IL-1β are known to be secreted directly from myoblasts after LPS stimulation [51], treatment condition limitations and detection methods may affect the differential results in TNF-α and IL-1β. Thus, more studies are still required to explore the effects of CM-SD on other proinflammatory cytokines in C2C12 cells.
Several studies assessed the inhibitory effect of curcumin against inflammation by modulating the macrophage response to LPS, which is a well-established, commonly used, and effective model for assessing inflammatory response in vitro. For instance, it was revealed that curcumin (1–10 μM) inhibited the expression of IL-1β and TNF-α in a dose-dependent manner in RAW 264.7 cells, further suggesting that curcumin could be a potential anti-inflammation agent [55]. Furthermore, LPS-induced TNF-α expression level was reduced by the pretreatment of 10 μM curcumin in RAW 264.7 cells [56]. Parallel with these findings, results from the current study confirm that pretreatment with 3–30 μg/mL of CM-SD potently inhibited expression of LPS-induced inflammatory cytokines, implying the prominent role of curcumin in the prevention of inflammation-related muscle illnesses.
Previously, it was reported that IsoR downregulates inflammatory responses via inhibition of c-Jun N-terminal protein kinase, Akt, and NF-κB in LPS-stimulated macrophages [57]. In addition, IsoR protects cells from oxidative stress by activating Nrf2-dependent antioxidant genes as well as AMP-activated protein kinase [58,59]. Moreover, IsoR induces extracellular regulated protein kinase-dependent Nrf2 activation and thereby scavenges H2O2-mediated oxidative stress in C2C12 cells [22]. Therefore, in the present study, IsoR (30 μM) was used as a reference drug to compare the beneficial effects of CM-SD in C2C12 cells. IsoR significantly reduced H2O2-mediated cytotoxicity, increased mRNA expression of Nrf2-dependent antioxidant genes, and decreased H2O2-mediated ROS and MDA productions in C2C12 cells. On the contrary, the present results indicate that IsoR alone showed severe cytotoxicity in C2C12 cells, while Choi [22] reported treatment with 30 μM of IsoR alone for 24 h did not change the cell viability. In addition, IsoR did not affect SOD activity, restore H2O2-mediated GSH depletion, or block LPS-inducible IL-6 production. Therefore, Nrf2-independent mechanisms as well as polymorphism of reference drug may differentially influence antioxidant and anti-inflammatory activities of IsoR in C2C12 cells. These issues should be further clarified for using IsoR as an antioxidant and anti-inflammatory nutraceutical in C2C12 cells.
4. Conclusions
The present study demonstrated that CM-SD in C2C12 myoblast cells showed in vitro antioxidant and anti-inflammatory activities. Specifically, CM-SD enhanced DPPH radical scavenging activity in a dose-dependent manner, and it did not affect the cell viability of C2C12 cells with concentrations up to 30 μg/mL. Furthermore, CM-SD significantly suppressed H2O2-induced cell toxicity, ROS generation, and lipid peroxidation and restored GSH depletion, which involved an increase of antioxidant genes through Nrf2 activation, providing evidence that CM-SD can protect cells by reducing oxidative stress. Besides, CM-SD significantly inhibited LPS-mediated IL-6 productions. Therefore, CM-SD could be a useful candidate against oxidative stress and inflammation-related muscle disorders.
Author Contributions
Conceptualization, S.-K.K. and S.-M.S.; Data curation, D.-Y.L., Y.-S.C., J.-K.K., and J.-O.L.; Funding acquisition, Y.-S.C., J.-K.K., and J.-O.L.; Investigation, D.-Y.L., Y.-S.C., and J.-K.K.; Methodology, Y.-J.L. and S.-K.K.; Project administration, S.-K.K.; Resources, Y.-J.L.; Supervision, S.-M.S.; Validation, D.-Y.L.; Writing—original draft, D.-Y.L.; and Writing—Review and editing, S.-M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Phillips, S.M. Nutritional supplements in support of resistance exercise to counter age-related sarcopenia. Adv. Nutr. 2015, 6, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Choi, W.H. Clinical and physiopathological mechanism of sarcopenia. Korean J. Med. 2012, 83, 444–454. [Google Scholar] [CrossRef]
- Meng, S.-J.; Yu, L.-J. Oxidative stress, molecular inflammation and sarcopenia. Int. J. Mol. Sci. 2010, 11, 1509–1526. [Google Scholar] [CrossRef] [PubMed]
- Dirks, A.; Leeuwenburgh, C. Apoptosis in skeletal muscle with aging. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R519–R527. [Google Scholar] [CrossRef] [PubMed]
- Pistilli, E.E.; Jackson, J.R.; Alway, S.E. Death receptor-associated pro-apoptotic signaling in aged skeletal muscle. Apoptosis 2006, 11, 2115. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.; Marrone, A.; Smith, J. Programmed cell death and senescence in skeletal muscle stem cells. Ann. N. Y. Acad. Sci. 2000, 908, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Janssen, I. The epidemiology of sarcopenia. Clin. Geriatr. Med. 2011, 27, 355–363. [Google Scholar] [CrossRef]
- Kido, K.; Sato, K.; Makanae, Y.; Ato, S.; Hayashi, T.; Fujita, S. Herbal supplement Kamishimotsuto augments resistance exercise-induced mTORC1 signaling in rat skeletal muscle. Nutrition 2016, 32, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Takanche, J.S.; Kim, J.E.; Jeong, S.H.; Han, S.H.; Yi, H.K. Schisandra chinensis extract ameliorates age-related muscle wasting and bone loss in ovariectomized rats. Phytother. Res. 2019, 33, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Kishida, Y.; Kagawa, S.; Arimitsu, J.; Nakanishi, M.; Sakashita, N.; Otsuka, S.; Yoshikawa, H.; Hagihara, K. Go-sha-jinki-Gan (GJG), a traditional Japanese herbal medicine, protects against sarcopenia in senescence-accelerated mice. Phytomedicine 2015, 22, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kim, O.S. Jaeumganghwa-Tang, a traditional herbal formula, improves muscle function and attenuates muscle loss in aged mice. J. Exerc. Nutr. Biochem. 2017, 21, 48. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Tsuiji, K.; Li, B.; Tadakawa, M.; Yaegashi, N. Proliferative effect of Hachimijiogan, a Japanese herbal medicine, in C2C12 skeletal muscle cells. Clin. Interv. Aging 2015, 10, 445. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, O.-K.; Yoon, H.-G.; Park, J.; You, Y.; Kim, K.; Lee, Y.-H.; Choi, K.-C.; Lee, J.; Jun, W. Anti-obesity effect of extract from fermented Curcuma longa L. through regulation of adipogenesis and lipolysis pathway in high-fat diet-induced obese rats. Food Nutr. Res. 2016, 60, 30428. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Choi, J.-H. Korean Curcuma longa L. induces lipolysis and regulates leptin in adipocyte cells and rats. Nutr. Res. Pract. 2016, 10, 487–493. [Google Scholar] [CrossRef][Green Version]
- Bradford, P.G. Curcumin and obesity. Biofactors 2013, 39, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Ammon, H.; Wahl, M. Pharmacology of Curcuma longa. Planta Med. 1991, 57, 1–7. [Google Scholar] [CrossRef]
- Willenbacher, E.; Khan, S.Z.; Mujica, S.C.A.; Trapani, D.; Hussain, S.; Wolf, D.; Willenbacher, W.; Spizzo, G.; Seeber, A. Curcumin: New insights into an ancient ingredient against cancer. Int. J. Mol. Sci. 2019, 20, 1808. [Google Scholar] [CrossRef]
- Wang, D.; Yang, Y.; Zou, X.; Zheng, Z.; Zhang, J. Curcumin ameliorates CKD-induced mitochondrial dysfunction and oxidative stress through inhibiting GSK-3β activity. J. Nutr. Biochem. 2020, 83, 108404. [Google Scholar] [CrossRef]
- Kim, D.-O.; Chun, O.K.; Kim, Y.J.; Moon, H.-Y.; Lee, C.Y. Quantification of polyphenolics and their antioxidant capacity in fresh plums. J. Agric. Food Chem. 2003, 51, 6509–6515. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, B.; Ye, M.; Liang, P.; Zhangfang, Y.; Huang, J.; Liu, M.; Songyang, Z.; Ma, W. Ccndbp1 is a new positive regulator of skeletal myogenesis. J. Cell Sci. 2016, 129, 2767–2777. [Google Scholar] [CrossRef]
- Moon, S.Y.; Lee, J.-H.; Choi, H.Y.; Cho, I.J.; Kim, S.C.; Kim, Y.W. Tryptanthrin protects hepatocytes against oxidative stress via activation of the extracellular signal-regulated kinase/NF-E2-related factor 2 pathway. Biol. Pharm. Bull. 2014, 37, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H. The cytoprotective effect of isorhamnetin against oxidative stress is mediated by the upregulation of the Nrf2-dependent HO-1 expression in C2C12 myoblasts through scavenging reactive oxygen species and ERK inactivation. Gen. Physiol. Biophys. 2016, 35, 145. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.R.; Cho, I.J.; Jung, S.J.; Kim, J.K.; Park, S.M.; Lee, D.G.; Ku, S.K.; Park, K.M. Lemon balm and dandelion leaf extract synergistically alleviate ethanol-induced hepatotoxicity by enhancing antioxidant and anti-inflammatory activity. J. Food Biochem. 2020, 44, e13232. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, J.E.; Jung, E.H.; Jung, J.Y.; Jung, D.H.; Ku, S.K.; Cho, I.J.; Kim, S.C. Hemistepsin A ameliorates acute inflammation in macrophages via inhibition of nuclear factor-κB and activation of nuclear factor erythroid 2-related factor 2. Food Chem. Toxicol. 2018, 111, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Park, S.M.; Kim, S.W.; Jung, E.H.; Ko, H.L.; Im, C.K.; Lee, J.R.; Byun, S.H.; Ku, S.K.; Kim, S.C.; Park, C.A. Sipjeondaebo-tang alleviates oxidative stress-mediated liver injury through activation of the CaMKK2-AMPK signaling pathway. Evid. Based Complement. Alternat. Med. 2018, 2018. [Google Scholar] [CrossRef]
- Borra, S.K.; Gurumurthy, P.; Mahendra, J. Antioxidant and free radical scavenging activity of curcumin determined by using different in vitro and ex vivo models. J. Med. Plant Res. 2013, 7, 2680–2690. [Google Scholar] [CrossRef]
- Acharya, S.; Stark, T.D.; Oh, S.T.; Jeon, S.; Pak, S.C.; Kim, M.; Hur, J.; Matsutomo, T.; Hofmann, T.; Hill, R.A. (2R,3S,2″R,3″R)-manniflavanone protects proliferating skeletal muscle cells against oxidative stress and stimulates myotube formation. J. Agric. Food Chem. 2017, 65, 3636–3646. [Google Scholar] [CrossRef]
- Kang, J.S.; Choi, I.-W.; Han, M.H.; Hong, S.H.; Kim, S.O.; Kim, G.-Y.; Hwang, H.J.; Kim, B.W.; Choi, B.T.; Kim, C.M. Sargassum horneri methanol extract rescues C2C12 murine skeletal muscle cells from oxidative stress-induced cytotoxicity through Nrf2-mediated upregulation of heme oxygenase-1. BMC Complement. Altern. Med. 2015, 15, 17. [Google Scholar] [CrossRef]
- Kang, J.S.; Han, M.H.; Kim, G.-Y.; Kim, C.M.; Kim, B.W.; Hwang, H.J.; Choi, Y.H. Nrf2-mediated HO-1 induction contributes to antioxidant capacity of a Schisandrae Fructus ethanol extract in C2C12 myoblasts. Nutrients 2014, 6, 5667–5678. [Google Scholar] [CrossRef]
- Ono, Y.; Sakamoto, K. Lipopolysaccharide inhibits myogenic differentiation of C2C12 myoblasts through the Toll-like receptor 4-nuclear factor-κB signaling pathway and myoblast-derived tumor necrosis factor-α. PLoS ONE 2017, 12, e0182040. [Google Scholar] [CrossRef]
- Adler, V.; Yin, Z.; Tew, K.D.; Ronai, Z.E. Role of redox potential and reactive oxygen species in stress signaling. Oncogene 1999, 18, 6104–6111. [Google Scholar] [CrossRef]
- Cantoni, O.; Cattabeni, F.; Stocchi, V.; Meyn, R.E.; Cerutti, P.; Murray, D. Hydrogen peroxide insult in cultured mammalian cells: Relationships between DNA single-strand breakage, poly (ADP-ribose) metabolism and cell killing. Biochim. Biophys. Acta Mol. Cell Res. 1989, 1014, 1–7. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Sivandzade, F.; Prasad, S.; Bhalerao, A.; Cucullo, L. NRF2 and NF-қB interplay in cerebrovascular and neurodegenerative disorders: Molecular mechanisms and possible therapeutic approaches. Redox Biol. 2019, 21, 101059. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yamamoto, M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Li, J.; Johnson, D.A.; Stein, T.D.; Kraft, A.D.; Calkins, M.J.; Jakel, R.J.; Johnson, J.A. Nrf2, a multi-organ protector? The FASEB J. 2005, 19, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Itoh, K.; Yamamoto, M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002, 348, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Tong, K.I.; Yamamoto, M. Molecular mechanism activating Nrf2–Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 2004, 36, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, M.; Wang, L.; Yu, T.; Jiang, S.; Jiang, P.; Sun, Y.; Pi, J.; Zhao, R.; Guan, D. Activation of cannabinoid type 2 receptor protects skeletal muscle from ischemia-reperfusion injury partly via Nrf2 signaling. Life Sci. 2019, 230, 55–67. [Google Scholar] [CrossRef]
- Surh, Y.-J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer. 2003, 3, 768–780. [Google Scholar] [CrossRef]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, S.A.; Ogborne, R.M.; Charalambos, C.A.; O’Connell, M.A. Role of protein kinase C δ in curcumin-induced antioxidant response element-mediated gene expression in human monocytes. Biochem. Biophys. Res. Commun. 2006, 341, 1007–1016. [Google Scholar] [CrossRef]
- Zimmermann, K.; Baldinger, J.; Mayerhofer, B.; Atanasov, A.G.; Dirsch, V.M.; Heiss, E.H. Activated AMPK boosts the Nrf2/HO-1 signaling axis—A role for the unfolded protein response. Free Radic. Biol. Med. 2015, 88, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Katori, M.; Anselmo, D.M.; Busuttil, R.W.; Kupiec-Weglinski, J.W. A novel strategy against ischemia and reperfusion injury: Cytoprotection with heme oxygenase system. Transpl. Immunol. 2002, 9, 227–233. [Google Scholar] [CrossRef]
- Chen, Y.; Dong, H.; Thompson, D.; Shertzer, H.; Nebert, D.; Vasiliou, V. Glutathione defense mechanism in liver injury: Insights from animal models. Food Chem. Toxicol. 2013, 60, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Kepa, J.K.; Winski, S.L.; Beall, H.D.; Anwar, A.; Siegel, D. NAD(P)H:quinone oxidoreductase 1 (NQO1): Chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem. Biol. Interact. 2000, 129, 77–97. [Google Scholar] [CrossRef]
- Somani, S.M. Exercise, drugs and tissue specific antioxidant system. In Pharmacology in Exercise and Sports; Somani, S.M., Ed.; CRC Press: Boca Raton, FL, USA, 1995; Volume 32, pp. 57–95. [Google Scholar]
- Barton, G.M.; Medzhitov, R. Toll-like receptor signaling pathways. Science 2003, 300, 1524–1525. [Google Scholar] [CrossRef]
- Frost, R.A.; Nystrom, G.J.; Lang, C.H. Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R698–R709. [Google Scholar] [CrossRef]
- Lang, C.H.; Silvis, C.; Deshpande, N.; Nystrom, G.; Frost, R.A. Endotoxin stimulates in vivo expression of inflammatory cytokines tumor necrosis factor alpha, interleukin-1β,-6, and high-mobility-group protein-1 in skeletal muscle. Shock 2003, 19, 538–546. [Google Scholar] [CrossRef]
- Jonsdottir, I.; Schjerling, P.; Ostrowski, K.; Asp, S.; Richter, E.A.; Pedersen, B. Muscle contractions induce interleukin-6 mRNA production in rat skeletal muscles. J. Physiol. 2000, 528, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Kurek, J.; Austin, L.; Cheema, S.; Bartlett, P.; Murphy, M. Up-regulation of leukaemia inhibitory factor and interleukin-6 in transected sciatic nerve and muscle following denervation. Neuromuscul. Disord. 1996, 6, 105–114. [Google Scholar] [CrossRef]
- Ostrowski, K.; Rohde, T.; Zacho, M.; Asp, S.; Pedersen, B. Evidence that interleukin-6 is produced in human skeletal muscle during prolonged running. J. Physiol. 1998, 508, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Nie, M.; Fan, M.-w.; Bian, Z. Anti-inflammatory activity of curcumin in macrophages stimulated by lipopolysaccharides from Porphyromonas gingivalis. Pharmacology 2008, 82, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, M.R.; Leite, F.R.M.; Spolidorio, L.C.; Kirkwood, K.L.; Rossa, C., Jr. Curcumin abrogates LPS-induced pro-inflammatory cytokines in RAW 264.7 macrophages. Evidence for novel mechanisms involving SOCS-1,-3 and p38 MAPK. Arch. Oral Biol. 2013, 58, 1309–1317. [Google Scholar] [CrossRef]
- Yang, J.H.; Kim, S.C.; Shin, B.Y.; Jin, S.H.; Jo, M.J.; Jegal, K.H.; Kim, Y.W.; Lee, J.R.; Ku, S.K.; Cho, I.J. O-methylated flavonol isorhamnetin prevents acute inflammation through blocking of NF-κB activation. Food Chem. Toxicol. 2013, 59, 362–372. [Google Scholar] [CrossRef]
- Dong, G.-Z.; Lee, J.-H.; Ki, S.H.; Yang, J.H.; Cho, I.J.; Kang, S.H.; Zhao, R.J.; Kim, S.C.; Kim, Y.W. AMPK activation by isorhamnetin protects hepatocytes against oxidative stress and mitochondrial dysfunction. Eur. J. Pharmacol. 2014, 740, 634–640. [Google Scholar] [CrossRef]
- Yang, J.H.; Shin, B.Y.; Han, J.Y.; Kim, M.G.; Wi, J.E.; Kim, Y.W.; Cho, I.J.; Kim, S.C.; Shin, S.M.; Ki, S.H. Isorhamnetin protects against oxidative stress by activating Nrf2 and inducing the expression of its target genes. Toxicol. Appl. Pharmacol. 2014, 274, 293–301. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).