Val16A SOD2 Polymorphism Promotes Epithelial–Mesenchymal Transition Antagonized by Muscadine Grape Skin Extract in Prostate Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Reagents, and Antibodies
2.2. Site-Directed Mutagenesis
2.3. SOD2 SNP Stable Overexpression
2.4. ROS Assay
2.5. Immunofluorescence
2.6. Treatments
2.7. Annexin V/Dead Cell Apoptosis Assay
2.8. Cell Viability Assay
2.9. Cell Migration Assay
2.10. Western Blot Analysis
2.11. SOD2 Genotyping by Pyrosequencing
2.12. Quantitative Real-Time RT-PCR Analysis
2.13. Statistical Analysis
3. Results
3.1. SOD2 Protein Expression and Genotype in Prostate Cancer Cell Lines
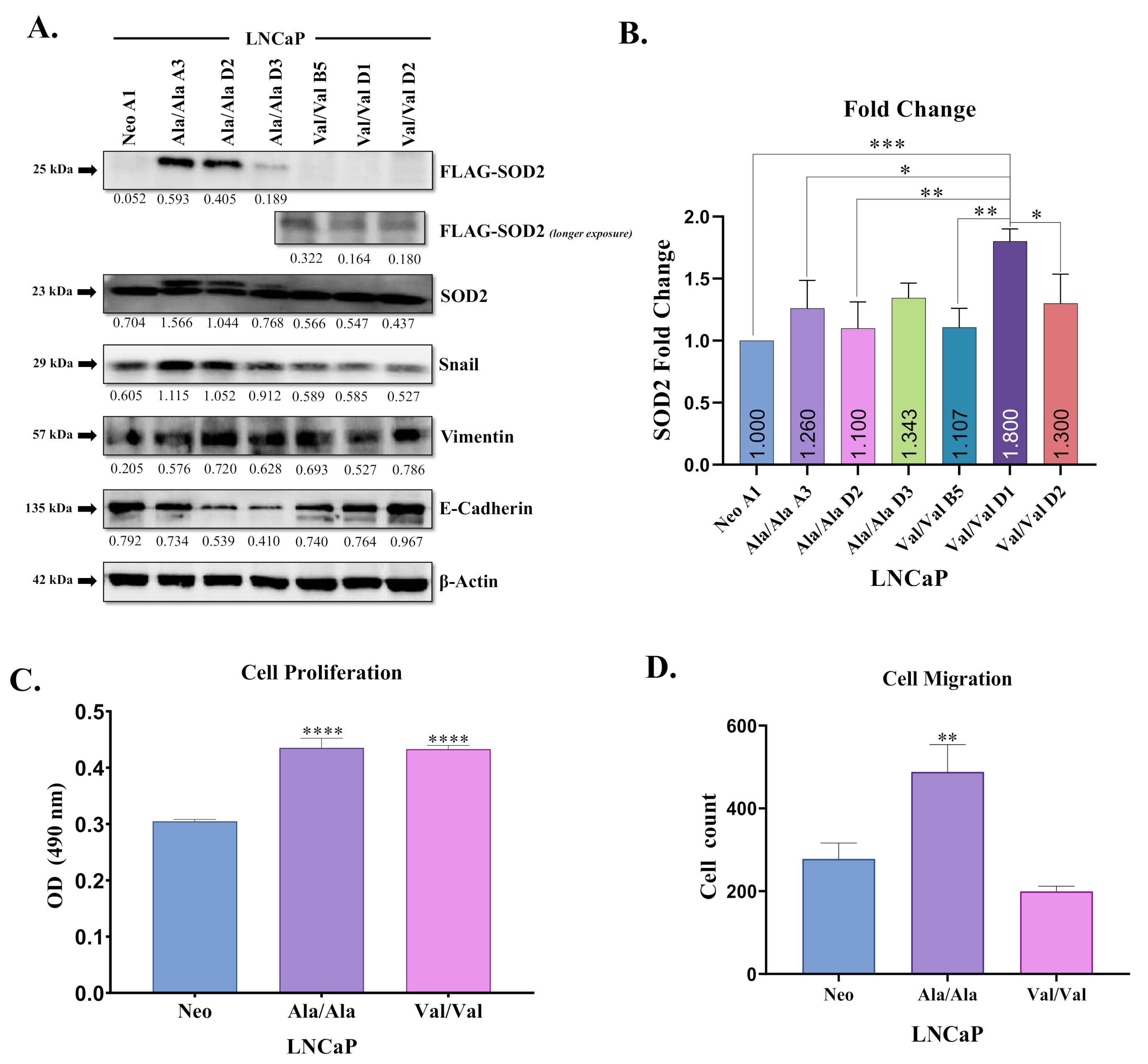
3.2. Ala-SOD2 SNP Promotes EMT
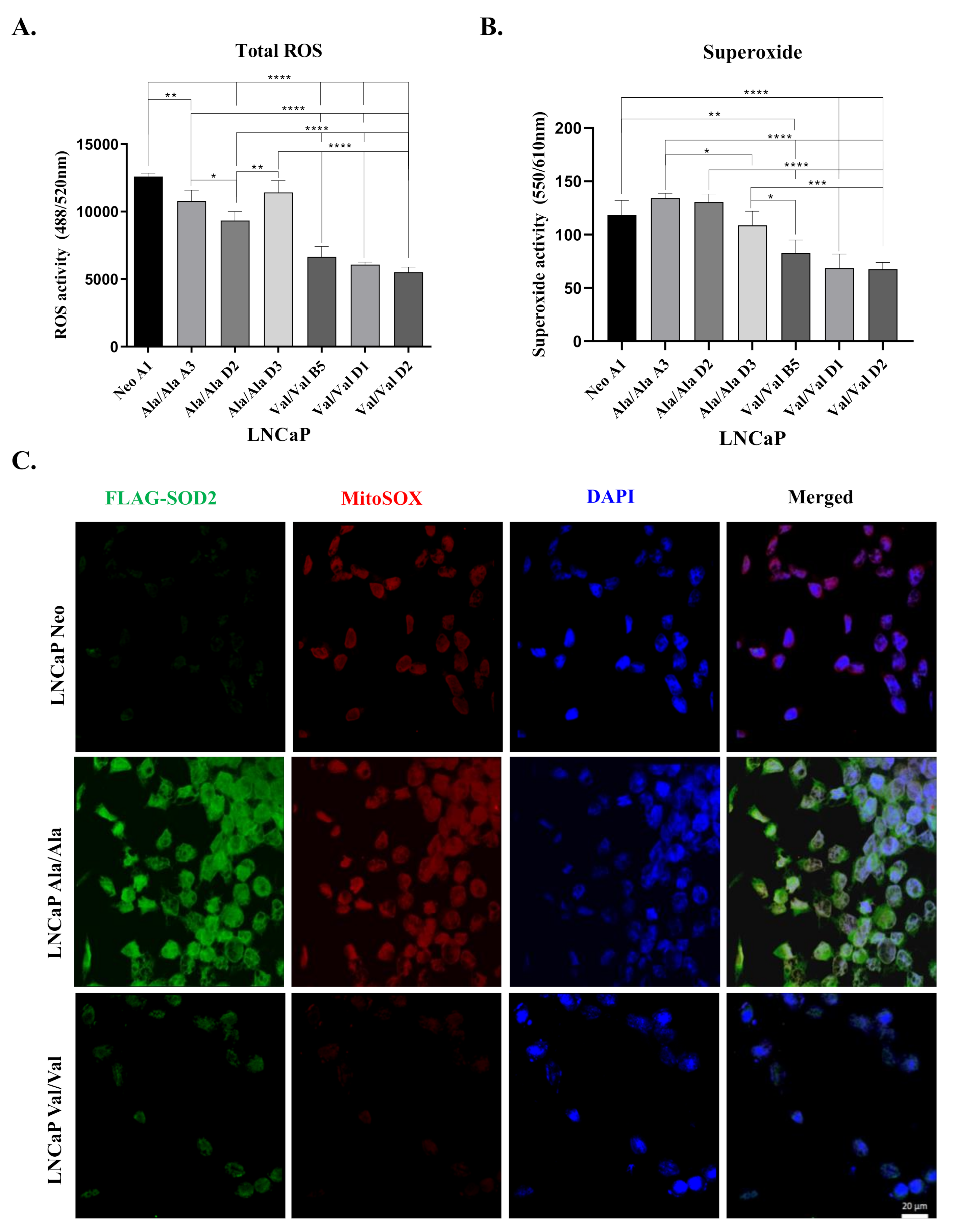
3.3. Overexpression of Ala-SOD2 SNP Leads to Increased Levels of Total ROS and Superoxide Compared to Val-SOD2 SNP
3.4. MSKE Inhibits SOD2 SNP-Mediated EMT
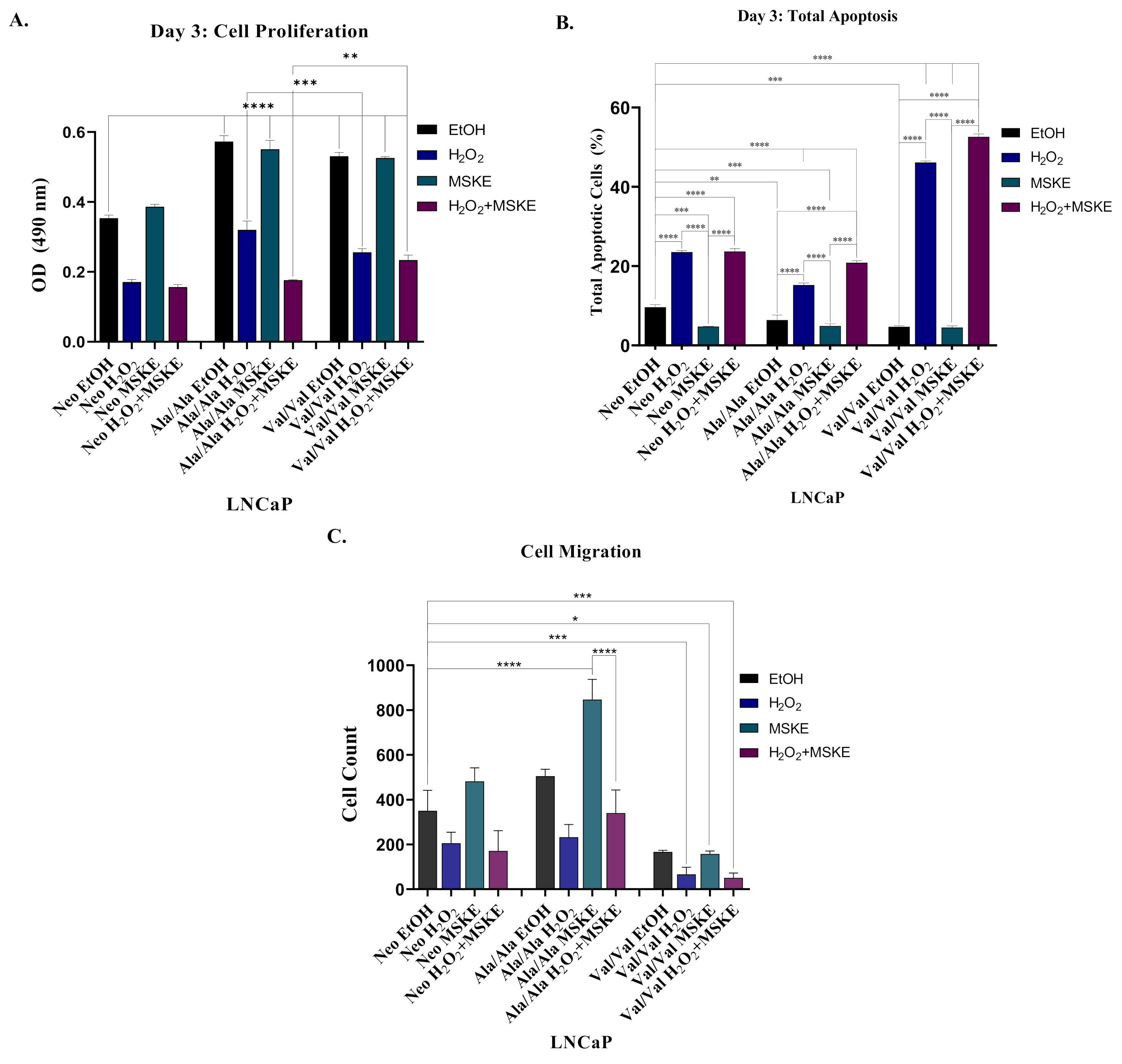
3.5. Co-Treatment with H2O2 and MSKE Decreases Cell Proliferation and Increases Apoptotic Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Montanari, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Malzone, M.G.; Vanacore, D.; di Franco, R.; la Mantia, E.; Iovane, G.; Piscitelli, R.; et al. Epithelial-mesenchymal transition in prostate cancer: An overview. Oncotarget 2017, 8, 35376–35389. [Google Scholar] [CrossRef]
- Ganguly, S.S.; Eli, X.; Miranti, C.K. The Host Microenvironment Influences Prostate Cancer Invasion, Systemic Spread, Bone Colonization, and Osteoblastic Metastasis. Front. Oncol. 2014, 4, 364. [Google Scholar] [CrossRef]
- Lu, W.; Kang, Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef]
- Antognelli, C.; Cecchetti, R.; Riuzzi, F.; Peirce, M.J.; Talesa, V.N. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J. Cell. Mol. Med. 2018, 22, 2865–2883. [Google Scholar] [CrossRef]
- Odero-Marah, V.; Hawsawi, O.; Henderson, V.; Sweeney, J. Epithelial-Mesenchymal Transition (EMT) and Prostate Cancer. Adv. Exp. Med. Biol. 2018, 1095, 101–110. [Google Scholar] [CrossRef]
- Nisticò, P.; Bissell, M.J.; Radisky, D.C. Epithelial-Mesenchymal Transition: General Principles and Pathological Relevance with Special Emphasis on the Role of Matrix Metalloproteinases. Cold Spring Harb. Perspect. Biol. 2012, 4, a011908. [Google Scholar] [CrossRef]
- Smith, B.N.; Odero-Marah, V.A. The role of Snail in prostate cancer. Cell Adhes. Migr. 2012, 6, 433–441. [Google Scholar] [CrossRef]
- Khandrika, L.; Kumar, B.; Koul, S.; Maroni, P.; Koul, H.K. Oxidative stress in prostate cancer. Cancer Lett. 2009, 282, 125–136. [Google Scholar] [CrossRef]
- Oh, B.; Figtree, G.; Costa, D.; Eade, T.; Hruby, G.; Lim, S.; ElFiky, A.; Martine, N.; Rosenthal, D.; Clarke, S.; et al. Oxidative stress in prostate cancer patients: A systematic review of case control studies. Prostate Int. 2016, 4, 71–87. [Google Scholar] [CrossRef]
- Freitas, M.; Baldeiras, I.; Proença, T.; Alves, V.; Mota-Pinto, A.; Sarmento-Ribeiro, A.B. Oxidative stress adaptation in aggressive prostate cancer may be counteracted by the reduction of glutathione reductase. FEBS Open Bio 2012, 2, 119–128. [Google Scholar] [CrossRef]
- Candas, D.; Li, J.J. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid. Redox Signal. 2014, 20, 1599–1617. [Google Scholar] [CrossRef]
- Miriyala, S.; Spasojevic, I.; Tovmasyan, A.; Salvemini, D.; Vujaskovic, Z.; Clair, D.K.S.; Batinic-Haberle, I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 794–814. [Google Scholar] [CrossRef]
- Mikhak, B.D.J.; Hunter, D.; Spiegelman, E.A.; Platz, K.; Wu, J.W.; Erdman, J.; Giovannucci, E. Manganese superoxide dismutase (MnSOD) gene polymorphism, interactions with carotenoid levels and prostate cancer risk. Carcinogenesis 2008, 29, 2335–2340. [Google Scholar] [CrossRef]
- Borrelli, A.; Schiattarella, A.; Bonelli, P.; Tuccillo, F.M.; Buonaguro, L.; Mancini, A. The Functional Role of MnSOD as a Biomarker of Human Diseases and Therapeutic Potential of a New Isoform of a Human Recombinant MnSOD. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Margalit, D.N.; Jordahl, K.M.; Werner, L.; Wang, X.; Lee, M.G.-S.; Penney, K.L.; Batista, J.L.; Martin, N.E.; Chan, J.M.; Kantoff, P.W.; et al. Germline Variation in Superoxide Dismutase-2 (SOD2) and Survival Outcomes after Radiation Therapy for Prostate Cancer: Results from a Test and Validation Set Analysis. Clin. Genitourin. Cancer 2015, 13, 370–377. [Google Scholar] [CrossRef]
- Shimoda-Matsubayashi, S.; Matsumine, H.; Kobayashi, T.; Nakagawa-Hattori, Y.; Shimizu, Y.; Mizuno, Y. Structural dimorphism in the mitochondrial targeting sequence in the human manganese superoxide dismutase gene. A predictive evidence for conformational change to influence mitochondrial transport and a study of allelic association in Parkinson’s disease. Biochem Biophys Res Commun 1996, 226, 561–565. [Google Scholar] [CrossRef]
- Sutton, A.; Imbert, A.; Igoudjil, A.; Descatoire, V.; Cazanave, S.; Pessayre, D.; Degoul, F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharm. Genom. 2005, 15, 311–319. [Google Scholar] [CrossRef]
- Sutton, A.; Khoury, H.; Prip-Buus, C.; Cepanec, C.; Pessayre, D.; Degoul, F. The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics 2003, 13, 145–157. [Google Scholar] [CrossRef]
- Atoum, M.; Abdel-Fattah, M.; Nimer, N.; Abdel-Rahman, S.; Abdeldayem, S.A. Association of Alanine-Valine Manganese Superoxide Dismutase Gene Polymorphism and Microheterogeneity Manganese Superoxide Dismutase Activity in Breast Cancer and Benign Breast Tissue. J. Breast Cancer 2012, 15, 157–161. [Google Scholar] [CrossRef]
- Li, H.; Kantoff, P.W.; Giovannucci, E.; Leitzmann, M.F.; Gaziano, J.M.; Stampfer, M.J.; Ma, J. Manganese Superoxide Dismutase Polymorphism, Prediagnostic Antioxidant Status, and Risk of Clinical Significant Prostate Cancer. Cancer Res. 2005, 65, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, L.-F.; Xu, K.; Tang, B.-W.; Zhang, W.; Yuan, W.; Yue, C.; Shi, L.; Mi, Y.-Y.; Zuo, L.; Zhu, L.-J. Association between SOD2 V16A variant and urological cancer risk. Aging 2020, 12, 825–843. [Google Scholar] [CrossRef]
- Iguchi, T.; Wang, C.Y.; Delongchamps, N.B.; Kato, M.; Tamada, S.; Yamasaki, T.; de la Roza, G.; Nakatani, T.; Haas, G.P. Association of MnSOD AA Genotype with the Progression of Prostate Cancer. PLoS ONE 2015, 10, e0131325. [Google Scholar] [CrossRef]
- Cai, Q.; Shu, X.-O.; Wen, W.; Cheng, J.-R.; Dai, Q.; Gao, Y.-T.; Zheng, W. Genetic polymorphism in the manganese superoxide dismutase gene, antioxidant intake, and breast cancer risk: Results from the Shanghai Breast Cancer Study. Breast Cancer Res. 2004, 6, R647–R655. [Google Scholar] [CrossRef]
- Kang, D.; Lee, K.M.; Park, S.K.; Berndt, S.I.; Peters, U.; Reding, D.; Chatterjee, N.; Welch, R.; Chanock, S.; Huang, W.-Y.; et al. Functional Variant of Manganese Superoxide Dismutase (SOD2 V16A) Polymorphism Is Associated with Prostate Cancer Risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Study. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1581–1586. [Google Scholar] [CrossRef]
- Hudson, T.S.; Hartle, D.K.; Hursting, S.D.; Nunez, N.P.; Wang, T.T.; Young, H.A.; Arany, P.; Green, J.E. Inhibition of Prostate Cancer Growth by Muscadine Grape Skin Extract and Resveratrol through Distinct Mechanisms. Cancer Res. 2007, 67, 8396–8405. [Google Scholar] [CrossRef]
- Burton, L.J.; Rivera, M.; Hawsawi, O.; Zou, J.; Hudson, T.; Wang, G.; Zhang, Q.; Cubano, L.; Boukli, N.; A Odero-Marah, V. Muscadine Grape Skin Extract Induces an Unfolded Protein Response-Mediated Autophagy in Prostate Cancer Cells: A TMT-Based Quantitative Proteomic Analysis. PLoS ONE 2016, 11, e0164115. [Google Scholar] [CrossRef]
- Paller, C.; Rudek, M.A.; Zhou, X.C.; Wagner, W.D.; Hudson, T.S.; Anders, N.M.; Hammers, H.J.; Dowling, D.; King, S.; Antonarakis, E.S.; et al. A phase I study of muscadine grape skin extract in men with biochemically recurrent prostate cancer: Safety, tolerability, and dose determination. Prostate 2015, 75, 1518–1525. [Google Scholar] [CrossRef]
- Paller, C.; Zhou, X.C.; Heath, E.I.; Taplin, M.-E.; Mayer, T.M.; Stein, M.N.; Bubley, G.J.; Pili, R.; Hudson, T.S.; Kakarla, R.; et al. Muscadine Grape Skin Extract (MPX) in Men with Biochemically Recurrent Prostate Cancer: A Randomized, Multicenter, Placebo-Controlled Clinical Trial. Clin. Cancer Res. 2018, 24, 306–315. [Google Scholar] [CrossRef]
- Burton, L.J.; Hawsawi, O.; Loyd, Q.; Henderson, V.; Howard, S.; Harlemon, M.; Ragin, C.; Roberts, R.; Bowen, N.; Gacii, A.; et al. Association of Epithelial Mesenchymal Transition with prostate and breast health disparities. PLoS ONE 2018, 13, e0203855. [Google Scholar] [CrossRef]
- Burton, L.J.; Barnett, P.; Smith, B.; Arnold, R.S.; Hudson, T.S.; Kundu, K.; Murthy, N.; A Odero-Marah, V. Muscadine grape skin extract reverts snail-mediated epithelial mesenchymal transition via superoxide species in human prostate cancer cells. BMC Complement. Altern. Med. 2014, 14, 97. [Google Scholar] [CrossRef]
- Vilema-Enríquez, G.; Arroyo, A.; Grijalva, M.; Amador-Zafra, R.I.; Camacho, J. Molecular and Cellular Effects of Hydrogen Peroxide on Human Lung Cancer Cells:Potential Therapeutic Implications. Oxidative Med. Cell. Longev. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Mollsten, A.; Jorsal, A.; Lajer, M.; Vionnet, N.; Tarnow, L. The V16A polymorphism in SOD2 is associated with increased risk of diabetic nephropathy and cardiovascular disease in type 1 diabetes. Diabetologia 2009, 52, 2590–2593. [Google Scholar] [CrossRef]
- Pourvali, K.; Abbasi, M.; Mottaghi, A. Role of Superoxide Dismutase 2 Gene Ala16Val Polymorphism and Total Antioxidant Capacity in Diabetes and its Complications. Avicenna J. Med. Biotechnol. 2016, 8, 48–56. [Google Scholar]
- Jones, D.; Prior, S.; Tang, T.; Bain, S.; Hurel, S.; Humphries, S.; Stephens, J.W. Association between the rs4880 superoxide dismutase 2 (C>T) gene variant and coronary heart disease in diabetes mellitus. Diabetes Res. Clin. Pr. 2010, 90, 196–201. [Google Scholar] [CrossRef]
- Wiener, H.W.; Perry, R.T.; Chen, Z.; Harrell, L.E.; Go, R.C. A polymorphism in SOD2 is associated with development of Alzheimer’s disease. Genes Brain Behav. 2007, 6, 770–775. [Google Scholar] [CrossRef]
- Glynn, S.A.; Boersma, B.J.; Howe, T.M.; Edvardsen, H.; Geisler, S.B.; Goodman, J.E.; Ridnour, L.A.; Lønning, P.E.; Børresen-Dale, A.-L.; Naume, B.; et al. A Mitochondrial Target Sequence Polymorphism in Manganese Superoxide Dismutase Predicts Inferior Survival in Breast Cancer Patients Treated with Cyclophosphamide. Clin. Cancer Res. 2009, 15, 4165–4173. [Google Scholar] [CrossRef]
- Pan, Y.; Kytölä, S.; Farnebo, F.; Wang, N.; Lui, W.; Nupponen, N.; Isola, J.; Visakorpi, T.; Bergerheim, U.; Larsson, C. Characterization of chromosomal abnormalities in prostate cancer cell lines by spectral karyotyping. Cytogenet. Cell Genet. 1999, 87, 225–232. [Google Scholar] [CrossRef]
- Beheshti, B.; Karaskova, J.; Park, P.C.; Squire, J.A.; Beatty, B.G. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential giemsa banding and spectral karyotyping. Mol. Diagn. 2000, 5, 23–32. [Google Scholar] [CrossRef]
- van Bokhoven, A.; Caires, A.; di Maria, M.; Schulte, A.P.; Lucia, M.S.; Nordeen, S.K.; Miller, G.J.; Varella-Garcia, M. Spectral karyotype (SKY) analysis of human prostate carcinoma cell lines. Prostate 2003, 57, 226–244. [Google Scholar] [CrossRef]
- Li, W.; Cao, L.; Han, L.; Xu, L.; Ma, Q. Superoxide dismutase promotes the epithelial-mesenchymal transition of pancreatic cancer cells via activation of the H2O2/ERK/NF-kappaB axis. Int. J. Oncol. 2015, 46, 2613–2620. [Google Scholar] [CrossRef]
- McAtee, B.L.; Yager, J.D. Manganese superoxide dismutase: Effect of the ala16val polymorphism on protein, activity, and mRNA levels in human breast cancer cell lines and stably transfected mouse embryonic fibroblasts. Mol. Cell. Biochem. 2009, 335, 107–118. [Google Scholar] [CrossRef][Green Version]
- Martin, R.C.G.; Li, Y.; Liu, Q.; Jensen, N.S.; Barker, D.F.; Doll, M.A.; Hein, D.W. Manganese Superoxide Dismutase V16A Single-Nucleotide Polymorphism in the Mitochondrial Targeting Sequence Is Associated with Reduced Enzymatic Activity in Cryopreserved Human Hepatocytes. DNA Cell Biol. 2009, 28, 3–7. [Google Scholar] [CrossRef]
- Burton, L.J.; Smith, B.A.; Smith, B.N.; Loyd, Q.; Nagappan, P.; McKeithen, D.; Wilder, C.L.; Platt, M.O.; Hudson, T.; A Odero-Marah, V. Muscadine grape skin extract can antagonize Snail-cathepsin L-mediated invasion, migration and osteoclastogenesis in prostate and breast cancer cells. Carcinogenesis 2015, 36, 1019–1027. [Google Scholar] [CrossRef]
- Woodson, K.; Tangrea, J.A.; Lehman, T.A.; Modali, R.; Taylor, K.M.; Snyder, K.; Taylor, P.R.; Virtamo, J.; Albanes, D. Manganese superoxide dismutase (MnSOD) polymorphism, α-tocopherol supplementation and prostate cancer risk in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (Finland). Cancer Causes Control. 2003, 14, 513–518. [Google Scholar] [CrossRef]
- Loo, S.Y.; Hirpara, J.L.; Pandey, V.; Tan, T.Z.; Yap, C.T.; Lobie, P.E.; Thiery, J.P.; Goh, B.C.; Pervaiz, S.; Clément, M.-V.; et al. Manganese Superoxide Dismutase Expression Regulates the Switch Between an Epithelial and a Mesenchymal-Like Phenotype in Breast Carcinoma. Antioxid. Redox Signal. 2016, 25, 283–299. [Google Scholar] [CrossRef]
- Mitrunen, K.; Sillanpää, P.; Kataja, V.; Eskelinen, M.; Kosma, V.-M.; Benhamou, S.; Uusitupa, M.; Hirvonen, A. Association between manganese superoxide dismutase (MnSOD) gene polymorphism and breast cancer risk. Carcinogenesis 2001, 22, 827–829. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Zhou, J.; Ye, L.; Chen, N.; Zhu, M.; Wang, C.-D. There is no relationship between SOD2 Val-16Ala polymorphism and breast cancer risk or survival. Mol. Clin. Oncol. 2017, 7, 579–590. [Google Scholar] [CrossRef]
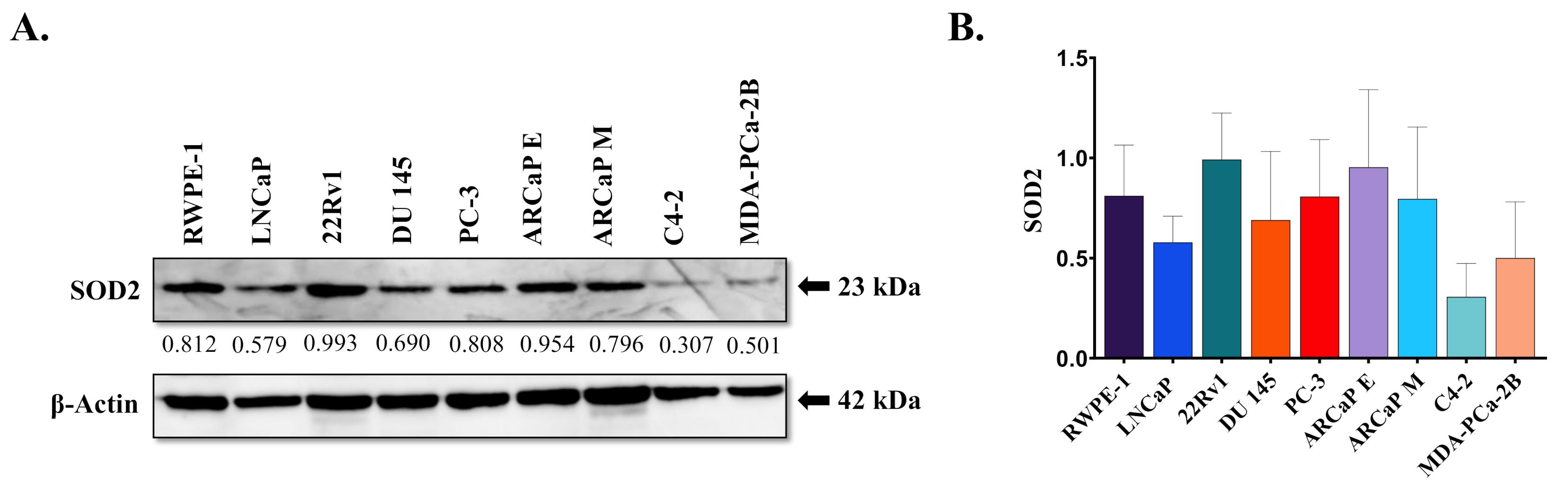

| Cell Line | Origin | AR | Tumorigenic Potential | SOD2 Pyrosequencing Results | Genotype |
|---|---|---|---|---|---|
| RWPE-1 | Caucasian | + | None | Heterozygous C/T | Ala/Val |
| LNCaP | Caucasian | + | Low | Heterozygous C/T | Val/Val/Ala |
| PC-3 | Caucasian | − | High | Heterozygous C/T | Ala/Val |
| ARCaP E | Caucasian | + | High | Heterozygous C/T | Ala/Ala/Val |
| ARCaP M | Caucasian | + | Very high | Heterozygous C/T | Ala/Val/Val |
| C4-2 | Caucasian | + | High | Heterozygous C/T | Val/Val/Ala |
| MDA-PCa-2a | African American | + | High | Homozygous WT C/C | Ala/Ala |
| MDA-PCa-2b | African American | + | High | Homozygous WT C/C | Ala/Ala |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sweeney, J.D.; Debeljak, M.; Riel, S.; Millena, A.C.; Eshleman, J.R.; Paller, C.J.; Odero-Marah, V. Val16A SOD2 Polymorphism Promotes Epithelial–Mesenchymal Transition Antagonized by Muscadine Grape Skin Extract in Prostate Cancer Cells. Antioxidants 2021, 10, 213. https://doi.org/10.3390/antiox10020213
Sweeney JD, Debeljak M, Riel S, Millena AC, Eshleman JR, Paller CJ, Odero-Marah V. Val16A SOD2 Polymorphism Promotes Epithelial–Mesenchymal Transition Antagonized by Muscadine Grape Skin Extract in Prostate Cancer Cells. Antioxidants. 2021; 10(2):213. https://doi.org/10.3390/antiox10020213
Chicago/Turabian StyleSweeney, Janae D., Marija Debeljak, Stacy Riel, Ana Cecilia Millena, James R. Eshleman, Channing J. Paller, and Valerie Odero-Marah. 2021. "Val16A SOD2 Polymorphism Promotes Epithelial–Mesenchymal Transition Antagonized by Muscadine Grape Skin Extract in Prostate Cancer Cells" Antioxidants 10, no. 2: 213. https://doi.org/10.3390/antiox10020213
APA StyleSweeney, J. D., Debeljak, M., Riel, S., Millena, A. C., Eshleman, J. R., Paller, C. J., & Odero-Marah, V. (2021). Val16A SOD2 Polymorphism Promotes Epithelial–Mesenchymal Transition Antagonized by Muscadine Grape Skin Extract in Prostate Cancer Cells. Antioxidants, 10(2), 213. https://doi.org/10.3390/antiox10020213







