Exploring the Leukemogenic Potential of GATA-1S, the Shorter Isoform of GATA-1: Novel Insights into Mechanisms Hampering Respiratory Chain Complex II Activity and Limiting Oxidative Phosphorylation Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transient Transfection
2.3. Protein Extraction
2.4. Western Blot Analysis
2.5. Total RNA Extraction
2.6. Real-Time PCR Analysis
2.7. Measurement of the Enzymatic Activity of Complex II Succinate-Ubiquinone Oxidoreductase
2.8. Seahorse Assay for Measurement of Cellular Respiration
2.9. Mitochondrial Mass Measurement
2.10. AML Patient Samples
2.11. Statistical Analysis
3. Results
3.1. Correlation between GATA-1 Isoforms and Expression Levels of SDHC Isoforms
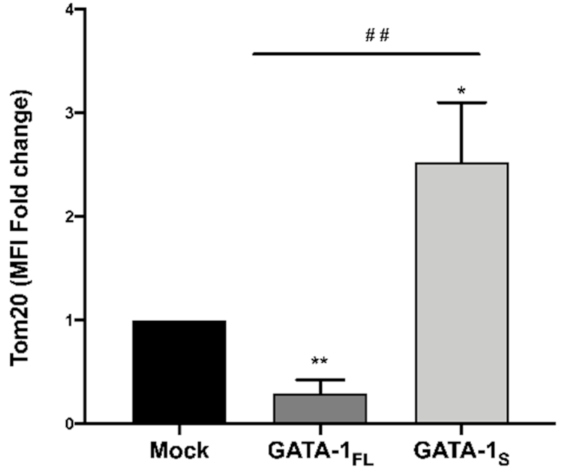
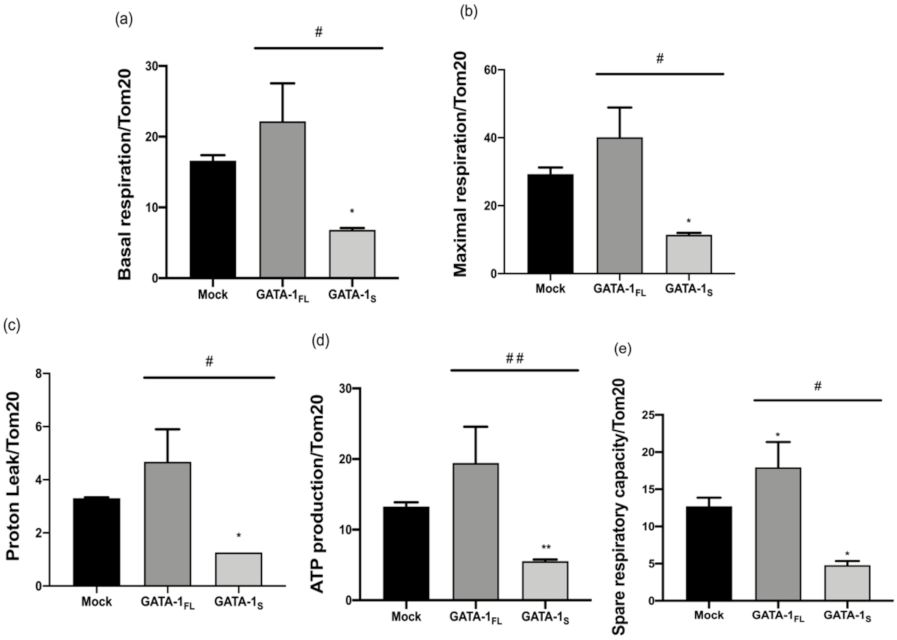
3.2. Effects of GATA-1 Isoforms on Mitochondrial Metabolism
3.2.1. Measurement of SQR Activity
3.2.2. Evaluation of Cellular Energy Metabolism
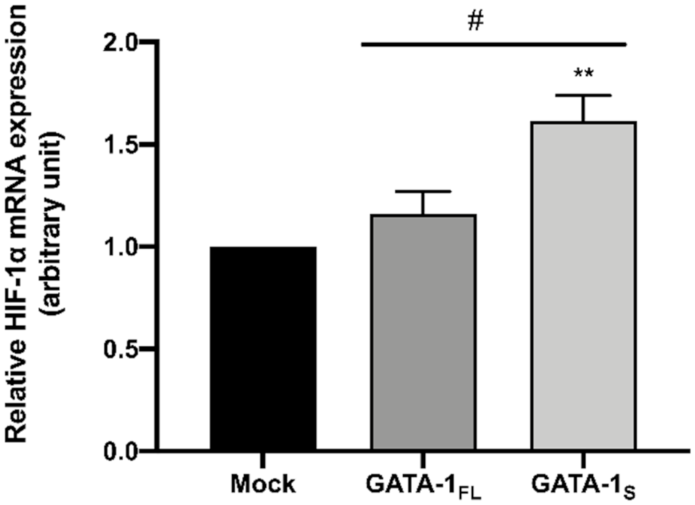
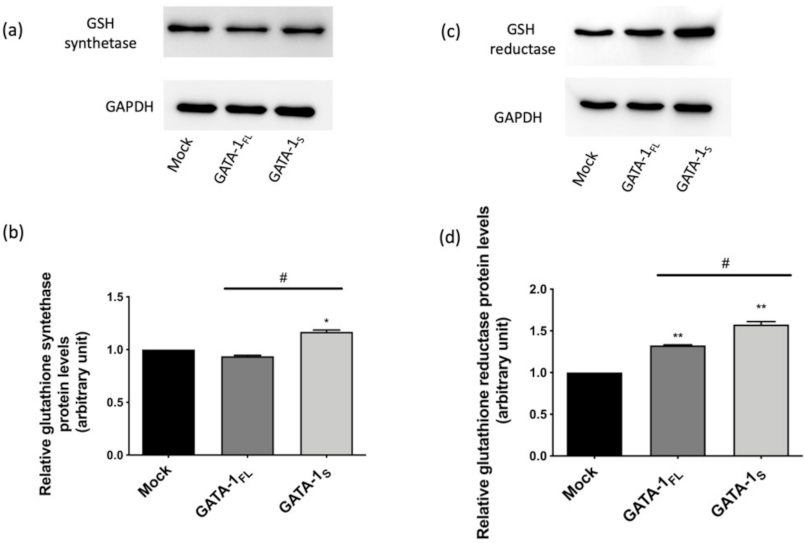
3.3. Effects of GATA-1 Isoforms on Glutathione (GSH) Biosynthesis
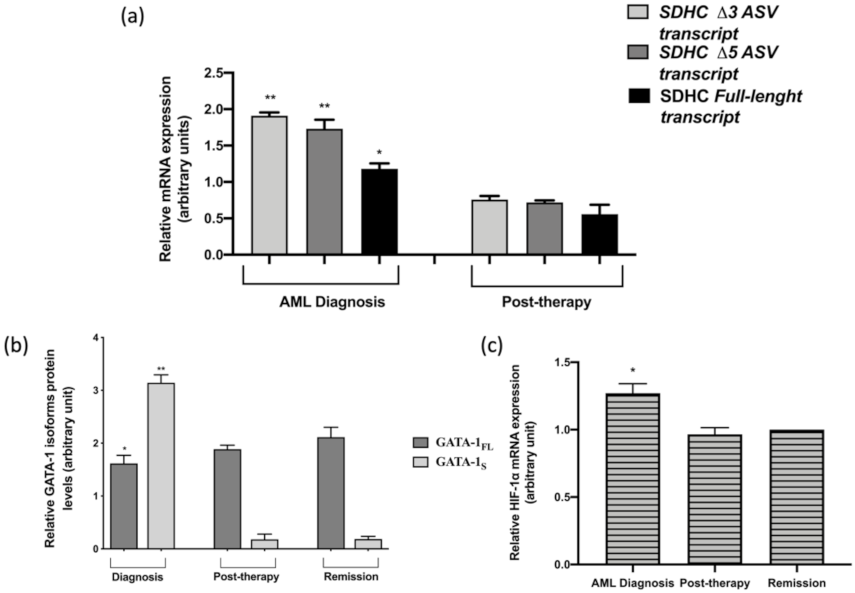
3.4. Expression Levels of SDHC ASV Isoforms in an AML Patient
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al Ageeli, E. Alterations of mitochondria and related metabolic pathways in leukemia: A narrative review. Saudi J. Med. Med. Sci. 2020, 8, 3–11. [Google Scholar] [CrossRef]
- Panuzzo, C.; Jovanovski, A.; Pergolizzi, B.; Pironi, L.; Stanga, S.; Fava, C.; Cilloni, D. Mitochondria: A Galaxy in the Hematopoietic and Leukemic Stem Cell Universe. Int. J. Mol. Sci. 2020, 21, 3928. [Google Scholar] [CrossRef]
- Trombetti, S.; Cesaro, E.; Catapano, R.; Sessa, R.; Bianco, A.L.; Izzo, P.; Grosso, M. Oxidative Stress and ROS-Mediated Signaling in Leukemia: Novel Promising Perspectives to Eradicate Chemoresistant Cells in Myeloid Leukemia. Int. J. Mol. Sci. 2021, 22, 2470. [Google Scholar] [CrossRef] [PubMed]
- Sillar, J.R.; Germon, Z.P.; De Iuliis, G.N.; Dun, M.D. The Role of Reactive Oxygen Species in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2019, 20, 6003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juntilla, M.; Patil, V.D.; Calamito, M.; Joshi, R.P.; Birnbaum, M.; Koretzky, G.A. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 2010, 115, 4030–4038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccio, P.; Sessa, R.; de Nicola, S.; Petruzziello, F.; Trombetti, S.; Menna, G.; Pepe, G.; Maddalena, P.; Izzo, P.; Grosso, M. GATA-1 isoforms differently contribute to the production and compartmentation of reactive oxygen species in the myeloid leukemia cell line K. J. Cell. Physiol. 2019, 234, 20829–20846. [Google Scholar] [CrossRef] [Green Version]
- Panina, S.; Baran, N.; da Costa, F.H.B.; Konopleva, M.; Kirienko, N.V. A mechanism for increased sensitivity of acute myeloid leukemia to mitotoxic drugs. Cell Death Dis. 2019, 10, 617. [Google Scholar] [CrossRef] [Green Version]
- Lemarie, A.; Grimm, S. Mitochondrial respiratory chain complexes: Apoptosis sensors mutated in cancer? Oncogene 2011, 30, 3985–4003. [Google Scholar] [CrossRef] [Green Version]
- Nelson, M.A.M.; McLaughlin, K.L.; Hagen, J.T.; Coalson, H.S.; Schmidt, C.; Kassai, M.; Kew, K.A.; McClung, J.M.; Neufer, P.D.; Brophy, P.; et al. Intrinsic OXPHOS limitations underlie cellular bioenergetics in leukemia. eLife 2021, 10, 63104. [Google Scholar] [CrossRef]
- Ye, Z.-W.; Zhang, J.; Townsend, D.M.; Tew, K.D. Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim. Biophys. Acta (BBA) Gen. Subj. 2015, 1850, 1607–1621. [Google Scholar] [CrossRef] [Green Version]
- Farge, T.; Saland, E.; De Toni, F.; Aroua, N.; Hosseini, M.; Perry, R.; Bosc, C.; Sugita, M.; Stuani, L.; Fraisse, M.; et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017, 7, 716–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, L.V.R.; Bremner, L.; Barros, M.H. Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae. Life 2020, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, V.; Ciccarese, F.; Ciminale, V. Oncogenic pathways and the electron transport chain: A dangeROS liaison. Br. J. Cancer 2020, 122, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Tretter, L.; Patocs, A.; Chinopoulos, C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta (BBA) Bioenergy 2016, 1857, 1086–1101. [Google Scholar] [CrossRef]
- Dröse, S. Differential effects of complex II on mitochondrial ROS production and their relation to cardioprotective pre- and postconditioning. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1827, 578–587. [Google Scholar] [CrossRef] [Green Version]
- Bezawork-Geleta, A.; Rohlena, J.; Dong, L.; Pacak, K.; Neuzil, J. Mitochondrial Complex II: At the Crossroads. Trends Biochem. Sci. 2017, 42, 312–325. [Google Scholar] [CrossRef]
- Quinlan, C.L.; Orr, A.L.; Perevoshchikova, I.V.; Treberg, J.; Ackrell, B.A.; Brand, M. Mitochondrial Complex II Can Generate Reactive Oxygen Species at High Rates in Both the Forward and Reverse Reactions. J. Biol. Chem. 2012, 287, 27255–27264. [Google Scholar] [CrossRef] [Green Version]
- Al Rasheed, M.R.H.; Tarjan, G. Succinate Dehydrogenase Complex: An Updated Review. Arch. Pathol. Lab. Med. 2018, 142, 1564–1570. [Google Scholar] [CrossRef] [Green Version]
- Lemarie, A.; Grimm, S. Mutations in the heme b-binding residue of SDHC inhibit assembly of respiratory chain complex II in mammalian cells. Mitochondrion 2009, 9, 254–260. [Google Scholar] [CrossRef]
- Pozza, E.D.; Dando, I.; Pacchiana, R.; Liboi, E.; Scupoli, M.T.; Donadelli, M.; Palmieri, M. Regulation of succinate dehydrogenase and role of succinate in cancer. Semin. Cell Dev. Biol. 2020, 98, 4–14. [Google Scholar] [CrossRef]
- Rutter, J.; Winge, D.R.; Schiffman, J.D. Succinate dehydrogenase—Assembly, regulation and role in human disease. Mitochondrion 2010, 10, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Amar, L.; Pacak, K.; Steichen, O.; Akker, S.A.; Aylwin, S.J.B.; Baudin, E.; Buffet, A.; Burnichon, N.; Clifton-Bligh, R.J.; Dahia, P.L.M.; et al. International consensus on initial screening and follow-up of asymptomatic SDHx mutation carriers. Nat. Rev. Endocrinol. 2021, 17, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Andrews, K.A.; Ascher, D.B.; Pires, D.E.V.; Barnes, D.R.; Vialard, L.; Casey, R.T.; Bradshaw, N.; Adlard, J.; Aylwin, S.; Brennan, P.; et al. Tumour risks and genotype–phenotype correlations associated with germline variants in succinate dehydrogenase subunit genes SDHB, SDHC and SDHD. J. Med. Genet. 2018, 55, 384–394. [Google Scholar] [CrossRef] [Green Version]
- Laukka, T.; Mariani, C.J.; Ihantola, T.; Cao, J.Z.; Hokkanen, J.; Kaelin, W.G., Jr.; Godley, L.A.; Koivunen, P. Fumarate and Succinate Regulate Expression of Hypoxia-inducible Genes via TET Enzymes. J. Biol. Chem. 2016, 291, 4256–4265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer-Morales, K.L.; Heer, C.D.; Mapuskar, K.A.; Domann, F.E. Succinate Accumulation Links Mitochondrial MnSOD Depletion to Aberrant Nuclear DNA Methylation and Altered Cell Fate. J. Exp. Pathol. 2020, 1, 19. [Google Scholar] [CrossRef]
- Ishii, T.; Yasuda, K.; Akatsuka, A.; Hino, O.; Hartman, P.S.; Ishii, N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res. 2005, 65, 203–209. [Google Scholar]
- Slane, B.G.; Aykin-Burns, N.; Smith, B.J.; Kalen, A.L.; Goswami, P.C.; Domann, F.E.; Spitz, D.R. Mutation of Succinate Dehydrogenase Subunit C Results in Increased O2−, Oxidative Stress, and Genomic Instability. Cancer Res. 2006, 66, 7615–7620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, N.; Ishii, T.; Hartman, P.S. The role of the electron transport gene SDHC on lifespan and cancer. Exp. Gerontol. 2006, 41, 952–956. [Google Scholar] [CrossRef]
- De Sousa, S.M.C.; Toubia, J.; Hardy, T.S.E.; Feng, J.; Wang, P.; Schreiber, A.W.; Geoghegan, J.; Hall, R.; Rawlings, L.; Buckland, M.; et al. Aberrant Splicing of SDHC in Families with Unexplained Succinate Dehydrogenase-Deficient Paragangliomas. J. Endocr. Soc. 2020, 4, bvaa071. [Google Scholar] [CrossRef]
- Satoh, N.; Yokoyama, C.; Itamura, N.; Miyajima-Nakano, Y.; Hisatomi, H. Alternative splicing isoform in succinate dehydrogenase complex, subunit C causes downregulation of succinate-coenzyme Q oxidoreductase activity in mitochondria. Oncol. Lett. 2015, 9, 330–334. [Google Scholar] [CrossRef]
- Garnett, C.; Hernandez, D.C.; Vyas, P. GATA1 and cooperating mutations in myeloid leukaemia of Down syndrome. IUBMB Life 2019, 72, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Crispino, J.D. GATA1 in normal and malignant hematopoiesis. Semin. Cell Dev. Biol. 2005, 16, 137–147. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Caballero, N.; Fernández-Calleja, L.; Karkoulia, E.; Strouboulis, J. Regulation of GATA1 levels in erythropoiesis. IUBMB Life 2019, 72, 89–105. [Google Scholar] [CrossRef]
- Burda, P.; Laslo, P.; Stopka, T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 2010, 24, 1249–1257. [Google Scholar] [CrossRef] [Green Version]
- Meinders, M.; Hoogenboezem, M.; Scheenstra, M.R.; De Cuyper, I.M.; Papadopoulos, P.; Németh, T.; Mócsai, A.; Berg, T.K.V.D.; Kuijpers, T.W.; Gutiérrez, L. Repercussion of Megakaryocyte-Specific Gata1 Loss on Megakaryopoiesis and the Hematopoietic Precursor Compartment. PLoS ONE 2016, 11, e0154342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K. Recent advances in the understanding of transient abnormal myelopoiesis in Down syndrome. Pediatr. Int. 2019, 61, 222–229. [Google Scholar] [CrossRef] [Green Version]
- Kanezaki, R.; Toki, T.; Terui, K.; Xu, G.; Wang, R.; Shimada, A.; Hama, A.; Kanegane, H.; Kawakami, K.; Endo, M.; et al. Down syndrome and GATA1 mutations in transient abnormal myeloproliferative disorder: Mutation classes correlate with progression to myeloid leukemia. Blood 2010, 116, 4631–4638. [Google Scholar] [CrossRef]
- Grimm, J.; Heckl, D.; Klusmann, J.-H. Molecular Mechanisms of the Genetic Predisposition to Acute Megakaryoblastic Leukemia in Infants with Down Syndrome. Front. Oncol. 2021, 11, 636633. [Google Scholar] [CrossRef] [PubMed]
- de Castro, C.P.M.; Cadefau, M.; Cuartero, S. The Mutational Landscape of Myeloid Leukaemia in Down Syndrome. Cancers 2021, 13, 4144. [Google Scholar] [CrossRef]
- Sarnelli, G.; Grosso, M.; Palumbo, I.; Pesce, M.; D’Alessandro, A.; Zaninotto, G.; Annese, V.; Petruzzelli, R.; Izzo, P.; Sepulveres, R.; et al. Allele-specific transcriptional activity of the variable number of tandem repeats of the inducible nitric oxide synthase gene is associated with idiopathic achalasia. United Eur. Gastroenterol. J. 2017, 5, 200–207. [Google Scholar] [CrossRef]
- Cesaro, E.; Pastore, A.; Polverino, A.; Manna, L.; Divisato, G.; Quintavalle, C.; Di Sanzo, M.; Faniello, M.C.; Grosso, M.; Costanzo, P. ZNF224 is a mediator of TGF-β pro-oncogenic function in melanoma. Hum. Mol. Genet. 2021, ddab173, 1–10. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford Method for Protein Quantitation. In The Protein Protocols Handbook, 3rd ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 17–24. [Google Scholar]
- Montano, G.; Vidovic, K.; Palladino, C.; Cesaro, E.; Sodaro, G.; Quintarelli, C.; De Angelis, B.; Errichiello, S.; Pane, F.; Izzo, P.; et al. WT1-mediated repression of the proapoptotic transcription factor ZNF224 is triggered by the BCR-ABL oncogene. Oncotarget 2015, 6, 28223–28237. [Google Scholar] [CrossRef] [Green Version]
- Sodaro, G.; Cesaro, E.; Montano, G.; Blasio, G.; Fiorentino, F.; Romano, S.; Jacquel, A.; Aurberger, P.; Costanzo, P. Role of ZNF224 in c-Myc repression and imatinib responsiveness in chronic myeloid leukemia. Oncotarget 2017, 9, 3417–3431. [Google Scholar] [CrossRef]
- Xu, G. HIF-1-mediated expression of Foxo1 serves an important role in the proliferation and apoptosis of osteoblasts derived from children’s iliac cancellous bone. Mol. Med. Rep. 2018, 17, 6621–6631. [Google Scholar] [CrossRef] [Green Version]
- Sodaro, G.; Blasio, G.; Fiorentino, F.; Auberger, P.; Costanzo, P.; Cesaro, E. ZNF224 is a transcriptional repressor of AXL in chronic myeloid leukemia cells. Biochimie 2018, 154, 127–131. [Google Scholar] [CrossRef]
- Rinaldi, L.; Sepe, M.; Donne, R.D.; Conte, K.; Arcella, A.; Borzacchiello, D.; Amente, S.; De Vita, F.; Porpora, M.; Garbi, C.; et al. Mitochondrial AKAP1 supports mTOR pathway and tumor growth. Cell Death Dis. 2017, 8, e2842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamuri, S.S.V.P.; Sperling, J.A.; Sure, V.N.; Dholakia, M.H.; Peterson, N.R.; Rutkai, I.; Mahalingam, P.S.; Satou, R.; Katakam, P.V.G. Measurement of respiratory function in isolated cardiac mitochondria using Seahorse XFe24 Analyzer: Applications for aging research. GeroScience 2018, 40, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Kirschberg, M.; Heuser, S.; Marcuzzi, G.P.; Hufbauer, M.; Seeger, J.M.; Đukić, A.; Tomaić, V.; Majewski, S.; Wagner, S.; Wittekindt, C.; et al. ATP synthase modulation leads to an increase of spare respiratory capacity in HPV associated cancers. Sci. Rep. 2020, 10, 17339. [Google Scholar] [CrossRef]
- Marchetti, P.; Fovez, Q.; Germain, N.; Khamari, R.; Kluza, J. Mitochondrial spare respiratory capacity: Mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020, 34, 13106–13124. [Google Scholar] [CrossRef]
- Fonseca, T.B.; Sánchez-Guerrero, Á.; Milosevic, I.; Raimundo, N. Mitochondrial fission requires DRP1 but not dynamins. Nature 2019, 570, E34–E42. [Google Scholar] [CrossRef]
- Rudat, S.; Pfaus, A.; Cheng, Y.Y.; Holtmann, J.; Ellegast, J.; Bühler, C.; Di Marcantonio, D.; Martinez, E.; Göllner, S.; Wickenhauser, C.; et al. RET-mediated autophagy suppression as targetable co-dependence in acute myeloid leukemia. Leukemia 2018, 32, 2189–2202. [Google Scholar] [CrossRef]
- Bachhawat, A.K.; Yadav, S. The glutathione cycle: Glutathione metabolism beyond the γ-glutamyl cycle. IUBMB Life 2018, 70, 585–592. [Google Scholar] [CrossRef] [Green Version]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocabas, F.; Xie, L.; Xie, J.; Yu, Z.; DeBerardinis, R.J.; Kimura, W.; Thet, S.; Elshamy, A.F.; Abouellail, H.; Muralidhar, S.; et al. Hypoxic metabolism in human hematopoietic stem cells. Cell Biosci. 2015, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, D.; Gaete, D.; Rodriguez, D.; Hoogewijs, D.; Rauner, M.; Sormendi, S.; Wielockx, B. Hypoxia Pathway Proteins are Master Regulators of Erythropoiesis. Int. J. Mol. Sci. 2020, 21, 8131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-L.; Shen, G.-M.; Liu, X.-L.; Wang, F.; Zhao, Y.-Z.; Zhang, J.-W. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J. Cell. Mol. Med. 2012, 16, 1889–1899. [Google Scholar] [CrossRef]
- Xie, Y.; Li, W.; Feng, J.; Wu, T.; Li, J. MicroRNA-363 and GATA-1 are regulated by HIF-1α in K562 cells under hypoxia. Mol. Med. Rep. 2016, 14, 2503–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Li, M.; Xu, X.-J.; Xuan, L.; Huang, G.-N.; Song, X.-L.; Liu, Q.-F. HIF-1α and GLUT1 Gene Expression is Associated with Chemoresistance of Acute Myeloid Leukemia. Asian Pac. J. Cancer Prev. 2014, 15, 1823–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhoseiny, S.M. Hypoxia-Inducible Factor 1 Alpha (HIF-1α) and Its Prognostic Value in Acute Myeloid Leukemia. Hematol. Transfus. Int. J. 2017, 4, 19–25. [Google Scholar] [CrossRef]
- Hasegawa, A.; Shimizu, R. GATA1 Activity Governed by Configurations of cis-Acting Elements. Front. Oncol. 2017, 6, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halsey, C.; Docherty, M.; McNeill, M.; Gilchrist, D.; Le Brocq, M.; Gibson, B.; Graham, G. The GATA1s isoform is normally down-regulated during terminal haematopoietic differentiation and over-expression leads to failure to repress MYB, CCND2 and SKI during erythroid differentiation of K562 cells. J. Hematol. Oncol. 2012, 5, 45. [Google Scholar] [CrossRef]
- Khan, I.; Malinge, S.; Crispino, J.D. Myeloid Leukemia in Down Syndrome. Crit. Rev. Oncog. 2011, 16, 25–36. [Google Scholar] [CrossRef]
- Kaneko, H.; Kobayashi, E.; Yamamoto, M.; Shimizu, R. N- and C-terminal Transactivation Domains of GATA1 Protein Coordinate Hematopoietic Program. J. Biol. Chem. 2012, 287, 21439–21449. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, M.; Sanchez-Ferras, O.; Bouchard, M. GATA transcription factors in development and disease. Development 2018, 145, dev164384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lentjes, M.H.F.M.; Niessen, H.E.C.; Akiyama, Y.; De Bruine, A.P.; Melotte, V.; Van Engeland, M. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fu, H.; Gao, L.; Wang, L.; Wang, W.; Li, J.; Li, Y.; Dou, L.; Gao, X.; Luo, X.; et al. BCR-ABL/GATA1/miR-138 mini circuitry contributes to the leukemogenesis of chronic myeloid leukemia. Oncogene 2012, 33, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Movahed, Z.G.; Rastegari-Pouyani, M.; Mohammadi, M.H.; Mansouri, K. Cancer cells change their glucose metabolism to overcome increased ROS: One step from cancer cell to cancer stem cell? Biomed. Pharm. 2019, 112, 108690. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, T.; Palumbo, I.; Grosso, M.; Slater, N.; Miles, C.G. WT1 regulates murine hematopoiesis via maintenance of VEGF isoform ratio. Blood 2013, 122, 188–192. [Google Scholar] [CrossRef] [Green Version]
- Cesaro, E.; Sodaro, G.; Montano, G.; Grosso, M.; Lupo, A.; Costanzo, P. The Complex Role of the ZNF224 Transcription Factor in Cancer. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 107, pp. 191–222. [Google Scholar]
- Shimizu, R.; Yamamoto, M. Contribution of GATA1 dysfunction to multi-step leukemogenesis. Cancer Sci. 2012, 103, 2039–2044. [Google Scholar] [CrossRef]
- Halsey, C.; Tunstall, O.; Gibson, B.; Roberts, I.; Graham, G. Role of GATA-1s in early hematopoiesis and differences between alternative splicing in human and murine GATA-1. Blood 2010, 115, 3415–3416. [Google Scholar] [CrossRef] [Green Version]
- Grech, G.; Pollacco, J.; Portelli, M.; Sacco, K.; Baldacchino, S.; Grixti, J.M.; Saliba, C. Expression of different functional isoforms in haematopoiesis. Int. J. Hematol. 2014, 99, 4–11. [Google Scholar] [CrossRef] [PubMed]
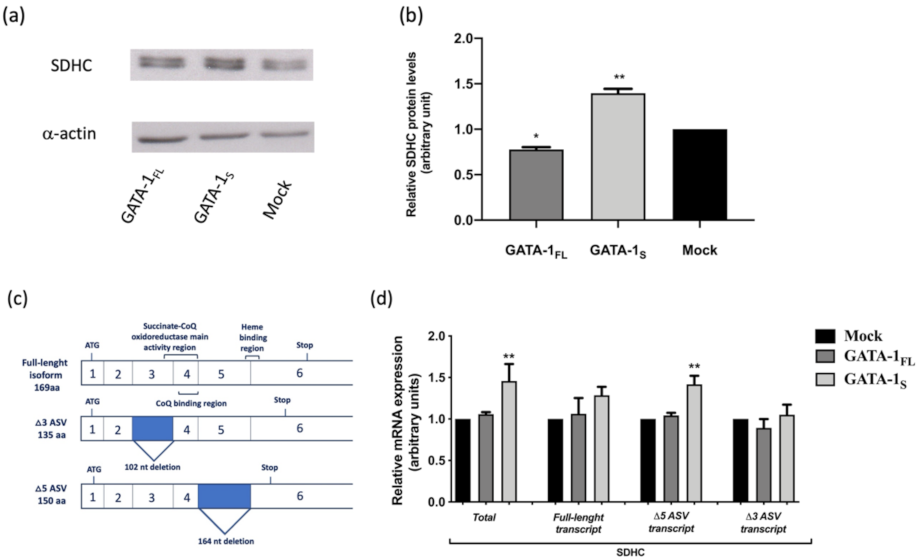
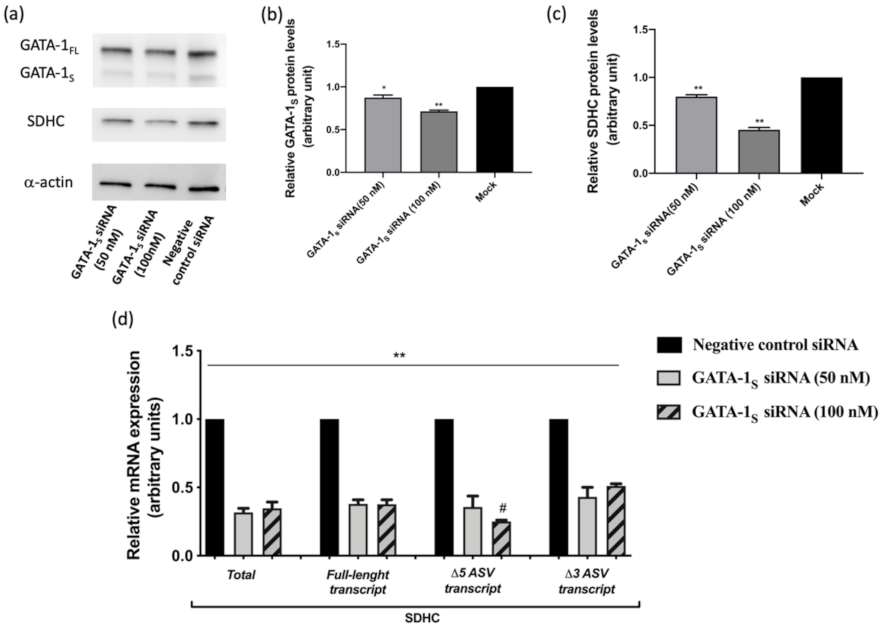


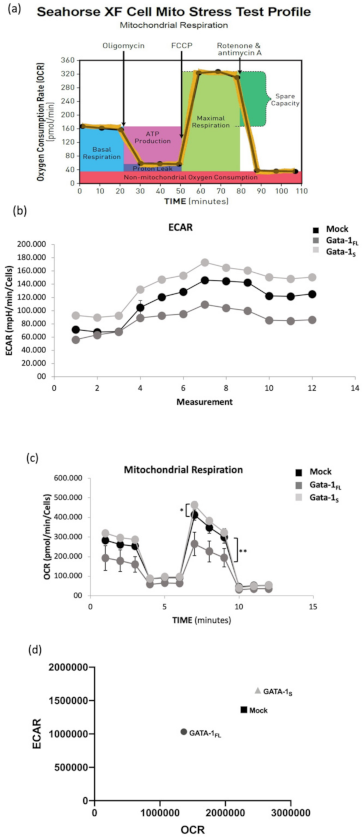
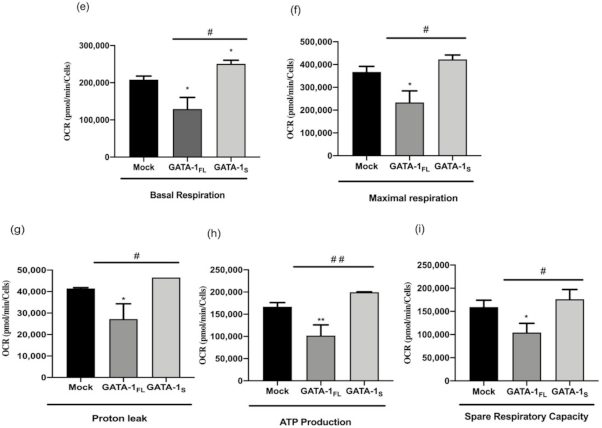
| Transcript | Accession Number | Primer | Sequence 5′-3′ | Amplicon Size |
|---|---|---|---|---|
| SDHC | NG_012767.1 | For 1 | CACTTCCGTCCAGACCGGA | 100 bp |
| Rev 1 | CTGATACAGAGCTGAGGGCTAA | |||
| SDHC full-length | NM_003001.5 | For 2 | TCTGTATCAGAAATGCTGTTCC | 183 bp |
| Rev 2 | GAGACCCCTGCACTCAAAGC | |||
| SDHC Δ 3 ASV | NM_001035512.2 (AB211234.1) | For 3 | GCTCTGTATCAGAAATTGGTCT | 250 bp |
| Rev 3 | GTCCCACATCAAGTGTCGGA | |||
| SDHC Δ 5 ASV | NM_001035511.2 (AB211235.1) | For 2 | TCTGTATCAGAAATGCTGTTCC | 187 bp |
| Rev 4 | GGTCCCACATCTGCACTCAA | |||
| HIF-1α | NM_001530.4 | For | TCCAAGAAGCCCTAACGTGT | 179 bp |
| Rev | TGATCGTCTGGCTGCTGTAA | |||
| GAPDH | NM_002046.7 | For | GAGCCACATCGCTCAGACAC | 116 bp |
| Rev | GGCAACAATATCCACTTTACCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trombetti, S.; Sessa, R.; Catapano, R.; Rinaldi, L.; Lo Bianco, A.; Feliciello, A.; Izzo, P.; Grosso, M. Exploring the Leukemogenic Potential of GATA-1S, the Shorter Isoform of GATA-1: Novel Insights into Mechanisms Hampering Respiratory Chain Complex II Activity and Limiting Oxidative Phosphorylation Efficiency. Antioxidants 2021, 10, 1603. https://doi.org/10.3390/antiox10101603
Trombetti S, Sessa R, Catapano R, Rinaldi L, Lo Bianco A, Feliciello A, Izzo P, Grosso M. Exploring the Leukemogenic Potential of GATA-1S, the Shorter Isoform of GATA-1: Novel Insights into Mechanisms Hampering Respiratory Chain Complex II Activity and Limiting Oxidative Phosphorylation Efficiency. Antioxidants. 2021; 10(10):1603. https://doi.org/10.3390/antiox10101603
Chicago/Turabian StyleTrombetti, Silvia, Raffaele Sessa, Rosa Catapano, Laura Rinaldi, Alessandra Lo Bianco, Antonio Feliciello, Paola Izzo, and Michela Grosso. 2021. "Exploring the Leukemogenic Potential of GATA-1S, the Shorter Isoform of GATA-1: Novel Insights into Mechanisms Hampering Respiratory Chain Complex II Activity and Limiting Oxidative Phosphorylation Efficiency" Antioxidants 10, no. 10: 1603. https://doi.org/10.3390/antiox10101603








