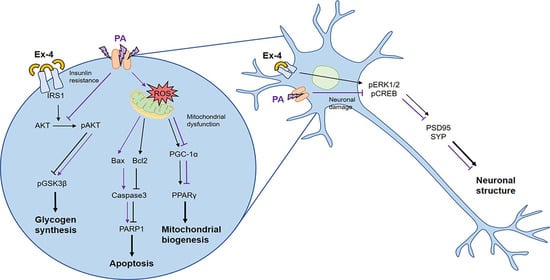
Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Drug Treatments and Preparation
2.2. SH-SY5Y Cell Culture and Differentiation
2.3. Rat Primary Cortical Neuron Culture and Transfection
2.4. Quantitative Real Time Polymerase Chain Reaction (PCR)
2.5. Western Blot Analyses
2.6. Reactive Oxygen Species (ROS) Assay
2.7. Transferase dUTP Nick End Labeling (TUNEL) Staining
2.8. Immunocytochemistry
2.9. Mitochondrial Activity Analyses
2.10. Neurite Outgrowth and Viability Measurement
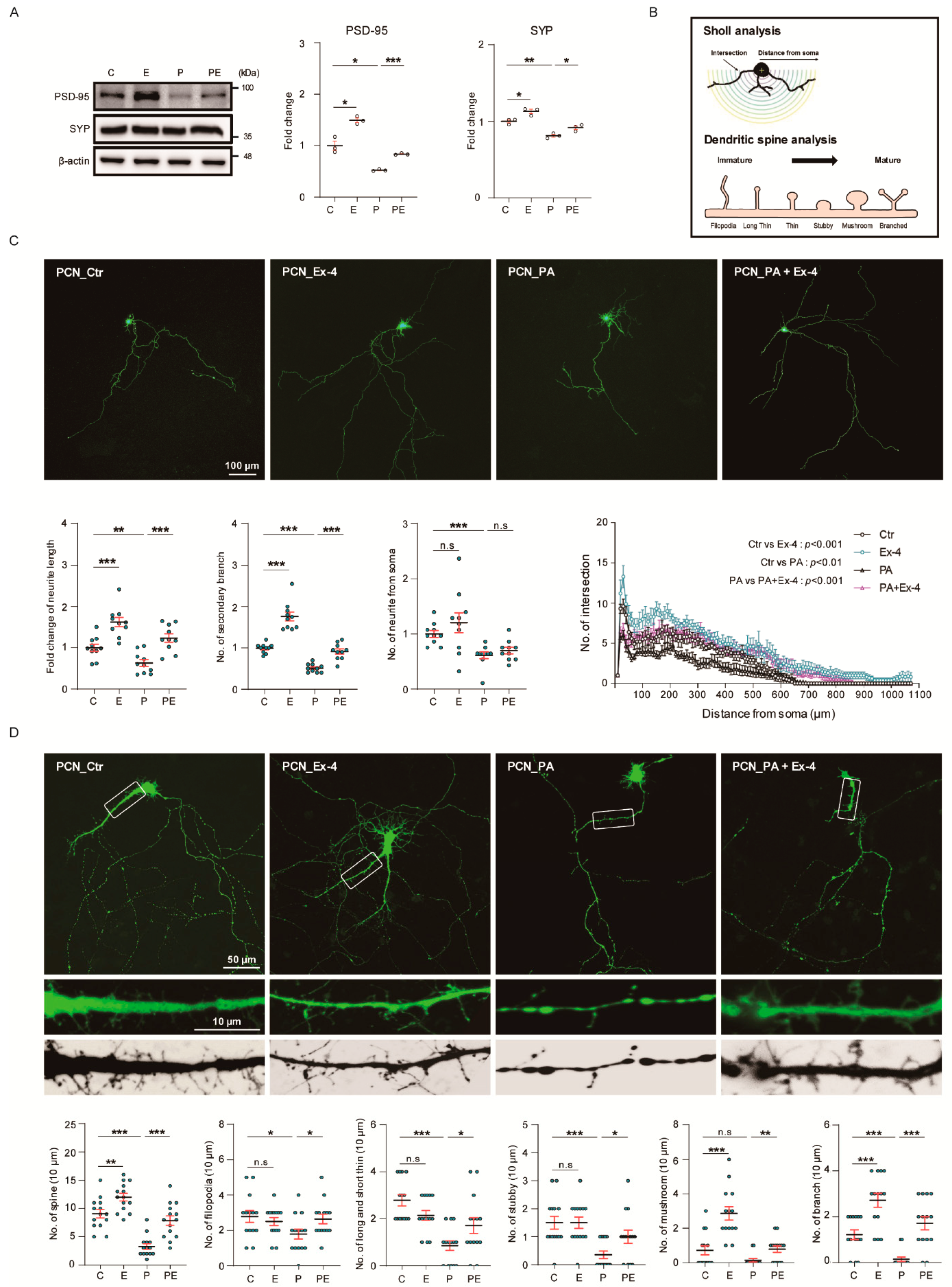
2.11. Neurite Complexity Analyses and Sholl Analyses
2.12. Dendritic Spine Analyses
2.13. Statistical Analyses
3. Results
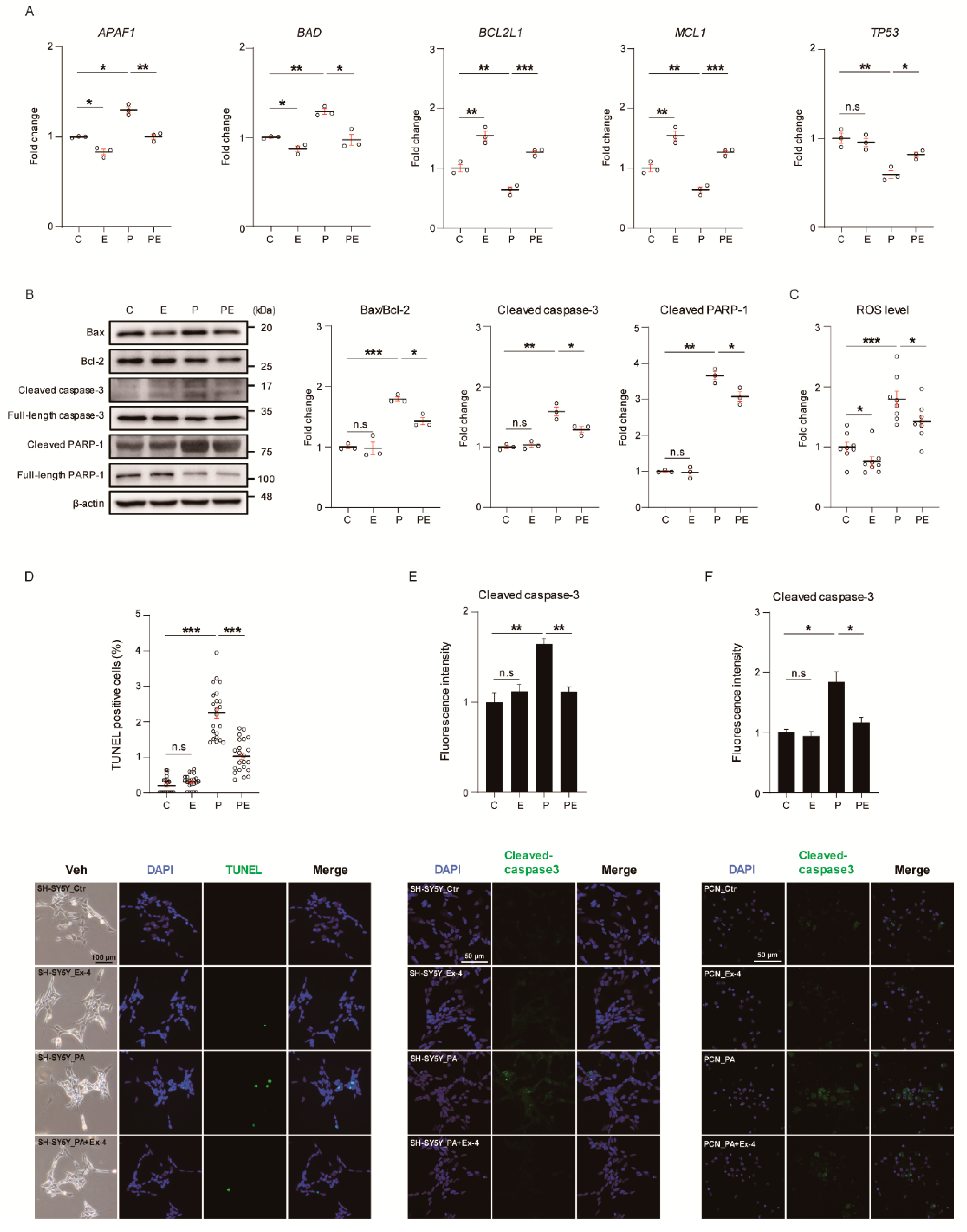
3.1. Ex-4 Suppressed Apoptosis in Neurons under PA-Induced Oxidative Stress
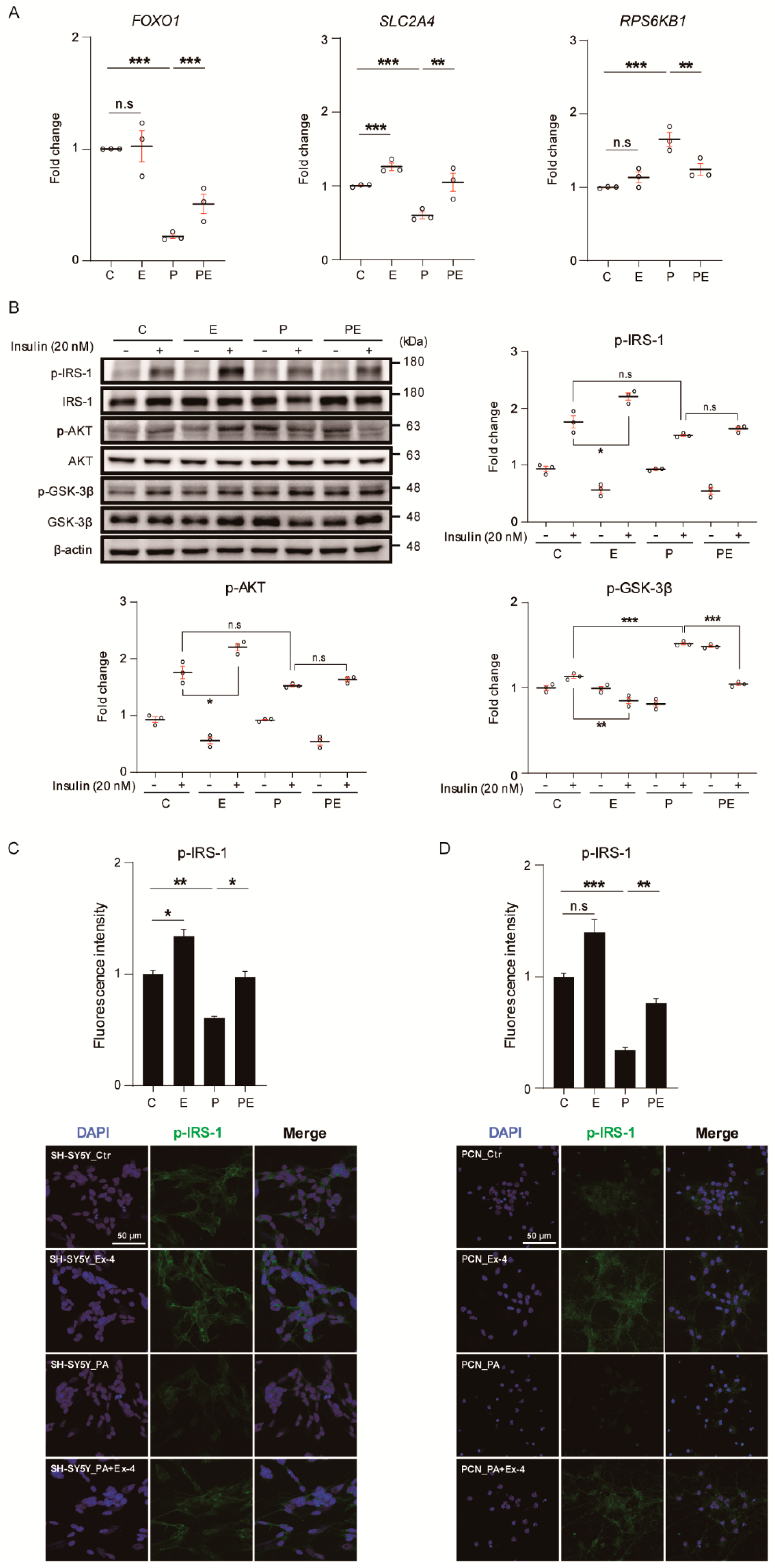
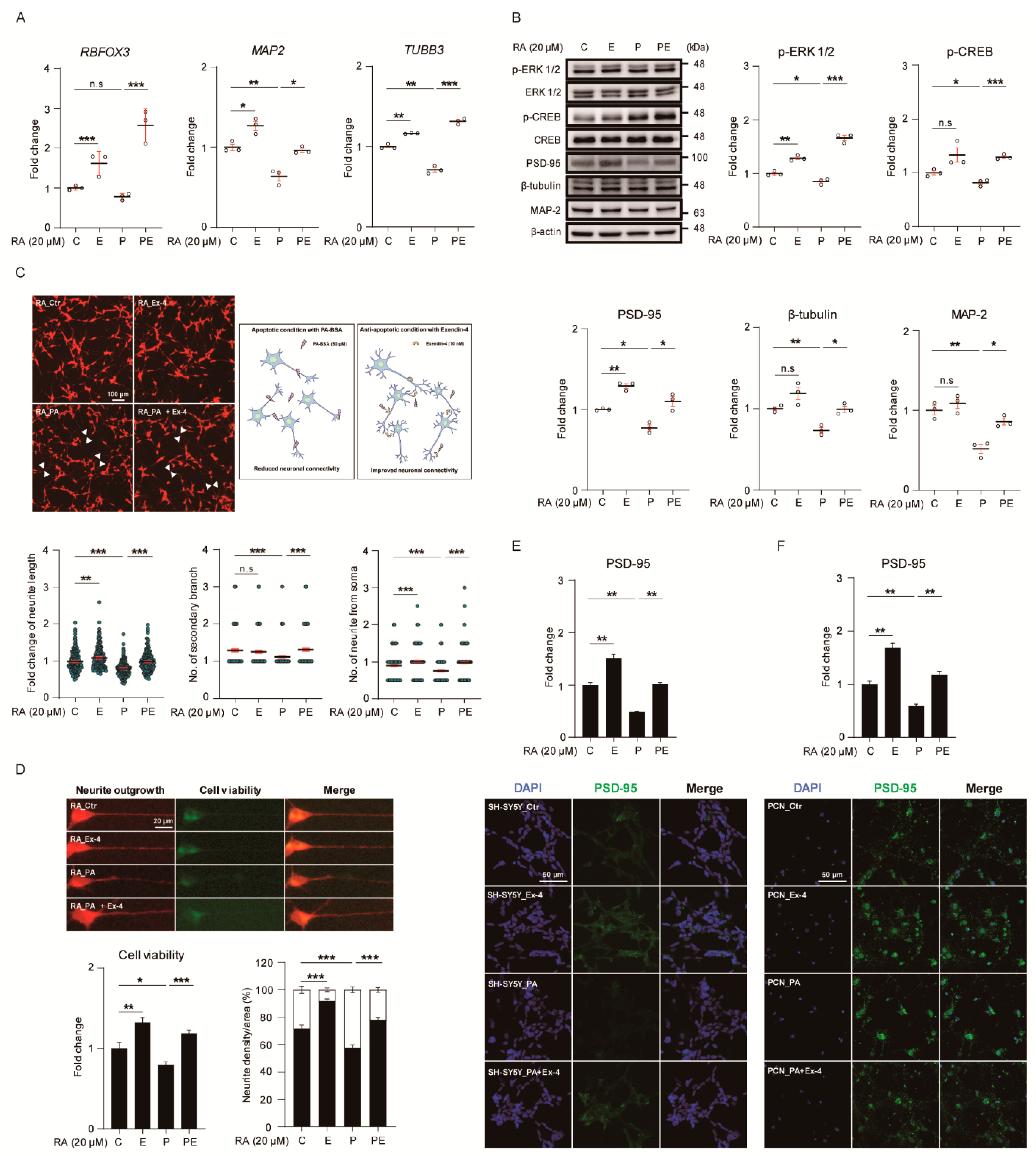
3.2. Ex-4 Improved Insulin Signaling in Neurons under PA-Induced Oxidative Stress
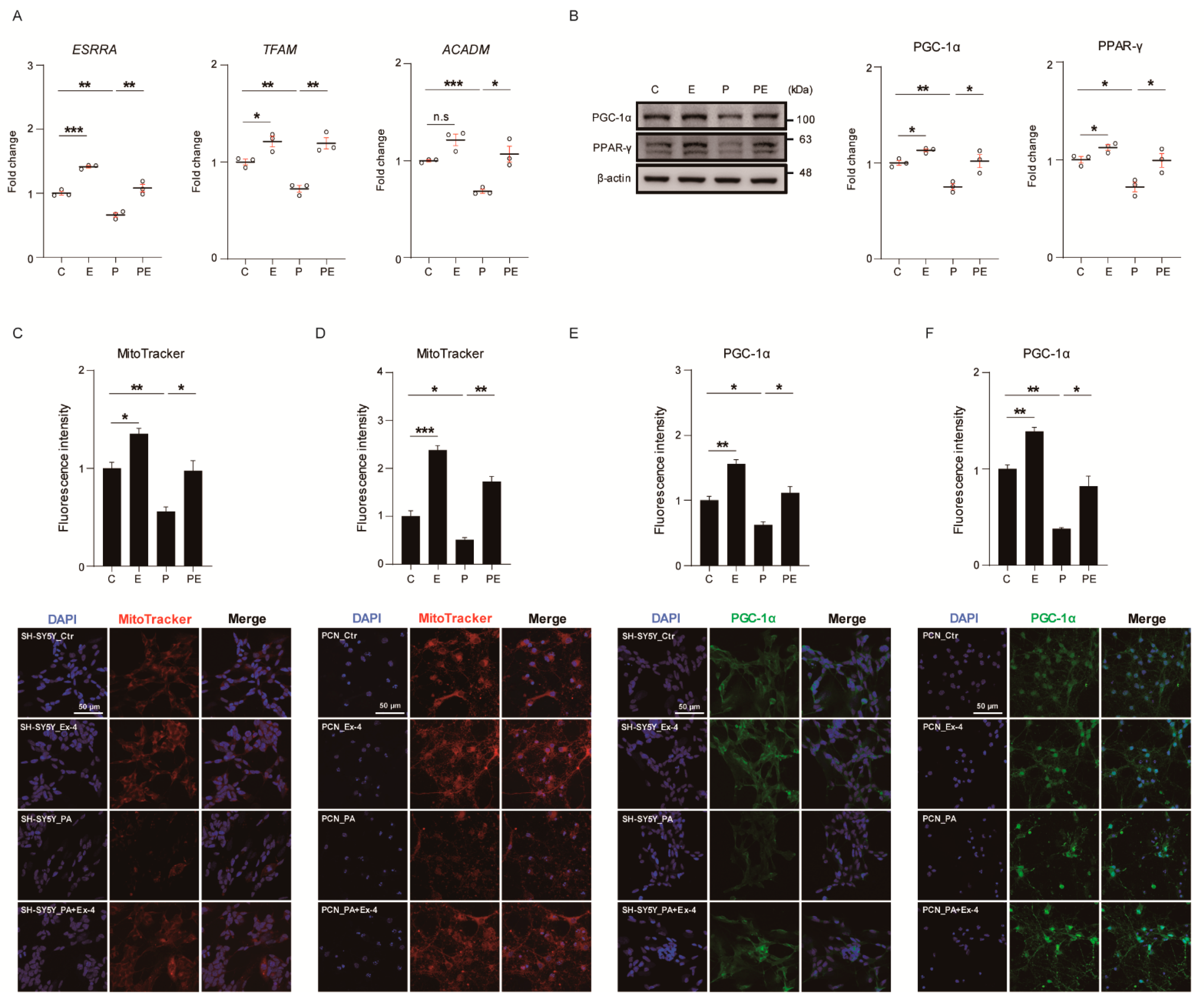
3.3. Ex-4 Improved Mitochondrial Function in Neurons against PA-Induced Oxidative Stress
3.4. Ex-4 Improved the Structure and Differentiation in Neurons under PA-Induced Oxidative Stress
3.5. Ex-4 Improved Synaptic Plasticity in Neurons under PA-Induced Oxidative Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GLP-1 | glucagon like peptide 1 |
| PA | palmitic acid |
| AD | Alzheimer’s disease |
| ER | endoplasmic reticulum |
| PCNs | primary cortical neurons |
| Ex-4 | exendin-4 |
| APAF1 | apoptotic peptidase activating factor 1 |
| BAD | BCL2-associated agonist of cell death |
| BCL2L1 | BCL2 like 1 |
| MCL1 | myeloid cell leukemia sequence 1 |
| TP53 | tumor protein p53 |
| FOXO1 | Forkhead Box O1 |
| RPS6KB1 | ribosomal protein S6 kinase B1 |
| IRS-1 | Insulin receptor substrate 1 |
| Bax | BCL2-associated X apoptosis regulator |
| Bcl-2 | B-cell lymphoma 2 |
| Caspase-3 | cysteine-aspartic acid protease 3 |
| AKT | phosphatidylinositol kinase (PI3K)/protein kinase B |
| GSK-3β | glycogen synthase kinase 3 beta |
| PPAR-γ | peroxisome proliferator-activated receptor gamma |
| ERK1/2 | p44/42 MAPK extracellular signal-regulated kinases 1/2 |
| CREB | cAMP-response element binding protein |
| PSD-95 | postsynaptic density protein 95 |
| PARP-1 | Cleaved-Poly [ADP-ribose] polymerase 1 |
| PGC-1α | peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| MAP-2 | microtubule-associated protein 2, ROS, reactive oxygen species |
| GFP | green fluorescent protein |
| ATP | adenosine triphosphate |
| ESRRA | estrogen-related receptor alpha |
| TFAM | transcription factor A, mitochondrial |
| ACADM | acyl-coenzyme A dehydrogenase |
| RBFOX | RNA binding fox-1 homolog 3 |
| TUBB3 | tubulin beta 3 III |
References
- Williams, E.P.; Mesidor, M.; Winters, K.; Dubbert, P.M.; Wyatt, S.B. Overweight and Obesity: Prevalence, Consequences, and Causes of a Growing Public Health Problem. Curr. Obes. Rep. 2015, 4, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Kivipelto, M.; Ngandu, T.; Fratiglioni, L.; Viitanen, M.; Kareholt, I.; Winblad, B.; Helkala, E.L.; Tuomilehto, J.; Soininen, H.; Nissinen, A. Obesity and vascular risk factors at midlife and the risk of dementia and Alzheimer disease. Arch Neurol. 2005, 62, 1556–1560. [Google Scholar] [CrossRef] [PubMed]
- Cordner, Z.A.; Tamashiro, K.L. Effects of high-fat diet exposure on learning & memory. Physiol. Behav. 2015, 152, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Francis, H.; Stevenson, R. The longer-term impacts of Western diet on human cognition and the brain. Appetite 2013, 63, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Anstey, K.J.; Cherbuin, N.; Budge, M.; Young, J. Body mass index in midlife and late-life as a risk factor for dementia: A meta-analysis of prospective studies. Obes. Rev. 2011, 12, e426–e437. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.A.; Spencer, S.J. Obesity and neuroinflammation: A pathway to cognitive impairment. Brain Behav. Immun. 2014, 42, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Karmi, A.; Iozzo, P.; Viljanen, A.; Hirvonen, J.; Fielding, B.A.; Virtanen, K.; Oikonen, V.; Kemppainen, J.; Viljanen, T.; Guiducci, L.; et al. Increased brain fatty acid uptake in metabolic syndrome. Diabetes 2010, 59, 2171–2177. [Google Scholar] [CrossRef]
- Marwarha, G.; Claycombe, K.; Schommer, J.; Collins, D.; Ghribi, O. Palmitate-induced Endoplasmic Reticulum stress and subsequent C/EBPalpha Homologous Protein activation attenuates leptin and Insulin-like growth factor 1 expression in the brain. Cell Signal. 2016, 28, 1789–1805. [Google Scholar] [CrossRef]
- Valdearcos, M.; Robblee, M.M.; Benjamin, D.I.; Nomura, D.K.; Xu, A.W.; Koliwad, S.K. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep. 2014, 9, 2124–2138. [Google Scholar] [CrossRef]
- Rodriguez-Navas, C.; Morselli, E.; Clegg, D.J. Sexually dimorphic brain fatty acid composition in low and high fat diet-fed mice. Mol. Metab. 2016, 5, 680–689. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, D.; Zhang, Q.; Wang, J.; Zhan, J.; Xian, X.; Du, Z.; Wang, X.; Hao, A. Palmitic acid affects proliferation and differentiation of neural stem cells in vitro. J. Neurosci. Res. 2014, 92, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhao, S.; Wang, F.; Zhang, H.; Chen, Z.J.; Wang, J.; Wang, Z.; Du, Z.; Ling, E.A.; Liu, Q.; et al. Palmitic acid increases apoptosis of neural stem cells via activating c-Jun N-terminal kinase. Stem. Cell Res. 2013, 10, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Melrose, J.; Chan, C. Involvement of astroglial ceramide in palmitic acid-induced Alzheimer-like changes in primary neurons. Eur. J. Neurosci. 2007, 26, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Park, H.R.; Kim, J.Y.; Park, K.Y.; Lee, J. Lipotoxicity of palmitic Acid on neural progenitor cells and hippocampal neurogenesis. Toxicol. Res. 2011, 27, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Martin, R.; Chan, C. Palmitate-activated astrocytes via serine palmitoyltransferase increase BACE1 in primary neurons by sphingomyelinases. Neurobiol. Aging 2013, 34, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 2017, 8, 2009. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, H.J.; Lee, S.J.; Jung, Y.H.; Yoo, D.Y.; Hwang, I.K.; Seong, J.K.; Ryu, J.M.; Han, H.J. Palmitic Acid-BSA enhances Amyloid-beta production through GPR40-mediated dual pathways in neuronal cells: Involvement of the Akt/mTOR/HIF-1alpha and Akt/NF-kappaB pathways. Sci. Rep. 2017, 7, 4335. [Google Scholar] [CrossRef]
- Yamamoto, H.; Lee, C.E.; Marcus, J.N.; Williams, T.D.; Overton, J.M.; Lopez, M.E.; Hollenberg, A.N.; Baggio, L.; Saper, C.B.; Drucker, D.J.; et al. Glucagon-like peptide-1 receptor stimulation increases blood pressure and heart rate and activates autonomic regulatory neurons. J. Clin. Investig. 2002, 110, 43–52. [Google Scholar] [CrossRef]
- Knauf, C.; Cani, P.D.; Kim, D.H.; Iglesias, M.A.; Chabo, C.; Waget, A.; Colom, A.; Rastrelli, S.; Delzenne, N.M.; Drucker, D.J.; et al. Role of central nervous system glucagon-like Peptide-1 receptors in enteric glucose sensing. Diabetes 2008, 57, 2603–2612. [Google Scholar] [CrossRef]
- During, M.J.; Cao, L.; Zuzga, D.S.; Francis, J.S.; Fitzsimons, H.L.; Jiao, X.; Bland, R.J.; Klugmann, M.; Banks, W.A.; Drucker, D.J.; et al. Glucagon-like peptide-1 receptor is involved in learning and neuroprotection. Nat. Med. 2003, 9, 1173–1179. [Google Scholar] [CrossRef]
- Kastin, A.J.; Akerstrom, V.; Pan, W. Interactions of glucagon-like peptide-1 (GLP-1) with the blood-brain barrier. J. Mol. Neurosci. 2002, 18, 7–14. [Google Scholar] [CrossRef]
- Yoon, G.; Kim, Y.K.; Song, J. Glucagon-like peptide-1 suppresses neuroinflammation and improves neural structure. Pharmacol. Res. 2020, 152, 104615. [Google Scholar] [CrossRef] [PubMed]
- Seppa, K.; Toots, M.; Reimets, R.; Jagomae, T.; Koppel, T.; Pallase, M.; Hasselholt, S.; Krogsbaek Mikkelsen, M.; Randel Nyengaard, J.; Vasar, E.; et al. GLP-1 receptor agonist liraglutide has a neuroprotective effect on an aged rat model of Wolfram syndrome. Sci. Rep. 2019, 9, 15742. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; Glantz, L.A.; Pierri, J.N.; Sweet, R.A. Altered cortical glutamate neurotransmission in schizophrenia: Evidence from morphological studies of pyramidal neurons. Ann. N. Y. Acad. Sci. 2003, 1003, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Tonks, K.T.; Ng, Y.; Miller, S.; Coster, A.C.; Samocha-Bonet, D.; Iseli, T.J.; Xu, A.; Patrick, E.; Yang, J.Y.; Junutula, J.R.; et al. Impaired Akt phosphorylation in insulin-resistant human muscle is accompanied by selective and heterogeneous downstream defects. Diabetologia 2013, 56, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.G.; Wang, N.; Cao, D.J.; Sachan, N.; Morris, D.J.; Gerard, R.D.; Kuro, O.M.; Rothermel, B.A.; Hill, J.A. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc. Natl. Acad. Sci. USA 2007, 104, 20517–20522. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.M.; Lopes, A.B.; Crisma, A.R.; de Sa, R.C.C.; Kuwabara, W.M.T.; Curi, R.; de Andrade, P.B.M.; Alonso-Vale, M.I.C. Palmitoleic acid (16:1n7) increases oxygen consumption, fatty acid oxidation and ATP content in white adipocytes. Lipids Health Dis. 2018, 17, 55. [Google Scholar] [CrossRef]
- Almaguel, F.G.; Liu, J.W.; Pacheco, F.J.; Casiano, C.A.; De Leon, M. Activation and reversal of lipotoxicity in PC12 and rat cortical cells following exposure to palmitic acid. J. Neurosci. Res. 2009, 87, 1207–1218. [Google Scholar] [CrossRef]
- Wright, K.M.; Smith, M.I.; Farrag, L.; Deshmukh, M. Chromatin modification of Apaf-1 restricts the apoptotic pathway in mature neurons. J. Cell Biol. 2007, 179, 825–832. [Google Scholar] [CrossRef]
- Datta, S.R.; Ranger, A.M.; Lin, M.Z.; Sturgill, J.F.; Ma, Y.C.; Cowan, C.W.; Dikkes, P.; Korsmeyer, S.J.; Greenberg, M.E. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev. Cell 2002, 3, 631–643. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.E.; Hochegger, H.; Takeda, S.; Caldecott, K.W. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell Biol. 2007, 27, 5597–5605. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Niwa, M.; Iwai, T.; Nakashima, M.; Bunai, Y.; Uematsu, T.; Yoshimi, N.; Mori, H. Neuronal apoptosis studied by a sequential TUNEL technique: A method for tract-tracing. Brain Res. Brain Res. Protoc. 1999, 4, 140–146. [Google Scholar] [CrossRef]
- Samuel, M.A.; Morrey, J.D.; Diamond, M.S. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J. Virol. 2007, 81, 2614–2623. [Google Scholar] [CrossRef]
- Liemburg-Apers, D.C.; Willems, P.H.; Koopman, W.J.; Grefte, S. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch. Toxicol. 2015, 89, 1209–1226. [Google Scholar] [CrossRef]
- Wang, W.; Fang, H.; Groom, L.; Cheng, A.; Zhang, W.; Liu, J.; Wang, X.; Li, K.; Han, P.; Zheng, M.; et al. Superoxide flashes in single mitochondria. Cell 2008, 134, 279–290. [Google Scholar] [CrossRef]
- Olguin-Albuerne, M.; Moran, J. ROS produced by NOX2 control in vitro development of cerebellar granule neurons development. ASN Neuro. 2015, 7. [Google Scholar] [CrossRef]
- Hidalgo, C.; Arias-Cavieres, A. Calcium, Reactive Oxygen Species, and Synaptic Plasticity. Physiology (Bethesda) 2016, 31, 201–215. [Google Scholar] [CrossRef]
- Shemer, J.; Adamo, M.; Raizada, M.K.; Heffez, D.; Zick, Y.; LeRoith, D. Insulin and IGF-I stimulate phosphorylation of their respective receptors in intact neuronal and glial cells in primary culture. J. Mol. Neurosci. 1989, 1, 3–8. [Google Scholar] [CrossRef]
- Mayer, C.M.; Belsham, D.D. Central insulin signaling is attenuated by long-term insulin exposure via insulin receptor substrate-1 serine phosphorylation, proteasomal degradation, and lysosomal insulin receptor degradation. Endocrinology 2010, 151, 75–84. [Google Scholar] [CrossRef]
- Arnold, S.E.; Arvanitakis, Z.; Macauley-Rambach, S.L.; Koenig, A.M.; Wang, H.Y.; Ahima, R.S.; Craft, S.; Gandy, S.; Buettner, C.; Stoeckel, L.E.; et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: Concepts and conundrums. Nat. Rev. Neurol. 2018, 14, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Wang, H.Y.; Kazi, H.; Han, L.Y.; Bakshi, K.P.; Stucky, A.; Fuino, R.L.; Kawaguchi, K.R.; Samoyedny, A.J.; Wilson, R.S.; et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Investig. 2012, 122, 1316–1338. [Google Scholar] [CrossRef] [PubMed]
- Moll, L.; Schubert, M. The Role of Insulin and Insulin-Like Growth Factor-1/FoxO-Mediated Transcription for the Pathogenesis of Obesity-Associated Dementia. Curr. Gerontol. Geriatr. Res. 2012, 2012, 384094. [Google Scholar] [CrossRef] [PubMed]
- Cifarelli, V.; Lee, S.; Kim, D.H.; Zhang, T.; Kamagate, A.; Slusher, S.; Bertera, S.; Luppi, P.; Trucco, M.; Dong, H.H. FOXO1 mediates the autocrine effect of endothelin-1 on endothelial cell survival. Mol. Endocrinol. 2012, 26, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Irvine, E.E.; Drinkwater, L.; Radwanska, K.; Al-Qassab, H.; Smith, M.A.; O’Brien, M.; Kielar, C.; Choudhury, A.I.; Krauss, S.; Cooper, J.D.; et al. Insulin receptor substrate 2 is a negative regulator of memory formation. Learn. Mem. 2011, 18, 375–383. [Google Scholar] [CrossRef][Green Version]
- Smith, M.A.; Katsouri, L.; Irvine, E.E.; Hankir, M.K.; Pedroni, S.M.; Voshol, P.J.; Gordon, M.W.; Choudhury, A.I.; Woods, A.; Vidal-Puig, A.; et al. Ribosomal S6K1 in POMC and AgRP Neurons Regulates Glucose Homeostasis but Not Feeding Behavior in Mice. Cell Rep. 2015, 11, 335–343. [Google Scholar] [CrossRef]
- Bian, H.; Bian, W.; Lin, X.; Ma, Z.; Chen, W.; Pu, Y. RNA Interference Silencing of Glycogen Synthase Kinase 3beta Inhibites Tau Phosphorylation in Mice with Alzheimer Disease. Neurochem. Res. 2016, 41, 2470–2480. [Google Scholar] [CrossRef]
- Schubert, M.; Gautam, D.; Surjo, D.; Ueki, K.; Baudler, S.; Schubert, D.; Kondo, T.; Alber, J.; Galldiks, N.; Kustermann, E.; et al. Role for neuronal insulin resistance in neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 2004, 101, 3100–3105. [Google Scholar] [CrossRef]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef]
- Wang, X.; Su, B.; Zheng, L.; Perry, G.; Smith, M.A.; Zhu, X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009, 109 (Suppl. 1), 153–159. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 2002, 286, 81–89. [Google Scholar] [CrossRef]
- Rasbach, K.A.; Schnellmann, R.G. Signaling of mitochondrial biogenesis following oxidant injury. J. Biol. Chem. 2007, 282, 2355–2362. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, X.; Feng, Y.; Handschin, C.; Feng, Y.; Gullicksen, P.S.; Bare, O.; Labow, M.; Spiegelman, B.; Stevenson, S.C. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. USA 2006, 103, 14379–14384. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Handschin, C.; Arlow, D.; Xie, X.; St Pierre, J.; Sihag, S.; Yang, W.; Altshuler, D.; Puigserver, P.; Patterson, N.; et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 2004, 101, 6570–6575. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, J.; Drori, S.; Uldry, M.; Silvaggi, J.M.; Rhee, J.; Jager, S.; Handschin, C.; Zheng, K.; Lin, J.; Yang, W.; et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 2006, 127, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.H.; Satoh, H.; Herzig, S.; Lee, C.H.; Hedrick, S.; Kulkarni, R.; Evans, R.M.; Olefsky, J.; Montminy, M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 2004, 10, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Buckman, J.F.; Hernandez, H.; Kress, G.J.; Votyakova, T.V.; Pal, S.; Reynolds, I.J. MitoTracker labeling in primary neuronal and astrocytic cultures: Influence of mitochondrial membrane potential and oxidants. J. Neurosci. Methods 2001, 104, 165–176. [Google Scholar] [CrossRef]
- Miglio, G.; Rosa, A.C.; Rattazzi, L.; Collino, M.; Lombardi, G.; Fantozzi, R. PPARgamma stimulation promotes mitochondrial biogenesis and prevents glucose deprivation-induced neuronal cell loss. Neurochem. Int. 2009, 55, 496–504. [Google Scholar] [CrossRef]
- Fuenzalida, K.; Quintanilla, R.; Ramos, P.; Piderit, D.; Fuentealba, R.A.; Martinez, G.; Inestrosa, N.C.; Bronfman, M. Peroxisome proliferator-activated receptor gamma up-regulates the Bcl-2 anti-apoptotic protein in neurons and induces mitochondrial stabilization and protection against oxidative stress and apoptosis. J. Biol. Chem. 2007, 282, 37006–37015. [Google Scholar] [CrossRef]
- Lin, Y.S.; Wang, H.Y.; Huang, D.F.; Hsieh, P.F.; Lin, M.Y.; Chou, C.H.; Wu, I.J.; Huang, G.J.; Gau, S.S.; Huang, H.S. Neuronal Splicing Regulator RBFOX3 (NeuN) Regulates Adult Hippocampal Neurogenesis and Synaptogenesis. PLoS ONE 2016, 11, e0164164. [Google Scholar] [CrossRef]
- Latremoliere, A.; Cheng, L.; DeLisle, M.; Wu, C.; Chew, S.; Hutchinson, E.B.; Sheridan, A.; Alexandre, C.; Latremoliere, F.; Sheu, S.H.; et al. Neuronal-Specific TUBB3 Is Not Required for Normal Neuronal Function but Is Essential for Timely Axon Regeneration. Cell Rep. 2018, 24, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Korzhevskii, D.E.; Karpenko, M.N.; Kirik, O.V. Microtubule-associated proteins as markers of nerve cell differentiation and functional status. Morfologiia 2011, 139, 13–21. [Google Scholar] [PubMed]
- Chai, J.; Wang, Y.; Li, H.; He, W.; Zou, W.; Zhou, Y.; Hu, X.; Chai, Q. Distribution of postsynaptic density protein 95 (PSD95) and synaptophysin during neuronal maturation. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2016, 32, 1619–1622. [Google Scholar] [PubMed]
- Sweatt, J.D. The neuronal MAP kinase cascade: A biochemical signal integration system subserving synaptic plasticity and memory. J. Neurochem. 2001, 76, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, J.; Ye, Z.; Huang, J.; He, F.; Xiao, W.; Hu, X.; Luo, Z. CaMKII-Mediated CREB Phosphorylation Is Involved in Ca2+-Induced BDNF mRNA Transcription and Neurite Outgrowth Promoted by Electrical Stimulation. PLoS ONE 2016, 11, e0162784. [Google Scholar] [CrossRef]
- Li, M.; Li, S.; Li, Y. Liraglutide Promotes Cortical Neurite Outgrowth via the MEK-ERK Pathway. Cell Mol. Neurobiol. 2015, 35, 987–993. [Google Scholar] [CrossRef]
- Hlushchenko, I.; Koskinen, M.; Hotulainen, P. Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity. Cytoskeleton (Hoboken) 2016, 73, 435–441. [Google Scholar] [CrossRef]
- van Beuningen, S.F.; Hoogenraad, C.C. Neuronal polarity: Remodeling microtubule organization. Curr. Opin. Neurobiol. 2016, 39, 1–7. [Google Scholar] [CrossRef]
- Yau, K.W.; Schatzle, P.; Tortosa, E.; Pages, S.; Holtmaat, A.; Kapitein, L.C.; Hoogenraad, C.C. Dendrites In Vitro and In Vivo Contain Microtubules of Opposite Polarity and Axon Formation Correlates with Uniform Plus-End-Out Microtubule Orientation. J. Neurosci. 2016, 36, 1071–1085. [Google Scholar] [CrossRef]
- Pacheco, A.; Gallo, G. Actin filament-microtubule interactions in axon initiation and branching. Brain Res. Bull. 2016, 126, 300–310. [Google Scholar] [CrossRef]
- Zikopoulos, B.; Barbas, H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Front. Hum. Neurosci. 2013, 7, 609. [Google Scholar] [CrossRef] [PubMed]
- Armijo-Weingart, L.; Gallo, G. It takes a village to raise a branch: Cellular mechanisms of the initiation of axon collateral branches. Mol. Cell Neurosci. 2017, 84, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Sears, J.C.; Broihier, H.T. FoxO regulates microtubule dynamics and polarity to promote dendrite branching in Drosophila sensory neurons. Dev. Biol. 2016, 418, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Santiago, C.; Bashaw, G.J. Transcription factors and effectors that regulate neuronal morphology. Development 2014, 141, 4667–4680. [Google Scholar] [CrossRef] [PubMed]
- Bencsik, N.; Pusztai, S.; Borbely, S.; Fekete, A.; Dulk, M.; Kis, V.; Pesti, S.; Vas, V.; Szucs, A.; Buday, L.; et al. Dendritic spine morphology and memory formation depend on postsynaptic Caskin proteins. Sci. Rep. 2019, 9, 16843. [Google Scholar] [CrossRef]
- van der Zee, E.A. Synapses, spines and kinases in mammalian learning and memory, and the impact of aging. Neurosci. Biobehav. Rev. 2015, 50, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Gipson, C.D.; Olive, M.F. Structural and functional plasticity of dendritic spines-root or result of behavior? Genes Brain Behav. 2017, 16, 101–117. [Google Scholar] [CrossRef]
- Lopes, S.; Teplytska, L.; Vaz-Silva, J.; Dioli, C.; Trindade, R.; Morais, M.; Webhofer, C.; Maccarrone, G.; Almeida, O.F.X.; Turck, C.W.; et al. Tau Deletion Prevents Stress-Induced Dendritic Atrophy in Prefrontal Cortex: Role of Synaptic Mitochondria. Cereb. Cortex 2017, 27, 2580–2591. [Google Scholar] [CrossRef]
- Bello-Medina, P.C.; Flores, G.; Quirarte, G.L.; McGaugh, J.L.; Prado Alcala, R.A. Mushroom spine dynamics in medium spiny neurons of dorsal striatum associated with memory of moderate and intense training. Proc. Natl. Acad. Sci. USA 2016, 113, E6516–E6525. [Google Scholar] [CrossRef]
- Zhao, F.; Li, J.; Wang, R.; Xu, H.; Ma, K.; Kong, X.; Sun, Z.; Niu, X.; Jiang, J.; Liu, B.; et al. Exendin-4 promotes actin cytoskeleton rearrangement and protects cells from Nogo-A-Delta20 mediated spreading inhibition and growth cone collapse by down-regulating RhoA expression and activation via the PI3K pathway. Biomed. Pharmacother. 2019, 109, 135–143. [Google Scholar] [CrossRef]
- Sango, K.; Utsunomiya, K. Efficacy of glucagon-like peptide-1 mimetics for neural regeneration. Neural Regen. Res. 2015, 10, 1723–1724. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, D.; Yoon, G.; Song, J. Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress. Antioxidants 2021, 10, 78. https://doi.org/10.3390/antiox10010078
Jo D, Yoon G, Song J. Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress. Antioxidants. 2021; 10(1):78. https://doi.org/10.3390/antiox10010078
Chicago/Turabian StyleJo, Danbi, Gwangho Yoon, and Juhyun Song. 2021. "Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress" Antioxidants 10, no. 1: 78. https://doi.org/10.3390/antiox10010078
APA StyleJo, D., Yoon, G., & Song, J. (2021). Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress. Antioxidants, 10(1), 78. https://doi.org/10.3390/antiox10010078