Protein Microarrays with Novel Microfluidic Methods: Current Advances
Abstract
:1. Introduction
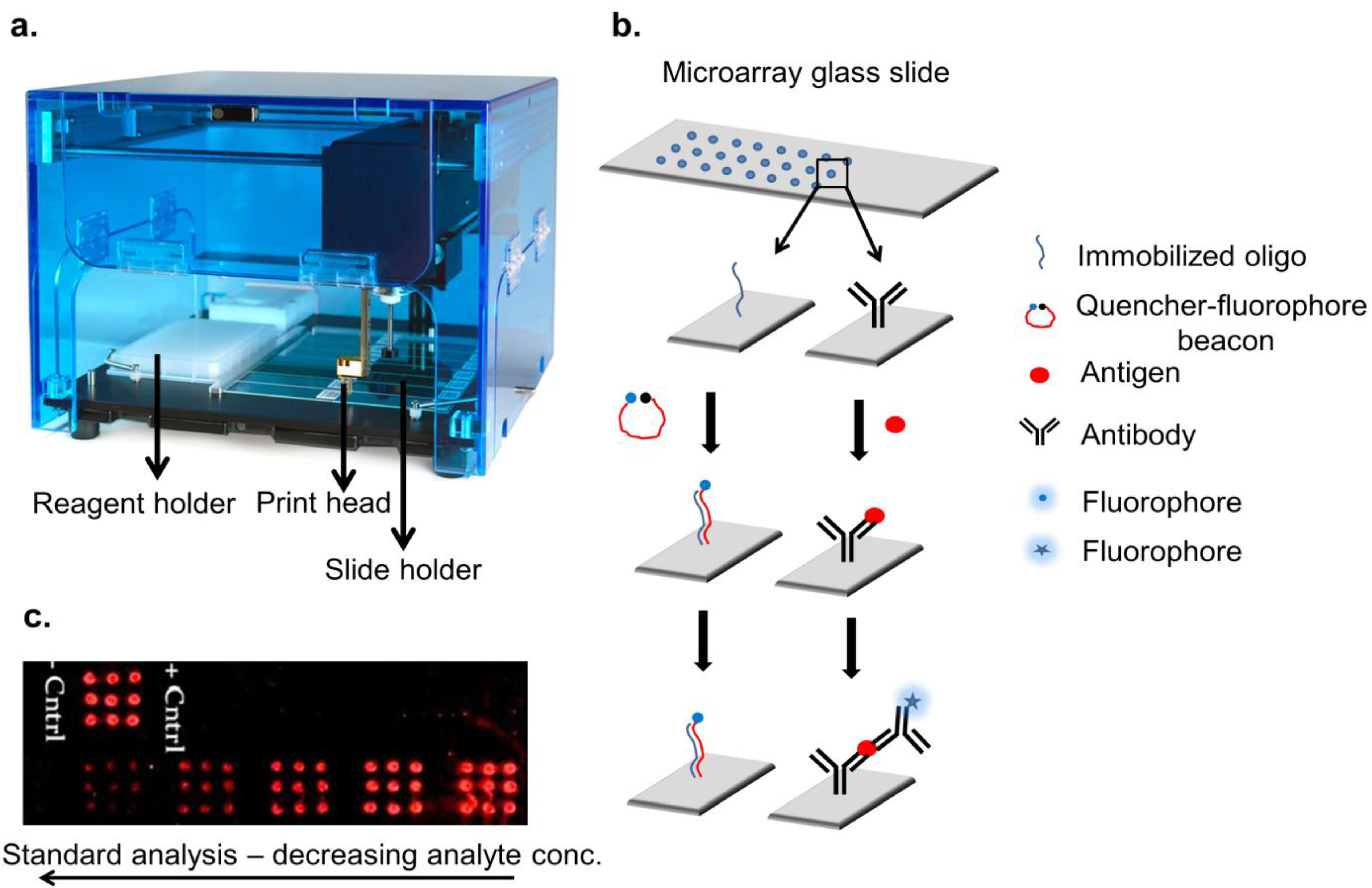
| Type of microarray | Method of development | Format | Density | Diagnostics application | References |
|---|---|---|---|---|---|
| DNA | Printing, in situ synthesis | Oligonucleotide, cDNA | Low-High | Respiratory, digestive tract infections | [17,18,19,20,21] |
| RNA | Printing | miRNA, total RNA | Low-High | Liver diseases, viral miRNA | [22,23,24,25] |
| Protein and Peptide | Printing, stamping | Immunoassays, enzymatic assays, label-free | Moderate | Biomarker discovery, bacterial antigen | [1,14,26,27,28,29,30,31,32,33] |
| Carbohydrate | Stamping, Drop-coating | Lectin assay | Moderate | Cell signaling, Biomarker discovery | [34,35,36,37] |
| Cellular | Droplet-coating | Immunoassays, protein assays, molecular assays | Low-moderate | Biomarker discovery, drug discovery, CTC identification | [38,39,40,41] |
2. Development of a Microarray
| Method | Principle/Description | Print density | Relative cost | References |
|---|---|---|---|---|
| Contact Printing | Reagent spotting on the desired surface either with a thin capillary-like nozzle/tip or with conformal contact of a biomolecule-coated stamp | |||
| Pin-based | A nozzle dispenses solution with a print head; dispensable volume varies (0.5–5 µL); spot sizes varies between 20–200 µm in diameter | Moderate | $$ | [42,43,44] |
| Nanotip-based | An AFM tip dispenses solution onto the surface; dispensable volume varies (0.1–0.3 µL); spot size varies between 30–100 nm in diameter | High | $$$ | [45,46] |
| Microstamping | A polymer cast with specific spot-patterns is dipped in desired protein solution that can replicate these protein spots on any surface by conformal contact | High | $ | [47,48,49,50] |
| Non-contact Printing | Spotting is performed without making a conformal contact with the surface; reagents are delivered either by spraying or localizing under the influence of various fields | |||
| Inkjet-based | A nozzle sprays the 4–8 nL volume of the reagent as a liquid jet; spot size is variable and poor resolution | Moderate | $ | [51,52,53,54,55,56,57] |
| Laser writing | Laser ablation of the thin film generates spatially localized evaporation that creates a confined droplet of sample/reagent placed parallel to the ablation zone; spot size 10–100 µm in diameter. | Moderate | $ | [58,59,60] |
| Solid Support | Inherent Chemical Nature | Functionalization Strategies |
|---|---|---|
| Plastic polymers | ||
| Polystyrene | Hydrophobic | For functionalizing surfaces:
|
| Polymethyl methacrylate | Hydrophobic | |
| Poly carbonate | Hydrophobic | |
| Cyclic poly-olefin | Hydrophobic | |
| Cellulose acetate | Hydrophilic | |
| Non-plastic polymers | ||
| Glass | Hydrophilic | |
| OSTE | Hydrophilic | |
| PDMS | Hydrophobic | |
| Attachment Method | Nature of Surface | Mechanism of Binding | Treatment for Binding | References |
|---|---|---|---|---|
| Adsorption (physisorption) | Hydrophobic (high density) Hydrophilic (low density) |
|
| [62,67] |
| Adsorption (Chemisorption) | Ionic bonds Coordination bonds |
|
| [62,67,68] |
| Covalent, random | Grafted with pendent amine, carboxyl, sulfhydryl, epoxy, and other functionalities |
|
| [62,63,64,67,68] |
| Covalent, site directed | Grafted with pendent amine, sulfhydryl, and carboxyl functionalities |
|
| [62,63,64] |
| Interaction-based oriented binding | Biofunctionalized surface with streptavidin, antibody-binding protein A, G, AG or L, FLAG tag, Ni+2-NTA tag, Enzyme-substrate reaction-mediated |
|
| [62,63,64,66,67,68] |
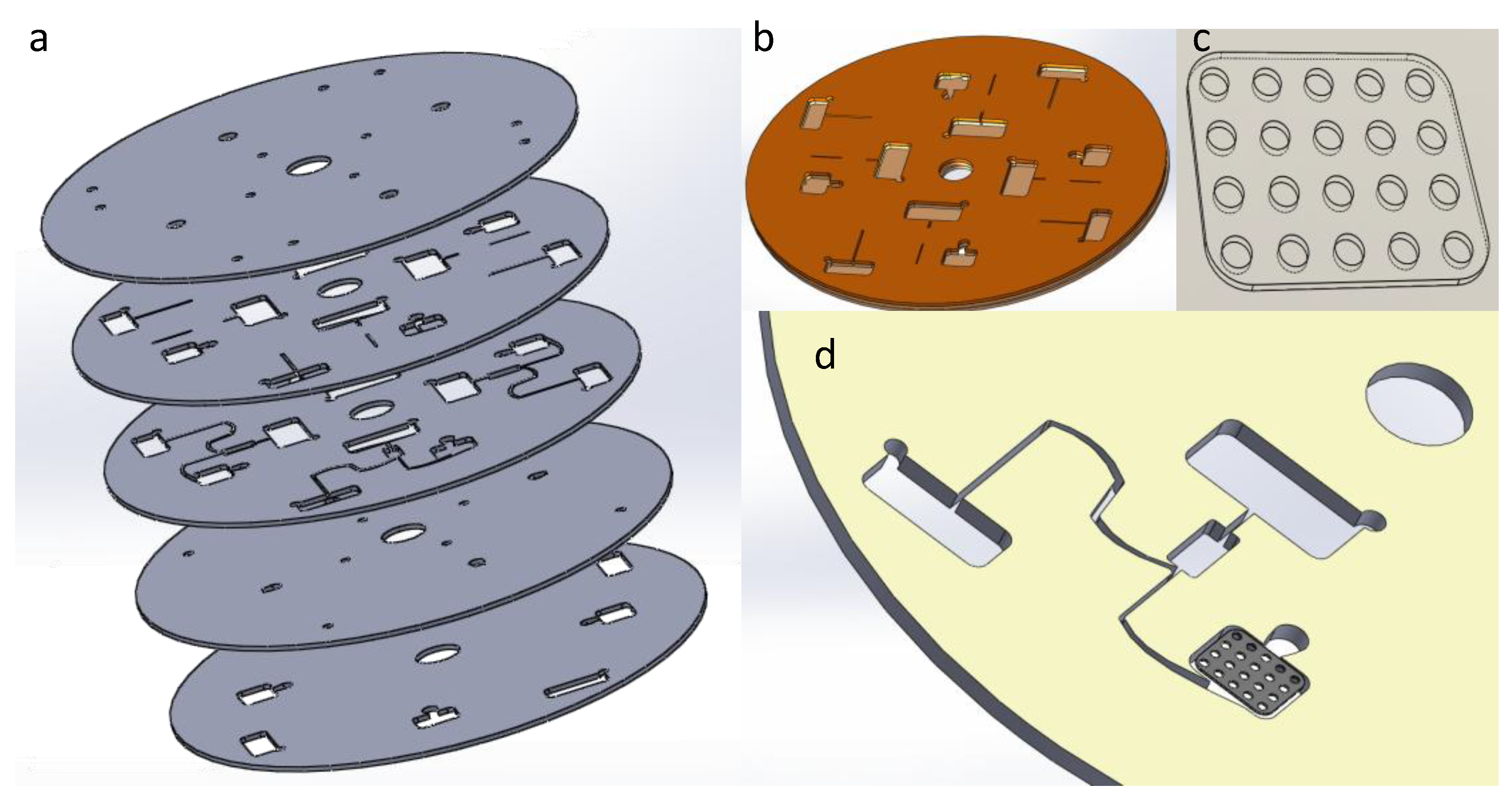
3. Microfluidic Networks (µFN)
3.1. Micromosaics
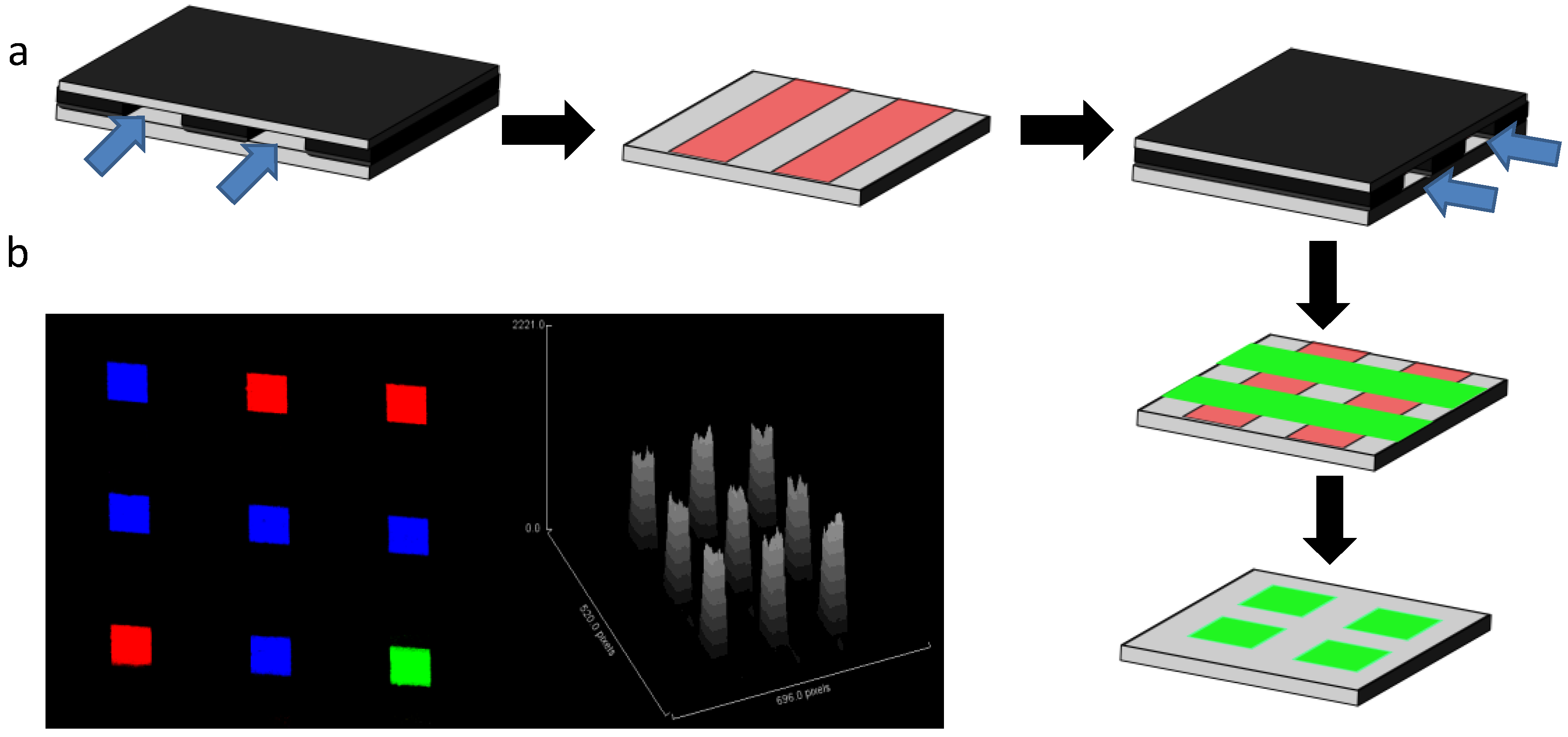
3.2. Other Networks
| Detection Method | Solid Support | Assay Format | Analyte | Sensitivity | Spots/Array (n × n) | Reference |
|---|---|---|---|---|---|---|
| Fluorescence | Silicon | Direct immunoassay | Guinea pig IgG | 6 ng/mL | 25 × 25 | [15] |
| Plasmon | PDMS | Hybridization | RNA/DNA | [87] | ||
| Fluorescence | Glass | Sandwich immunoassay | Bacteria | 6 × 6 | [88] | |
| Fluorescence | PDMS | Direct immunoassay | Gp41; gp120 | [72] | ||
| Fluorescence | PDMS | Sandwich immunoassay | Human TNF | 20 pg/mL | 10 × 17 | [73] |
| Bioluminescence | PDMS | Intracellular signal | Cells | 5 × 5 | [89] | |
| Fluorescence | PDMS | Sandwich immunoassay | C-reactive protein | 30 ng/mL | 7 × 7 | [90] |
| Fluorescence | PDMS | Direct immunoassay | Multiple Bacterial antigens | 1 × 5 | [74] | |
| Fluorescence | PDMS | Direct assay | Cells | 8 × 6 | [91] | |
| Fluorescence | PDMS | Direct immunoassay | Antibodies against bacteria in serum | 6 × 6 | [92] | |
| Fluorescence | Silicon nitride | Sandwich immunoassay | C-reactive protein | 2.5 µg/mL | 3 × 12 | [93] |
| Fluorescence | PDMS | Quantum dot-based sandwich immunoassay | Carcinoma embryonic antigen | 500 fM | 4 × 8 | [94] |
| Fluorescence | PDMS | Sandwich immunoassay | C-reactive protein | 1ng/mL | 5 × 14 | [78] |
| Fluorescence | PDMS | Sandwich immunoassay | Oxidative stress biomarkers 3-nitro tyrosine, Catalase Superoxide dismutase | 150 µM 5 ng/mL 0.5 ng/mL | 3 × 10 | [95] |
| Fluorescence | PVDF, PDMS | Direct immunoassay | 1 × 10 | [96] | ||
| Fluorescence | PDMS | Direct immunoassay | IgG | 5 ng/mL | 1 × 4 | [79] |
| Various | Various | Various | [97] (Patent number: US 8,075,854 B2) | |||
| Fluorescence | Polycarbonate and PDMS | Sandwich immunoassay | Rabbit IgG | 0.16 µM | 6 × 5 | [77] |
| Fluorescence | PDMS | Sandwich immunoassay | Panel of HIV associated antigens | 8 × 21 | [98] | |
| Fluorescence | PDMS | Sandwich immunoassay | Panel of HIV associated antigens | [99] |
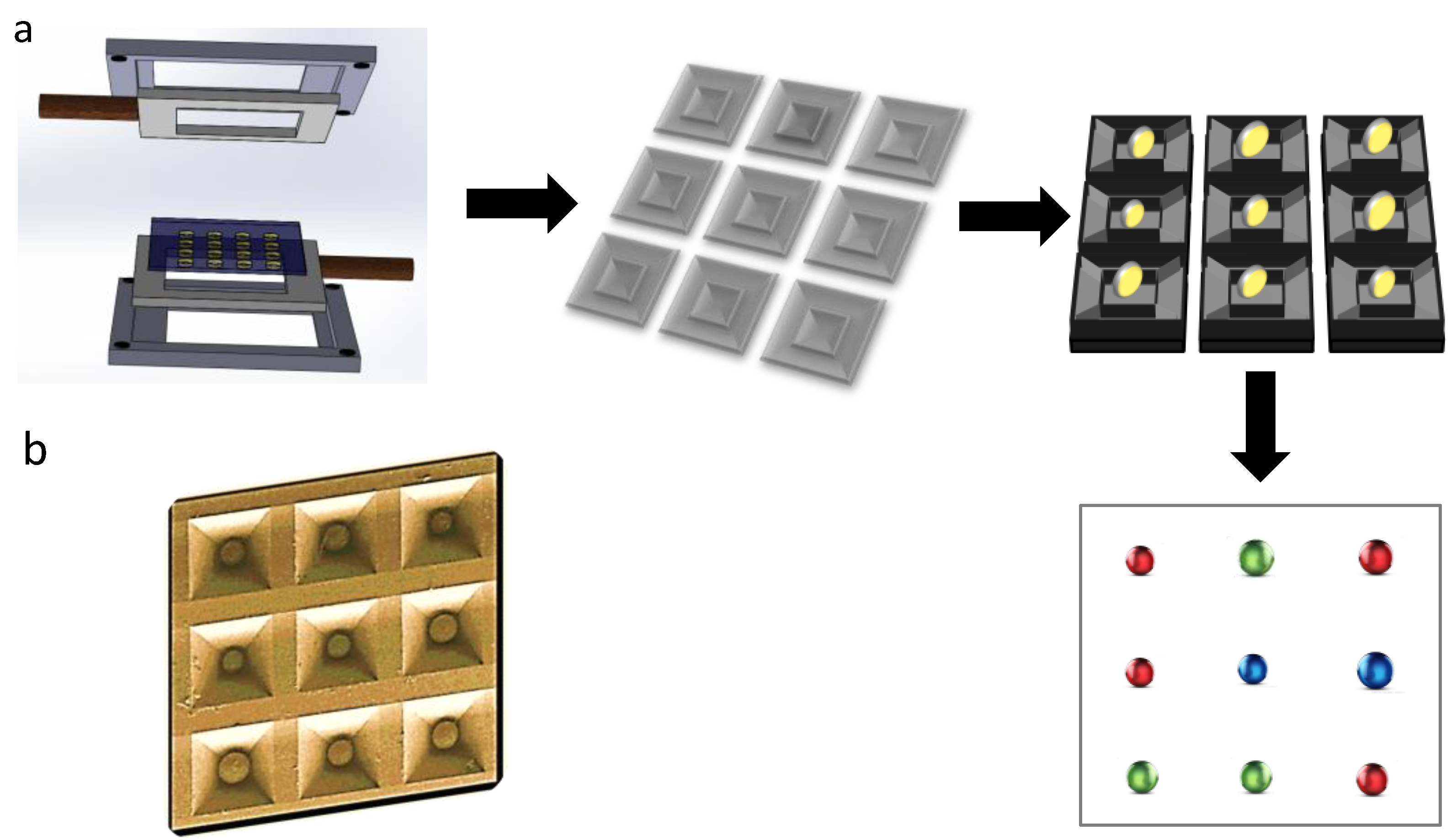
4. Microbeads
5. Perspective
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Desmet, C.; Blum, L.J.; Marquette, C.A. Multiplex microarray ELISA versus classical ELISA, a comparison study of pollutant sensing for environmental analysis. Environ. Sci. Process. Impacts 2013, 15, 1876–1882. [Google Scholar]
- Sun, H.; Chem, G.Y.J.; Yao, S.Q. Recent advances in microarray technology for proteomics. Chem. Biol. 2013, 20, 685–699. [Google Scholar] [CrossRef]
- Miller, M.B.; Tang, Y.W. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 2009, 22, 611–633. [Google Scholar]
- Gonzalez-Gonzalez, M.; Jara-Acevedo, R.; Matarraz, S.; Jara-Acevedo, M.; Paradinas, S.; Sayagues, J.M.; Orfao, A.; Fuentes, M. Nanotechniques in proteomics: Protein microarrays and novel detection platform. Eur. J. Pharm. Sci. 2012, 46, 499–506. [Google Scholar]
- South, S.T.; Lee, C.; Lamb, A.N.; Higgins, A.W.; Kearney, H.M. ACMG standards and guidelines for constitutional cytogenetic microarray analysis including postnatal and prenatal applications: Revision 2013. Genet. Med. 2012, 15, 901–909. [Google Scholar]
- Yu, X.; Schneiderhan-Marra, N.; Joos, T.O. Protein microarrays for personalized medicine. Clin. Chem. 2010, 56, 376–387. [Google Scholar] [CrossRef]
- Sutandy, F.X.R.; Qian, J.; Chen, C.-.S.; Zhu, H. Overview of protein microarrays. Curr. Protoc. Protein Sci. 2013, 72, 1–27. [Google Scholar]
- Koschmieder, A.; Zimmermann, K.; Tril, S.; Stoltmann, T.; Leser, U. Tools for managing and analyzing microarray data. Brief Bioinform. 2012, 13, 46–60. [Google Scholar] [CrossRef]
- Dupuy, A.; Simon, R.M. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J. Natl. Cancer Inst. 2007, 99, 147–157. [Google Scholar]
- Duarte, J.; Serufuri, J.-M.; Mulder, N.; Blackbum, J. Protein function microarrays: design, use and bioinformatic analysis in cancer biomarker discovery and quantitation. Bioinform. Hum. Proteom.: Transl. Bioinfor 2013, 3, 39–74. [Google Scholar] [CrossRef]
- Hartmann, M.; Roeraade, J.; Stoll, D.; Templin, M.F.; Joos, T.O. Protein microarrays for diagnostics assays. Anal. Bioanal. Chem. 2009, 393, 1407–1416. [Google Scholar] [CrossRef]
- Lo, R.C. Application of microfluidic in chemical engineering. Chem. Eng. Process Tech. 2013, 1, 1002–1005. [Google Scholar]
- Wang, T.; Zhang, M.; Dreher, D.D.; Zeng, Y. Ultrasensitive microfluidic solid-phase ELISA using an actuable microwell-patterned PDMS chip. Lab Chip 2013, 13, 4190–4197. [Google Scholar] [CrossRef]
- Thaitrong, N.; Charlermroj, R.; Himananto, O.; Seepiban, C.; Karoonuthaisiri, N. Implementation of microfluidic sandwich ELISA for superior detection of plant pathogens. PLoS One 2013, 8, e83231. [Google Scholar]
- Bernard, A.; Michel, B.; Delamarche, E. Micromosaic immunoassays. Anal. Chem. 2001, 73, 8–12. [Google Scholar] [CrossRef]
- Situma, C.; Hashimoto, M.; Soper, S.A. Merging microfluidics with microarray-based bioassays. Biomol. Eng. 2006, 23, 213–231. [Google Scholar] [CrossRef]
- Lucito, R.; Healy, J.; Alexander, J.; Reiner, A.; Esposito, D.; Chi, M.; Rodgers, L.; Brady, A.; Sebat, J.; Troge, J.; et al. Representational oligonucleotide microarray analysis: A high resolution number to detect genome copy number. Genome Res. 2003, 13, 2291–2305. [Google Scholar] [CrossRef]
- Ji, W.; Zhou, W.; Gregg, K.; Lindpaintner, K.; Davis, S. A method for gene expression analysis by oligonucleotide arrays from minute biological materials. Anal. Biochem. 2004, 331, 329–339. [Google Scholar]
- Guo, L.; Liu, Y.; Bai, Y.; Sun, Y.; Xiao, F.; Guo, Y. Gene expression profiling of drug-resistant small cell lung cancer cells. Eur. J. Cancer 2010, 46, 1692–1702. [Google Scholar] [CrossRef]
- Bae, E.K.; Lee, H.; Lee, J.S.; Noh, E.W. Isolation and characterization of osmotic stress-induced genes in popular cells by suppression subtractive hybridization and cDNA microarray analysis. Plant Physiol. Biochem. 2010, 48, 136–141. [Google Scholar] [CrossRef]
- Donatin, E.; Drancourt, M. DNA microarrays for the diagnosis of infectious diseases. Med. Maladies Infect. 2012, 42, 453–459. [Google Scholar] [CrossRef]
- Kogenaru, S.; Yan, Q.; Guo, Y.; Wang, N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 2012, 13, 629–644. [Google Scholar] [CrossRef]
- Van Roosbroeck, K.; Pollet, J.; Calin, G.A. miRNAs and long non-coding RNAs as biomarkers in human diseases. Expert Rev. Mol. Diag. 2013, 13, 183–204. [Google Scholar] [CrossRef]
- Murakami, Y.; Tanahashi, T. Analysis of circulating microRNA by microarray in liver disease. Methods Mol. Biol. 2013, 1024, 173–182. [Google Scholar]
- Filippone, C.; Marianneau, P.; Murri, S.; Mollard, N.; Avsic-Zupanc, T.; Chinikar, S.; Despres, P.; Caro, V.; Gessain, A.; Berthet, N.; et al. Molecular diagnostic and genetic characterization of highly pathogenic viruses: application during Crimean-Congo haemorrhagic fever virus outbreaks in Eastern Europe and Middle East. Clin. Microbiol. Infect. 2013, 19, E118–E128. [Google Scholar] [CrossRef]
- Hall, D.A.; Ptacek, J.; Snyder, M. Protein microarray technology. Mech Ageing Dev. 2007, 128, 161–167. [Google Scholar] [CrossRef]
- Borrebaeck, C.A.; Wingren, C. Design of high-density microarrays for disease proteomics: Key technological issues. J. Proteomics 2009, 72, 928–935. [Google Scholar] [CrossRef]
- Lee, K.; Shin, J.Y.; Yang, Y.S.; Shin, J.I.; Park, Y.C.; Seo, J.H.; Park, T.H.; Shin, C.S.; Jin, Y.S.; Kweon, D.H. Towards a microarray of functional membrane proteins: assembly of a surface-attachable, membrane-protein-anchored membrane structure using apolipoprotein A1. Enzyme Microb. Technol. 2009, 44, 217–222. [Google Scholar]
- Lian, W.; Wu, D.; Lim, D.V.; Jin, S. Sensitive detection of multiplex toxins using antibody microarray. Anal. Biochem. 2010, 401, 271–279. [Google Scholar] [CrossRef]
- Moschallski, M.; Baader, J.; Prucker, O.; Ruhe, J. Printed protein microarrays on unmodified plastic substrates. Anal. Chim. Acta 2010, 671, 92–98. [Google Scholar] [CrossRef]
- Zhong, L.; Zhang, W.; Zer, C.; Ge, K.; Gao, X.; Kernstine, K.H. Protein microarray: Sensitive and effective immunuodetection for drug residues. BMC Biotechnol. 2010, 10, 12–18. [Google Scholar]
- Gyorgy, A.B.; Walker, J.; Wingo, D.; Eidelman, O.; Pollard, H.B.; Molnar, A.; Agoston, D.V. Reverse phase protein microarray technology in traumatic brain injury. J. Neurosci. Methods 2010, 192, 96–101. [Google Scholar] [CrossRef]
- Liang, L.; Juarez, S.; Nga, T.V.T.; Dunstn, S.; Nakajima-Sasaki, R.; Davies, D.H.; McSorley, S.; Baker, S.; Felgner, P.L. Immune profiling with a Salmonella Typhi antigen microarray identifies new biomarkers of human typhoid. Sci. Rep. 2012, 3, 1043. [Google Scholar] [CrossRef]
- Park, S.; Gildersleeve, J.C.; Blixt, O.; Shin, I. Carbohydrate microarrays. Chem. Soc. Rev. 2013, 42, 4310–4326. [Google Scholar]
- Hirabayashi, J.; Yamada, M.; Kuno, A.; Tateno, H. Lectin microarrays: Concepts, principles, and applications. Chem. Soc. Rev. 2013, 42, 4443–4458. [Google Scholar] [CrossRef]
- Chandler, K.; Goldman, R. Glycoprotein disease markers and single protein omics. Mol. Cell Proteomics 2013, 12, 836–845. [Google Scholar] [CrossRef]
- Kerekgyarto, M.; Fekete, A.; Szurmai, Z.; Kerekgyarto, J.; Takacs, L.; Kurucz, I.; Guttman, A. Neoglycoproteins as carbohydrate antigens: Synthesis, analysis, and polyclonal antibody response. Electrophoresis 2013, 34, 2379–2386. [Google Scholar] [CrossRef]
- Yarmush, M.L.; King, K.R. Living cell microarrays. Annu. Rev. Biomed. Eng. 2009, 11, 235–257. [Google Scholar] [CrossRef]
- Lindquist, R.A.; Ottina, K.A.; Wheeler, D.B.; Hsu, P.P.; Thoreen, C.C.; Guertin, D.A.; Ali, S.M.; Sengupta, S.; Shaul, Y.D.; Lamprecht, M.R.; et al. Genome scale RNAi on living-cell microarrays identifies novel regulators of Drosophila melanogaster TORC1-S6K pathway signaling. Genome Res. 2011, 21, 433–446. [Google Scholar] [CrossRef] [Green Version]
- Rantala, J.K.; Mikela, R.; Aaltola, A.R.; Laasola, P.; Mpindi, J.P.; Nees, M.; Saviranta, P.; Kallioniemi, O. A cell spot microarray method for production of siRNA transfection microarrays. BMC Genomics 2011, 12, 162–175. [Google Scholar] [CrossRef]
- Yamamura, S.; Yatsushiro, S.; Abe, K.; Baba, Y.; Kataoka, M. Development of a cell microarray chip for detection of circulating tumor cells. J. Phys. Conf. Ser. 2012, 352, 012041. [Google Scholar] [CrossRef]
- Martinsky, T.; Haje, P. Microarray Tools, Kits, Reagents and Services. In Microarray Biochip Technology; Schena, M., Ed.; Eaton Publishing: Natick, MA, USA; pp. 201–220.
- MacBeath, G.; Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 2000, 289, 1760–1763. [Google Scholar]
- Weibel, C. The spotting acceleratorTM. J. Assoc. Lab Autom. 2002, 7, 91–96. [Google Scholar] [CrossRef]
- Piner, R.D.; Zhu, J.; Xu, F.; Hong, S.; Mirkin, C.A. Dip-pen nanolithography. Science 1999, 283, 661–663. [Google Scholar] [CrossRef]
- Lenhert, S.; Sun, P.; Wang, Y.; Fuchs, H.; Mirkin, C.A. Massively parallel dip-pen lithography of heterogeneous supported phospholipid multilayer patterns. Small 2007, 3, 71–75. [Google Scholar]
- Kim, E.; Xia, Y.N.; Whitesides, G.M. Polymer microstructures formed by molding in capillaries. Nature 1995, 376, 581–584. [Google Scholar]
- Kane, R.S.; Takayama, S.; Ostuni, E.; Ingber, D.E.; Whitesides, G.M. Patterning cells and proteins using soft lithography. Biomaterials 1999, 20, 2363–2376. [Google Scholar]
- Renault, J.P.; Bernard, A.; Bietsch, A.; Michael, B.; Bosshard, H.R.; Delamarche, E.; Kreiter, M.; Hecht, B.; Wild, U.P. Fabricating arrays of single molecule proteins on glass using microcontact printing. J. Phys. Chem. B 2003, 107, 703–711. [Google Scholar]
- Lin, S.C.; Tseng, F.G.; Huang, H.M.; Chen, Y.F.; Tsai, Y.C.; Ho, C.E.; Chieng, C.C. Simultaneous immobilization of protein microarrays by a micro stamper with a back filling reservoir. Sens. Actuators B 2004, 99, 174–185. [Google Scholar] [CrossRef]
- Okamoto, T.; Suzuki, T.; Yamamoto, N. Microarray fabrication with covalent attachment of DNA using bubble-jet technology. Nat. Biotechnol. 2000, 18, 438–441. [Google Scholar]
- Allain, L.R.; Askari, M.; Stokes, D.L.; Vo-Dinh, T.; Fresenius, J. Microarray sampling-platform fabrication using bubble-jet technology for a biochip system. Anal. Chem. 2001, 371, 146–150. [Google Scholar] [CrossRef]
- Tseng, F.G.; Kim, C.J.; Ho, C.M. A high-resolution high-frequency monolithic top-shooting microinjector free of satellite drops: part I. Concept, design, and model. J. Microelectromech. Syst. 2002, 11, 427–436. [Google Scholar] [CrossRef]
- Allain, L.R.; Stratis-Cullum, D.N.; Vo-Dinh, T. Investigation of microfabrication of biological sample arrays using piezoelectric and bubble-jet printing technologies. Anal. Chim. Acta 2004, 518, 77–85. [Google Scholar] [CrossRef]
- Lausted, C.G.; Warren, C.B.; Hood, L.E. A high-resolution high-frequency monolithic top-shooting microinjector free of satellite drops: Part I concept, design, and model. Methods Enzymol. 2006, 410, 168–189. [Google Scholar] [CrossRef]
- McWilliam, I.; Chong, K.M.; Hall, D. Inkjet printing for the production of protein microarrays. Methods Mol. Biol. 2011, 785, 345–361. [Google Scholar] [CrossRef]
- Yamada, M.; Imaishi, H.; Morigaki, K. Microarrays of phospholipid bilayers generated by inkjet printing. Langmuir 2013, 29, 6404–6408. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Wu, P.K.; Kim, H.; Pique, A.; Auyeung, R.Y.C.; Young, H.D.; Chrisey, D.B.; Krizman, D.B. Picoliter-scale protein microarrays by laser direct write. Biotechnol. Progr. 2002, 18, 1126–1129. [Google Scholar] [CrossRef]
- Tseng, F.G.; Lin, S.C.; Yao, D.J.; Huang, H.; Chieng, C.C. Technological Aspects of Protein Microarrays and Nanoarrays. In Protein Microarrays; Schena, M., Ed.; Jones and Barlett Publishers, Inc.: Sudbury, MA, USA; pp. 305–338.
- Palla-Papavlu, A.; Dinca, V.; Lippert, T.; Dinescu, M. Laser-induced forward transfer for material patterning. Roman. Rep. Phys. 2011, 63, 1285–1301. [Google Scholar]
- Guo, A.; Zhu, X.Y. The Critical Role of Surface Chemistry in Protein Microarray. In Functional Protein Microarrays in Drug Discovery, 1st ed.; Predki, P.F., Ed.; CRC Press: Boca Raton, FL, USA, 2007; pp. 53–71. [Google Scholar]
- Wong, L.S.; Khan, F.; Micklefield, J. Selective covalent protein immobilization: strategies and applications. Chem. Rev. 2009, 109, 4025–4053. [Google Scholar] [CrossRef]
- Makaraviciute, A.; Ramanaviciene, A. Site-directed antibody immobilization techniques for immunosensors. Biosens. Bioelectron. 2013, 50, 460–471. [Google Scholar] [CrossRef]
- Kohn, M. Immobilization strategies for small molecules, peptides and protein microarrays. J. Peptide Sci. 2009, 15, 393–397. [Google Scholar] [CrossRef]
- Dixit, C.K.; Vashist, S.K.; MacCraith, B.D.; O’Kennedy, R. Evaluation of apparent non-specific protein loss due to adsorption on sample tube surfaces and/or altered immunogenicity. Analyst 2011, 136, 1406–1411. [Google Scholar] [CrossRef]
- Lin, P.C.; Weinrich, D.; Waldmann, H. Protein biochips: Oriented surface immobilization of proteins. Macromol. Chem. Phys. 2010, 211, 136–144. [Google Scholar] [CrossRef]
- Dixit, C.K.; Vashist, S.K.; O’Neill, F.T.; O’Reilly, B.; MacCraith, B.D.; O’Kennedy, R. Development of a high-sensitivity rapid sandwich ELISA procedure and its comparison with the conventional approach. Anal. Chem. 2010, 82, 7049–7052. [Google Scholar]
- Vashist, S.K.; Dixit, C.K.; MacCraith, B.D.; O’Kennedy, R. Effect of antibody immobilization strategies on the analytical performance of surface plasmon resonance-based immunoassay. Analyst 2011, 136, 4431–4436. [Google Scholar] [CrossRef]
- Peterson, A.W.; Heaton, R.J.; Georgiadis, R.M. Effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001, 29, 5163–5168. [Google Scholar] [CrossRef]
- Kitsara, M.; Ducree, J. Integration of functional materials and surface modification for polymeric microfluidic systems. J. Micromech. Microeng. 2013, 23, 033001. [Google Scholar] [CrossRef]
- Jiang, X.; Ng, J.M.K.; Stroock, A.D.; Dertinger, S.K.W.; Whitesides, G.M. A miniaturized, parallel, serially diluted immunoassay for analyzing multiple antigens. J. Am. Chem. Soc. 2003, 125, 5294–5295. [Google Scholar] [CrossRef]
- Cesaro-Tadic, S.; Dernick, G.; Juncker, D.; Buurman, G.; Kropshofer, H.; Michel, B.; Fattinger, C.; Delamarche, E. High-sensitivity miniaturized immunoassays for tumor necrosis factor using microfluidic systems. Lab Chip 2004, 4, 563–569. [Google Scholar]
- Wei, C.W.; Cheng, J.Y.; Huang, C.T.; Yen, M.H.; Young, T.H. Using a microfluidic device for 1 microl DNA microarray hybridization in 500s. Nucleic Acids Res. 2005, 33, e78. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, G.; Lin, F.Y.H.; Sherman, P.M.; Li, D. An electrokinetically-controlled immunoassay for simultaneous detection of multiple microbial antigens. Biomed. Microdev. 2005, 7, 301–312. [Google Scholar] [CrossRef]
- Wolf, M.; Zimmermann, M.; Delamarche, E.; Hunziker, P. Screening of cell surface receptors using micromosaic immunoassay. Biomed. Microdev. 2007, 9, 135–141. [Google Scholar] [CrossRef]
- Chueh, B.-H.; Huh, D.; Kyrtsos, C.R.; Houssin, T.; Futai, N.; Takayama, S. Leakage-free bonding of porous membranes into layered microfluidic array systems. Anal. Chem. 2007, 79, 3504–3508. [Google Scholar]
- Liu, Y.; Yu, J.; Du, M.; Wang, W.; Zhang, W.; Wang, Z.; Jiang, X. Accelerating microfluidic immunoassays on filter membranes by applying vacuum. Biomed. Microdev. 2012, 14, 17–23. [Google Scholar] [CrossRef]
- Ziegler, J.; Zimmermann, M.; Hunziker, P.; Delamarche, E. High-performance immunoassays based in through-stencil patterned antibodies and capillary systems. Anal. Chem. 2008, 80, 1763–1769. [Google Scholar] [CrossRef]
- Shao, G.; Wang, J.; Li, Z.-H.; Saraf, L.; Wang, W.; Lin, Y. Poly (dimethysiloxane) microchip-based immunoassay with multiple reaction zones: toward on-chip multiplex detection platform. Sens. Actuators B 2011, 159, 44–50. [Google Scholar] [CrossRef]
- Peytavi, R.; Raymond, F.R.; Gagne, D.; Picard, F.J.; Jia, G.; Zoval, J.; Madou, M.; Boissinot, K.; Boissinot, M.; Bissonnette, L.; et al. Micfludic device for rapid (15 min) automated microarray hybridization. Clin. Chem. 2005, 51, 1836–1844. [Google Scholar] [CrossRef]
- Kartalov, E.P.; Zhong, J.F.; Scherez, A.; Quake, S.R.; Taylor, C.R.; Anderson, W.F. High-throughput multi-antigen microfluidic fluorescence immunoassay. BioTechnique 2006, 40, 85–90. [Google Scholar]
- Kartalov, E.P.; Lin, D.H.; Lee, D.T.; Anderson, W.F.; Taylor, C.R.; Scherer, A. Internally calibrated quantification of protein analyte in human serum by fluorescence immunoassays in disposable elastomeric microfluidic devices. Electrophoresis 2008, 29, 5010–5016. [Google Scholar]
- Yu, L.; Liu, Y.; Gan, Y.; Li, C.M. High-performance UV-curable epoxy resin-based microarray and microfluidic immunoassay devices. Biosens. Bioelectron. 2009, 24, 2997–3002. [Google Scholar] [CrossRef]
- Roy, S.; Soh, J.H.; Gao, Z. A microfluidic-assisted microarray for ultrasensitive detection of miRNA under an optical microscope. Lab Chip 2011, 11, 1886–1894. [Google Scholar] [CrossRef]
- Ahn, C.H.; Choi, J.-W.; Beaucage, G.; Nevin, J.H.; Lee, J.-B.; Puntambekar, A.; Lee, J.Y. Disposable smart lab on a chip for point-of-care clinical diagnostics. IEEE Proc. 2004, 92, 154–173. [Google Scholar] [CrossRef]
- Maoab, X.; Huang, T.J. Microfluidic diagnostics for the developing world. Lab Chip 2012, 12, 1412–1416. [Google Scholar] [CrossRef]
- Lee, H.J.; Goodrich, T.T.; Corn, R.M. Fabricating RNA microarrays RNA-DNA surface ligation chemistry. Anal. Chem. 2001, 73, 5525–5531. [Google Scholar] [CrossRef]
- Taitt, C.R.; Anderson, G.P.; Lingerfelt, B.M.; Feldstein, M.J.; Ligler, F.S. Nine-analyte detection using an array-based biosensor. Anal. Chem. 2002, 74, 6114–6120. [Google Scholar] [CrossRef]
- Tani, H.; Maehana, K.; Kamidate, T. Chip-based bioassay using bacterial sensor strains immobilized in three-dimensional microfluidic network. Anal. Chem. 2004, 76, 6693–6697. [Google Scholar] [CrossRef]
- Wolf, M.; Juncker, D.; Michel, B.; Hunziker, P.; Delamarche, E. Simultaneous detection of C-reactive protein and other cardiac marker in human plasma using micromosaic immunoassays and self-regulating microfluidic networks. Biosens. Bioelectron. 2004, 19, 1193–1202. [Google Scholar] [CrossRef]
- Chueh, B.-H.; Huh, D.; Kyrtsos, C.R.; Houssin, T.; Futai, N.; Takayama, S. Arrays of horizontally-oriented mini reservoirs generate steady microfluidic flows for continuous cell culture and gradient. Analyst 2004, 129, 1026–1031. [Google Scholar] [CrossRef]
- Gao, Y.; Sherman, P.M.; Sun, Y.; Li, D. Multiplexed high-throughput electrokinetically-controlled immunoassay for the detection of specific bacterial antibodies in human serum. Anal. Chim. Acta 2008, 606, 98–107. [Google Scholar] [CrossRef]
- Murphy, B.M.; He, X.; Dandy, D.; Henry, C.S. Competitive immunoassay methods for detection of metabolites and proteins using micromosaic patterning. Anal. Chem. 2008, 80, 444–450. [Google Scholar] [CrossRef]
- Yan, J.; Hu, M.; Li, D.; He, Y.; Zhao, R.; Jiang, X.; Song, S.; Wang, L.; Fan, C. A nano- and micro- integrated protein chip based on quantum dot probes and microfluidic network. Nano Res. 2008, 1, 490–496. [Google Scholar] [CrossRef]
- Murphy, B.M.; Dandy, D.S.; Henry, C.S. Analysis of oxidative stress biomarkers using a simultaneous competitive/non-competitive micromosiac immunoassay. Anal. Chim. Acta 2009, 640, 1–6. [Google Scholar] [CrossRef]
- Pan, W.; Chen, W.; Jiang, X. Microfluidic western blot. Anal. Chem. 2010, 82, 3974–3976. [Google Scholar] [CrossRef]
- Yiang, S.T.; Bi, Y.; Huang, W.C. Microfluidic Chips for Multiplexed Rapid ELISA. US Patent 8075854, 2011. [Google Scholar]
- Song, L.; Zhang, Y.; Wang, W.; Ma, L.; Liu, Y.; Hao, Y.; Shao, Y.; Zhang, W.; Jiang, X. Microfluidic assay without blocking for rapid HIV screening and confirmation. Biomed. Microdev. 2012, 14, 631–640. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Song, L.; Xu, C.; Ma, L.; Li, Z.; Xi, J.; Jiang, X. Matrix-localization for fast analysis of arrayed microfluidic immunoassays. Anal. Methods 2012, 4, 3466–3470. [Google Scholar] [CrossRef]
- Burger, R.; Ducree, J. Handling and analysis of cells and bioparticles on centrifugal microfluidic platform. Expert Rev. Mol. Diagn. 2012, 12, 407–421. [Google Scholar] [CrossRef]
- Bynum, M.A.; Gordon, G.B. Hybridization enhancement using microfluidic planetary centrifugal mixing. Anal Chem. 2004, 76, 7039–7044. [Google Scholar] [CrossRef]
- Noroozi, Z.; Kido, H.; Peytavi, R.; Nakajima-Sasaki, R.; Jasinskas, A.; Micic, M.; Felgner, P.L.; Madou, M.J. A multiplexed immunoassay system based upon reciprocating centrifugal microfluidic. Rev. Sci. Instrum. 2011, 82, 064303. [Google Scholar]
- Goodey, A.; Lavigne, J.J.; Savoy, S.M.; Rodriguez, M.D.; Curey, T.; Tsao, A.; Simmons, G.; Wright, J.; Yoo, S.-J.; Sohn, Y.; et al. Development of multianalyte sensor arrays of chemically derivatized polymeric microspheres localized in micromachined cavities. J. Am. Chem. Soc. 2001, 123, 2559–2570. [Google Scholar] [CrossRef]
- Christodoulides, N.; Tran, M.; Floriano, P.N.; Rodriguez, M.; Goodey, A.; Ali, M.; Neikirk, D.; McDevitt, J.T. A multichip-based multianalyte assay system for the assessment of cardiac risk. Anal. Chem. 2002, 74, 3030–3036. [Google Scholar] [CrossRef]
- Ali, M.F.; Kirby, R.; Goodey, A.P.; Rodrigues, M.D.; Ellington, A.D.; Neikirk, D.P.; McDevitt, J.T. DNA hybridization and discrimination of single nucleotide mismatches using chip-based microbead arrays. Anal. Chem. 2003, 75, 4732–4739. [Google Scholar] [CrossRef]
- Barbee, K.D.; Hsiao, A.P.; Roller, E.E.; Huang, X. Multiplexed protein detection using antibody-conjugated microbead arrays in a microfabricated electrophoretic device. Lab Chip 2010, 10, 3084–3093. [Google Scholar] [CrossRef]
- Yu, X.; Hartmann, M.; Wang, Q.; Poetz, O.; Schneiderhan-Marra, N.; Stoll, D.; Kazmaier, C.; Joos, T.O. µFBI: A microfluidic bead-based immunoassay for multiplexed detection of proteins from a µL sample volume. PLoS One 2010, 5, e13125. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, T.; Li, C.-.W.; Yang, M. A microfluidic device with microbead array for sensitive virus detection and genotyping using quantum dots as fluorescent labels. Biosens. Bioelectron. 2010, 25, 2402–2407. [Google Scholar] [CrossRef]
- Shirtcliffe, N.J.; Toon, R.; Roach, P. Surface treatment for microfluidic biocompatibility. Methods Mol. Biol. 2013, 949, 241–268. [Google Scholar] [CrossRef]
- Sin, M.L.Y.; Gau, V.; Liao, J.C.; Wong, P.K. A universal electrode approach for automated electrochemical molecular analysis. IEEE Nanotechnol. Mag. 2013, 7, 31–37. [Google Scholar]
- Johnson, A.S.; Anderson, K.B.; Halpin, S.T.; Kirkpatrick, D.C.; Spence, D.M.; Martin, R.S. Integration of multiple components in polystyrene-based microfluidic devices part I: Fabrication and characterization. Analyst 2013, 138, 129–136. [Google Scholar]
- Paguirigan, A.L.; Beebe, D.J. Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. BioEssays 2008, 30, 811–821. [Google Scholar] [CrossRef]
- Fang, C.; Wang, Y.; Vu, N.T.; Lin, W.-Y.; Hseih, Y.-T.; Rubbi, L.; Phelps, M.E.; Muschen, M.; Kim, Y.-M.; Chatziioannou, A.F.; et al. Integrated microfluidic and imaging platform for a kinase activity radioassay to analyze minute patient cancer samples. Cancer Res. 2010, 70, 8299–8308. [Google Scholar]
- Cooksey, G.A.; Atencia, J.; Forry, S.P. Measurement and validation of cell-based assays with microfluidics at the National Institute of Standards and Technology. Bioanalysis 2012, 4, 1849–1854. [Google Scholar] [CrossRef]
- Hughes, A.J.; Herr, A.E. Microfluidic western blotting. Proc. Nat. Am. Soc. 2012, 109, 21450–21455. [Google Scholar]
- Kim, D.; Herr, A.E. Protein immobilization techniques for microfluidic assays. Biomicrofluidics 2013, 7, 041501. [Google Scholar] [CrossRef]
- Pla-Roca, M.; Juncker, D. PDMS microfluidic capillary systems for patterning proteins on surfaces and performing miniaturized immunoassays. Methods Mol. Biol. Biol. Microarrays 2011, 671, 177–194. [Google Scholar] [CrossRef]
- Wellhausen, R.; Seitz, H. Facing current quantification challenges in protein microarrays. J. Biomed. Biotechnol. 2012, 2012, 831347. [Google Scholar]
- Karsenty, M.; Rubin, S.; Bercovici, M. Acceleration of surface-based hybridization reactions using isotachophoretic focusing. Anal. Chem. 2014, 86, 3028–3036. [Google Scholar] [CrossRef]
- Dixit, C.K.; Kaushik, A. Nano-structured arrays for multiplex analyses and lab-on-a-chip applications. Biochem. Biophys. Res. Commun. 2012, 419, 316–320. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dixit, C.K.; Aguirre, G.R. Protein Microarrays with Novel Microfluidic Methods: Current Advances. Microarrays 2014, 3, 180-202. https://doi.org/10.3390/microarrays3030180
Dixit CK, Aguirre GR. Protein Microarrays with Novel Microfluidic Methods: Current Advances. Microarrays. 2014; 3(3):180-202. https://doi.org/10.3390/microarrays3030180
Chicago/Turabian StyleDixit, Chandra K., and Gerson R. Aguirre. 2014. "Protein Microarrays with Novel Microfluidic Methods: Current Advances" Microarrays 3, no. 3: 180-202. https://doi.org/10.3390/microarrays3030180
APA StyleDixit, C. K., & Aguirre, G. R. (2014). Protein Microarrays with Novel Microfluidic Methods: Current Advances. Microarrays, 3(3), 180-202. https://doi.org/10.3390/microarrays3030180





