Vitamin D Receptor Gene Polymorphisms Associated with Childhood Autism
Abstract
1. Introduction
2. Materials and Methods
2.1. Control and Patient Characteristics
2.2. Polymorphism of VDR Genes in ASD and non-ASD Children with Autism
2.3. Vitamin D3 Concentration
2.4. Statistical Analysis
3. Results
3.1. Polymorphism of VDR Genes in non-ASD Control and ASD Children
3.2. Vitamin D3 Concentration
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- American Psychiatric Association (APA). Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 2000. [Google Scholar]
- Fombonne, E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009, 65, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Piskorz-Ogórek, K.; Ogórek, S.; Cieślińska, A.; Kostyra, E. Autism in Poland in comparison to other countries. Pol. Ann. Med. 2015, 22, 35–40. [Google Scholar] [CrossRef]
- Goines, P.; Van de Water, J. The immune system’s role in the biology of autism. Curr. Opin. Neurol. 2010, 23, 111. [Google Scholar] [CrossRef] [PubMed]
- Giulivi, C.; Zhang, Y.F.; Omanska-Klusek, A.; Ross-Inta, C.; Wong, S.; Hertz-Picciotto, I. Mitochondrial dysfunction in autism. JAMA 2010, 304, 2389–2396. [Google Scholar] [CrossRef] [PubMed]
- Buite, T. The relationship of autism and gluten. Clin. Ther. 2013, 35, 578–583. [Google Scholar]
- Muhle, R.; Trentacoste, S.V.; Rapin, I. The genetics of autism. Pediatrics 2004, 113, 472–486. [Google Scholar] [CrossRef]
- Heil, K.M.; Schaaf, C.P. The genetics of autism spectrum disorders—A guide for clinicians. Curr. Psychiatry Rep. 2013, 15, 334. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.; Feron, F.; Eyles, D. Vitamin D: The neglected neurosteroid? Trends Neurosci. 2001, 24, 570–572. [Google Scholar] [CrossRef]
- Garcion, E.; Wion-Barbot, N.; Montero-Menei, C.N.; Berger, F.; Wion, D. New clues about vitamin D functions in the nervous system. Trends Endocrinol. Metab. 2002, 13, 100–105. [Google Scholar] [CrossRef]
- Cannell, J.J. Autism and vitamin D. Med. Hypotheses 2008, 70, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Kočovská, E.; Fernell, E.; Billstedt, E.; Minnis, H.; Gillberg, C. Vitamin D and autism: Clinical review. Res. Dev. Disabil. 2012, 33, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Noriega, D.B.; Savelkoul, H.F. Immune dysregulation in autism spectrum disorder. Eur. J. Pediatr. 2014, 173, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Meguid, N.A.; Hashish, A.F.; Anwar, M.; Sidhom, G. Reduced serum levels of 25-hydroxy and 1,25-dihydroxy vitamin D in Egyptian children with autism. J. Altern. Complement. Med. 2010, 16, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, A.J.; Holt, B.J.; Serralha, M.; Holt, P.G.; Hart, P.H.; Kusel, M.M. Maternal vitamin D levels and the autism phenotype among offspring. J. Autism Devl. Disord. 2013, 43, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, molecular mechanism of action, and pleiotropic effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef] [PubMed]
- Haussler, M.R.; Whitfield, G.K.; Haussler, C.A.; Hsieh, J.C.; Thompson, P.D.; Selznick, S.H.; Dominguez, C.E.; Jurutka, P.W. The Nuclear Vitamin D Receptor: Biological and Molecular Regulatory Properties Revealed. J. Bone Miner. Res. 1998, 13, 325–349. [Google Scholar] [PubMed]
- Mehta, R.G.; Mehta, R.R. Vitamin D and cancer. J. Nutr. Biochem. 2002, 13, 252–264. [Google Scholar] [CrossRef]
- Mathieu, C.; van Etten, E.; Decallonne, B.; Guilietti, A.; Gysemans, C.; Bouillon, R.; Overbergh, L. Vitamin D and 1,25-dihydroxyvitamin D3 as modulators in the immune system. J. Steroid Biochem. Mol. Biol. 2004, 89, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Valdivielso, J.M.; Fernandez, E. Vitamin D receptor polymorphisms and diseases. Clin. Chim. Acta 2006, 1-2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, I.; Stein, D.G. Progesterone as a neuroprotective factor in traumatic and ischemic brain injury. Prog. Brain Res. 2009, 175, 219–237. [Google Scholar] [PubMed]
- Ramagopalan, S.V.; Heger, A.; Berlanga, A.J.; Maugeri, N.J.; Lincoln, M.R.; Burrell, A.; Handunnetthi, L.; Handel, A.E.; Disanto, G.; Orton, S.M. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010, 20, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Sigmundsdottir, H. From the bench to the clinic: New aspects on immunoregulation by vitamin D analogs. Dermatoendocrinology 2011, 3, 187–192. [Google Scholar] [CrossRef]
- Harms, L.R.; Burne, T.H.; Eyles, D.W.; McGrath, J.J. Vitamin D and the brain. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Eyles, D.W.; Smith, S.; Kinobe, R.; Hewison, M.; McGrath, J.J. Distribution of the vitamin D receptor and 1 alpha-hydroxylase in human brain. J. Chem. Neuroanat. 2005, 29, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Eyles, D.W.; Burne, T.H.; McGrath, J.J. Vitamin D, effects on brain development, adult brain function and the links between low levels of vitamin D and neuropsychiatric disease. Front. Neuroendocrinol. 2013, 34, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon., P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Kalueff, A.V.; Lou, Y.R.; Laaksi, I.; Tuohimaa, P. Increased grooming behavior in mice lacking vitamin D receptors. Physiol. Behav. 2004, 82, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Kalueff, A.V.; Keisala, T.; Minasyan, A.; Kuuslahti, M.; Miettinen, S.; Tuohimaa, P. Behavioural anomalies in mice evoked by “Tokyo” disruption of the vitamin D receptor gene. Neurosci. Res. 2006, 54, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Minasyan, A.; Keisala, T.; Zhang, Y.; Wang, J.H.; Lou, Y.R. Progressive hearing loss in mice with a mutated vitamin D receptor gene. Audiol. Neurotol. 2008, 13, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Faraco, J.H.; Morrison, N.A.; Baker, A.; Shine, J.; Frossard, P.M. ApaI dimorphism at the human vitamin D receptor gene locus. Nucleic Acids Res. 1989, 17, 2150. [Google Scholar] [CrossRef] [PubMed]
- Morrison, N.A.; Yeoman, R.; Kelly, P.J.; Eisman, J.A. Contribution of trans-acting factor alleles to normal physiological variability: Vitamin D receptor gene polymorphism and circulating osteocalcin. Proc. Natl. Acad. Sci. USA 1992, 89, 6665–6669. [Google Scholar] [CrossRef] [PubMed]
- Morrison, N.A.; Qi, J.C.; Tokita, A.; Kelly, P.J.; Crofts, L.; Nguyen, T.V.; Sambrook, P.N.; Eisman, J.A. Prediction of bone density from vitamin D receptor alleles. Nature 1994, 367, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Saijo, T.; Ito, M.; Takeda, E.; Huq, A.H.; Naito, E.; Yokota, I.; Sone, T.; Pike, J.W.; Kuroda, Y. A unique mutation in the vitamin D receptor gene in three Japanese patients with vitamin D-dependent rickets type II: Utility of single-strand conformation polymorphism analysis for heterozygous carrier detection. Am. J. Hum. Genet. 1991, 49, 668–673. [Google Scholar] [PubMed]
- Gross, C.; Eccleshall, T.R.; Malloy, P.J.; Villa, M.L.; Marcus, R.; Feldman, D. The presence of a polymorphism at the translation initiation site of the vitamin D receptor gene is associated with low bone mineral density in postmenopausal Mexican-American women. J. Bone Miner. Res. 1996, 11, 1850–1855. [Google Scholar] [CrossRef] [PubMed]
- Uitterlinden, A.G.; Fang, Y.; Van Meurs, J.B.; Pols, H.A.; Van Leeuwen, J.P. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Raby, B.A.; Lazarus, R.; Silverman, E.K.; Lake, S.; Lange, C.; Wjst, M.; Weiss, S.T. Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am. J. Respir. Crit. Care Med. 2004, 170, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.H.; Laprise, C.; Lemire, M.; Montpetit, A.; Sinnett, D.; Schurr, E.; Hudson, T.J. Association of vitamin D receptor genetic variants with susceptibility to asthma and atopy. Am. J. Respir. Crit. Care Med. 2004, 170, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Saadi, A.; Gao, G.; Li, H.; Wei, C.; Gong, Y.; Liu, Q. Association study between vitamin D receptor gene polymorphisms and asthma in the Chinese Han population: A case-control study. BMC Med. Genet. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Heine, G.; Hoefer, N.; Franke, A.; Nothling, U.; Schumann, R.R.; Hamann, L.; Worm, M. Association of vitamin D receptor gene polymorphisms with severe atopic dermatitis in adults. Br. J. Dermatol. 2013, 168, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Maalmi, H.; Sassi, F.H.; Berraies, A.; Ammara, J.; Hamzaoui, K.; Hamzaoui, A. Association of vitamin D receptor gene polymorphisms with susceptibility to asthma in Tunisian children: A case control study. Hum. Immunol. 2013, 74, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Knekt, P.; Kilkkinen, A.; Rissanen, H.; Marniemi, J.; Sääksjärvi, K.; Heliövaara, M. Serum vitamin D and the risk of Parkinson disease. Arch. Neurol. 2010, 67, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, Y.I.; Song, C.; Yoon, I.; Park, J.W.; Choi, Y.B.; Kim, H.T.; Lee, K.S. Association of vitamin D receptor gene polymorphism and Parkinson’s disease in Koreans. J. Korean Med. Sci. 2005, 20, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Maalmi, H.; Berraïes, A.; Tangour, E.; Ammar, J.; Abid, H.; Hamzaoui, K.; Hamzaoui, A. The impact of vitamin D deficiency on immune T cells in asthmatic children: A case-control study. J. Asthma Allergy 2012, 5, 11–19. [Google Scholar] [PubMed]
- Pani, M.A.; Knapp, M.; Donner, H.; Braun, J.; Baur, M.P.; Usadel, K.H.; Badenhoop, K. Vitamin D receptor allele combinations influence genetic susceptibility to type 1 diabetes in Germans. Diabetes 2000, 49, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Grafodatskaya, D.; Chung, B.; Szatmari, P.; Weksberg, R. Autism spectrum disorders and epigenetics. J. Am. Acad. Child Adolesct. Psychiatry 2010, 49, 794–809. [Google Scholar] [CrossRef] [PubMed]
- Nowotny, P.; Kwon, J.M.; Goate, A.M. SNP analysis to dissect human traits. Curr. Opin. Neurobiol. 2001, 11, 637–641. [Google Scholar] [CrossRef]
- Dealberto, M.J. Prevalence of autism according to maternal immigrant status and ethnic origin. Acta Psychiatr. Scand. 2011, 123, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Gardener, H.; Spiegelman, D.; Buka, S.L. Prenatal risk factors for autism: Comprehensive meta-analysis. Br. J. Psychiatry 2009, 195, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.G. Autism, immune dysfunction and Vitamin D. Acta Psychiatr. Scand. 2011, 124, 74. [Google Scholar] [CrossRef] [PubMed]
- Eyles, D.; Burne, T.; McGrath, J. Vitamin D in fetal brain development. Semin. Cell Dev. Biol. 2011, 22, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.P.; Myers, S.M. Identification and evaluation of children with autism spectrum disorders. Pediatrics 2007, 120, 1183–1215. [Google Scholar] [CrossRef] [PubMed]
- Coşkun, S.; Şimşek, Ş.; Camkurt, M.A.; Çim, A.; Çelik, S.B. Association of polymorphisms in the vitamin D receptor gene and serum 25-hydroxyvitamin D levels in children with autism spectrum disorder. Gene 2016, 588, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Zhu, W.Y.; He, X.; Tang, M.M.; Dang, R.L.; Li, H.D.; Xue, Y.; Zhang, L.H.; Wu, Y.Q.; Cao, L.J. Association between vitamin D receptor gene polymorphisms with childhood temporal lobe epilepsy. Int. J. Environ. Res. Public Health 2015, 12, 13913–13922. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.L.; Lin, X.X.; Guo, M.D.; Zhang, D.G.; Zheng, S.Z.; Jiang, L.J.; Jin, J.; Lin, X.Q.; Ding, R.; Jiang, Y. Association of vitamin D receptor gene polymorphisms and serum 25-hydroxyvitamin D levels with Crohn’s disease in Chinese patients. J. Gastroenterol. Hepatol. 2016, 31, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Albanes, I.J.; Berndt, S.I.; Peters, U.; Chatteriee, N.; Freedman, N.D.; Abnet, C.C.; Huang, W.Y.; Kibel, A.S.; Crawford, E.D.; et al. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis 2009, 30, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Orton, S.M.; Morris, A.P.; Herrera, B.M.; Ramagopalan, S.V.; Lincoln, M.R.; Chao, M.J.; Vieth, R.; Sadovnick, A.D.; Ebers, G.C. Evidence for genetic regulation of vitamin D status in twins with multiplesclerosis. Am. J. Clin. Nutr. 2008, 88, 441–447. [Google Scholar] [PubMed]
- Eyles, D.; Brown, J.; Mackay-Sim, A.; McGrath, J.; Feron, F. Vitamin D3 and brain development. Neuroscience 2003, 118, 641–653. [Google Scholar] [CrossRef]
- Klepstad, P.; Rakvag, T.T.; Kaasa, S.; Holthe, M.; Dale, O.; Borchgrevink, P.C.; Baar, C.; Vikan, T.; Krokan, H.E.; Skorpen, F. The 118A/G polymorphism in the human micro-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol. Scand. 2004, 48, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Mura, E.; Govoni, S.; Racchi, M.; Carossa, V.; Ranzani, G.N.; Allegri, M.; van Schaik, R.H. Consequences of the 118A>G polymorphism in the OPRM1 gene: Translation from bench to bedside? J. Pain Res. 2013, 6, 331–353. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Miyamoto, K.; Taketani, Y.; Yamamoto, H.; Iemori, Y.; Morita, K.; Tonai, T.; Nishisho, T.; Mori, S.; Takeda, E. A vitamin D receptor gene polymorphism in the translation initiation codon: Effect on protein activity and relation to bone mineral density in Japanese women. J. Bone Miner. Res. 1997, 12, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.K.; Miao, D.; Bolivar, I.; Li, J.; Huo, R.; Hendy, G.N.; Goltzman, D. Inactivation of the 25-hydroxyvitamin D 1alpha-hydroxylase and vitamin D receptor demonstrates independent and interdependent effects of calcium and vitamin D on skeletal and mineral homeostasis. J. Biol. Chem. 2004, 279, 16754–16766. [Google Scholar] [CrossRef] [PubMed]
- Bikle, D.D. Vitamin D metabolism, mechanism of action, and clinical applications. Chem. Biol. 2014, 21, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Cieślińska, A.; Sienkiewicz-Szłapka, E.; Wasilewska, J.; Fiedorowicz, E.; Chwała, B.; Moszyńska-Dumara, M.; Cieśliński, T.; Bukało, M.; Kostyra, E. Influence of candidate polymorphisms on the dipeptidyl peptidase IV and μ-opioid receptor genes expression in aspect of the β-casomorphin-7 modulation functions in autism. Peptides 2015, 65, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Hollis, B.W. Vitamin D requirement during pregnancy and lactation. J. Bone Miner. Res. 2007, 22, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Hollis, B.W.; Wagner, C.L. Vitamin D deficiency during pregnancy: An ongoing epidemic. Am. J. Clin. Nutr. 2006, 84, 273. [Google Scholar] [PubMed]
- Johnson, D.D.; Wagner, C.L.; Hulsey, T.C.; McNeil, R.B.; Ebeling, M.; Hollis, B.W. Vitamin D deficiency and insufficiency is common during pregnancy. Am. J. Perinatol. 2011, 28, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. Photosynthesis of vitamin D in the skin: Effect of environmental and life-style variables. Fed. Proc. 1987, 46, 1876–1882. [Google Scholar] [PubMed]
- Cieslinska, A.; Simmelink, J.; Teodorowicz, G.; Verhoef, H.; Tobi, H.; Savelkoul, H.F. Distribution of month of birth of individuals with autism spectrum disorder differs from the general population in the Netherlands. In Autism—Paradigms, Recent Research and Clinical Applications; Fitzgerald, M., Jane Yip, J., Eds.; InTech: Rijeka, Croatia, 2017; Chapter 4; pp. 43–54. [Google Scholar]
- Humble, M.B.; Gustafsson, S.; Bejerot, S. Low serum levels of 25-hydroxyvitamin D (25-OHD) among psychiatric out-patients in Sweden: Relations with season, age, ethnic origin and psychiatric diagnosis. J. Steroid Biochem. Mol. Biol. 2010, 121, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Leventhal, B.L.; Koh, Y.J.; Fombonne, E.; Laska, E.; Lim, E.C.; Cheon, K.A.; Kim, S.J.; Kim, Y.K.; Lee, H.; et al. Prevalence of autism spectrum disorders in a total population sample. Am. J. Psychiatry 2011, 168, 904–912. [Google Scholar] [CrossRef] [PubMed]
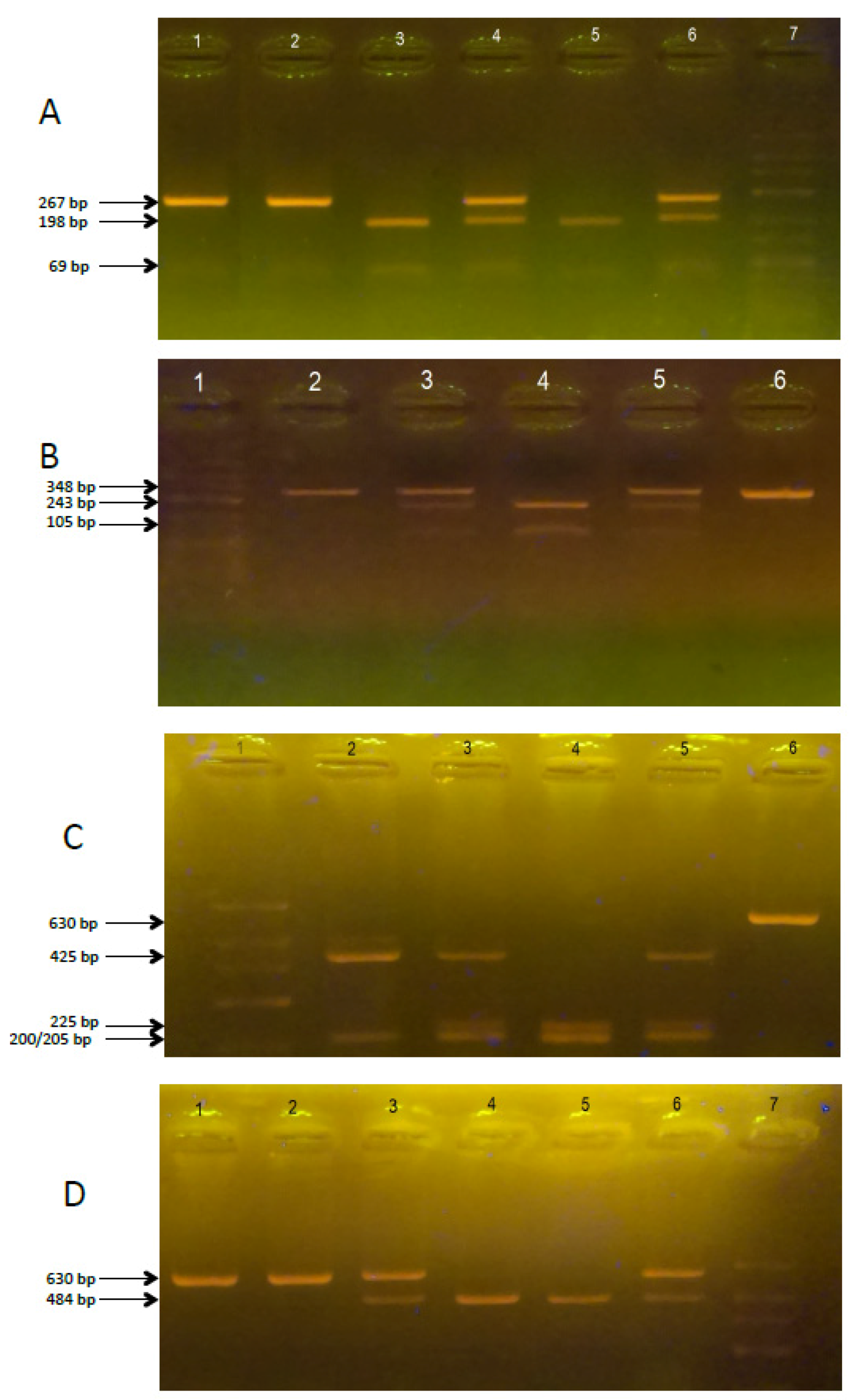
| SNP | Primer Sequence | Restriction Enzyme | PCR/RFLP Products (bp) |
|---|---|---|---|
| Fok-I | Fok1R: 5-ATGGAAACACCTTGCTTCTTCTCCCTC-3 Fok11F: 5-AGCTGGCCCTGGCACTGACTCtGGCTCT-3 | FastDigest Fok-I | ff: 198, 69 FF: 267 Ff: 267, 198, 69 PCR product: 267 |
| Bsm-I | ABsm1F: 5-CGGGGAGTATGAAGGACAAA-3 ABsm1R: 5-CCATCTCTCAGGCTCCAAAG-3 | FastDigest Bsm-I | bb: 243, 105 BB: 348 Bb: 348, 243, 105 PCR product: 348 |
| Taq-I | ATaq1F: 5-GGATCCTAAATGCACGGAGA-3 ATaq1R: 5-AGGAAAGGGGTTAGGTTGGA-3 | FastDigest Taq-I | tt: 225, 200, 205 TT: 425, 205 Tt: 425, 225, 200, 205 PCR product: 630 |
| Apa-I | ATaq1F: 5-GGATCCTAAATGCACGGAGA-3 ATaq1R: 5-AGGAAAGGGGTTAGGTTGGA-3 | FastDigest Apa-I | aa: 484, 146 AA: 630 Aa: 630, 484, 146 PCR product: 630 |
| Genotype/Allele | ASD n (%) | Control n (%) | OR (95% CI) Control vs. ASD | p-Value |
|---|---|---|---|---|
| Fok-I (rs2228570) FF | 38 (35) | 68 (35) | ||
| Ff | 51 (47) | 92 (47) | 0.99 (0.59–1.68) | 0.98 |
| ff | 19 (18) | 36 (18) | 0.94 (0.48–1.87) | 0.87 |
| F | 127 (58.8) | 228 (58.2) | 0.97 (0.7–1.4) | 0.88 |
| f | 89 (41.2) | 164 (41.8) | ||
| FF vs. Ff + ff | 0.97 (0.66–1.42) | 0.88 | ||
| Bsm-I (rs1544410) | ||||
| BB | 12 (11) | 24 (12) | ||
| Bb | 67 (62) | 118 (60) | 1.14 (0.53–2.42) | 0.74 |
| bb | 29 (27) | 54 (28) | 1.07 (0.47–2.46) | 0.86 |
| B | 91 (42.1) | 166 (42.3) | 1.01 (0.72–1.41) | 0.96 |
| b | 125 (57.9) | 226 (57.7) | ||
| BB vs. Bb + bb | 1.11 (0.65–1.9) | 0.71 | ||
| Taq-I (rs731236) TT | 33 (31) | 92 (47) | ||
| Tt | 61 (56) | 85 (43) | 2.00 (1.19–3.35) | 0.008 |
| tt | 14 (13) | 19 (10) | 2.05 (0.93–4.56) | 0.08 |
| T | 127 (58.8) | 269 (68.6) | 1.53 (1.08–2.16) | 0.02 |
| t | 89 (41.2) | 123 (31.4) | ||
| TT vs. Tt + TT | 2.08 (1.40–3.01) | 0.0004 | ||
| Apa-I (rs7975232) | ||||
| 22 (20) | 26 (13) | 3.32 (1.44–7.63) | 0.004 | |
| Aa | 73 (68) | 119 (61) | 2.41 (1.23–4.73) | 0.01 |
| aa | 13 (12) | 51 (26) | ||
| A | 117 (54.2) | 171 (43.6) | 1.53 (1.09–2.13) | 0.01 |
| a | 99 (45.8) | 221 (56.4) | ||
| aa vs. AA + Aa | 2.65 (1.6–4.3) | 0.0001 | ||
| ASD + non-ASD Control | OR (95% CI) Females vs. Males | p-Value | ||
|---|---|---|---|---|
| Female n (%) | Male n (%) | |||
| Fok-I (rs2228570) | ||||
| FF | 39 (34) | 67 (36) | FF vs. Ff + ff | 0.61 |
| Ff | 44 (47) | 88 (47) | 0.91 (0.62–1.32) | |
| Ff | 22 (19) | 33 (17) | ||
| Bsm-I (rs1544410) | ||||
| BB | 13 (11) | 23 (12) | BB vs. Bb + bb | |
| Bb | 67 (58) | 118 (63) | 0.9 (0.53–1.52) | 0.69 |
| Bb | 35 (30) | 48 (25) | ||
| Taq-I (rs731236) | ||||
| TT | 57 (50) | 68 (36) | TT vs. Tt +.tt | |
| Tt | 47 (41) | 99 (52) | 1.76 (1.2–2.6) | 0.0035 |
| Tt | 11 (9) | 22 (10) | ||
| Apa-I (rs7975232) | ||||
| AA | 11 (9) | 37 (20) | aa vs. AA + Aa | |
| Aa | 80 (70) | 112 (59) | 1.07 (1.07–1.65) | 0.75 |
| Aa | 24 (21) | 40 (21) | ||
| VDR Gene Polymorphism | No. of Patients | Concentration (ng/mL) | SEM | p-Value |
|---|---|---|---|---|
| Apa-I | ||||
| Aa | 9 | 41.6 (21.2–71.1) | 13.4 | 0.07 |
| AA | 10 | 57.2 (19.9–72.5) | 15.1 | |
| Taq-I | ||||
| TT | 4 | 48.8 (36.3–71.1) | 15.8 | 0.71 |
| Tt | 11 | 48.5 (19.9–72.5) | 14.7 | |
| Tt | 4 | 52.3 (21.2–71.0) | 17.1 | |
| Bsm-I | ||||
| BB | 5 | 48 (26.2–69.4) | 12.2 | 0.92 |
| Bb | 7 | 54.9 (21.2–72.5) | 21.2 | |
| Bb | 7 | 52.4 (31.3–72.2) | 18.8 | |
| Fok-I | ||||
| FF | 7 | 52.7 (31.3–72.5) | 13.1 | 0.21 |
| Ff | 8 | 50.3 (21.2–72.2) | 10.6 | |
| ff | 4 | 30.6 (19.9–45.5) | 11.7 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cieślińska, A.; Kostyra, E.; Chwała, B.; Moszyńska-Dumara, M.; Fiedorowicz, E.; Teodorowicz, M.; Savelkoul, H.F.J. Vitamin D Receptor Gene Polymorphisms Associated with Childhood Autism. Brain Sci. 2017, 7, 115. https://doi.org/10.3390/brainsci7090115
Cieślińska A, Kostyra E, Chwała B, Moszyńska-Dumara M, Fiedorowicz E, Teodorowicz M, Savelkoul HFJ. Vitamin D Receptor Gene Polymorphisms Associated with Childhood Autism. Brain Sciences. 2017; 7(9):115. https://doi.org/10.3390/brainsci7090115
Chicago/Turabian StyleCieślińska, Anna, Elżbieta Kostyra, Barbara Chwała, Małgorzata Moszyńska-Dumara, Ewa Fiedorowicz, Małgorzata Teodorowicz, and Huub F.J. Savelkoul. 2017. "Vitamin D Receptor Gene Polymorphisms Associated with Childhood Autism" Brain Sciences 7, no. 9: 115. https://doi.org/10.3390/brainsci7090115
APA StyleCieślińska, A., Kostyra, E., Chwała, B., Moszyńska-Dumara, M., Fiedorowicz, E., Teodorowicz, M., & Savelkoul, H. F. J. (2017). Vitamin D Receptor Gene Polymorphisms Associated with Childhood Autism. Brain Sciences, 7(9), 115. https://doi.org/10.3390/brainsci7090115








