Image Fusion of High-Resolution DynaCT and T2-Weighted MRI for Image-Guided Programming of dDBS
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Image Visualization and Initial Alignment
2.3. Manual Landmark-Based Alignment
2.4. Intensity-Based Image Registration
2.5. Evaluation Metrics
2.6. Image Processing
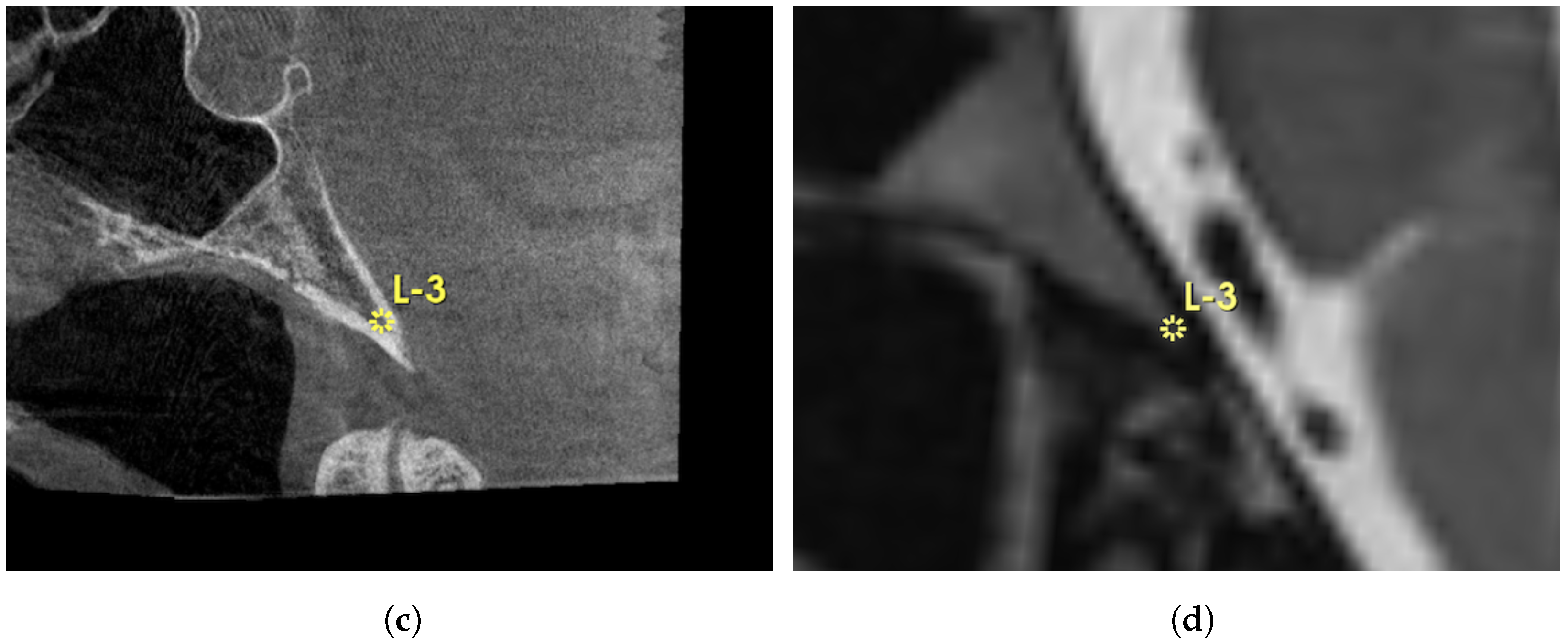
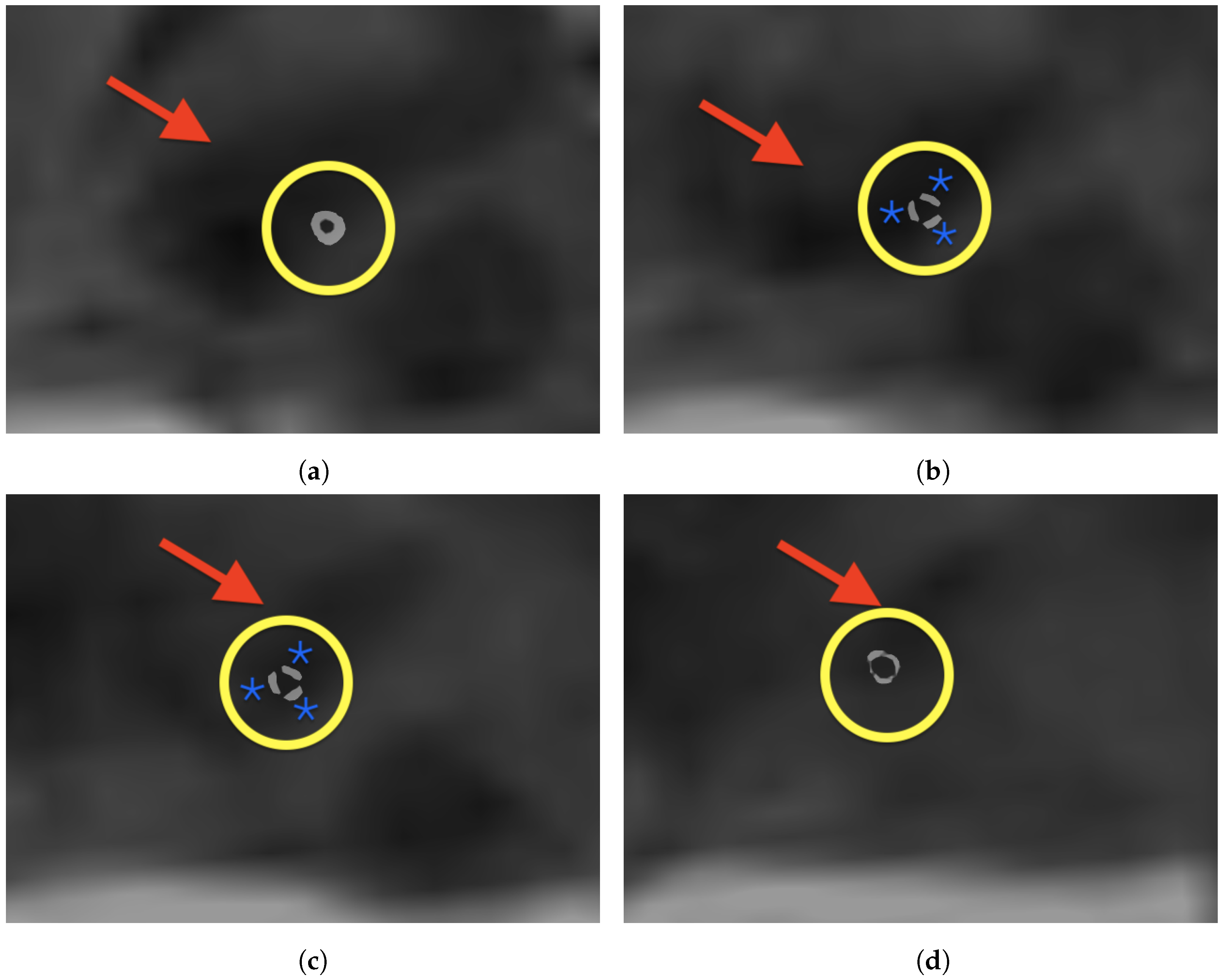
3. Results
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lozano, A.M.; Lipsman, N.; Bergman, H.; Brown, P.; Chabardes, S.; Chang, J.W.; Matthews, K.; McIntyre, C.C.; Schlaepfer, T.E.; Schulder, M.; et al. Deep brain stimulation: Current challenges and future directions. Nat. Rev. Neurol. 2019, 15, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Egger, K.; Rau, A.; Urbach, H.; Reisert, M.; Reinacher, P.C. 3D X-ray based visualization of directional deep brain stimulation lead orientation. J. Neuroradiol. 2022, 49, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Dembek, T.; Hoevels, M.; Hellerbach, A.; Horn, A.; Petry-Schmelzer, J.; Borggrefe, J.; Wirths, J.; Dafsari, H.; Barbe, M.; Visser-Vandewalle, V.; et al. Directional DBS leads show large deviations from their intended implantation orientation. Park. Relat. Disord. 2019, 67, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.M.; Buentjen, L.; Kaufmann, J.; Gruber, D.; Treuer, H.; Haghikia, A.; Voges, J. Deviation of the orientation angle of directional deep brain stimulation leads quantified by intraoperative stereotactic X-ray imaging. Neurosurg. Rev. 2022, 45, 2975–2982. [Google Scholar] [CrossRef] [PubMed]
- Al-Jaberi, F.; Moeskes, M.; Skalej, M.; Fachet, M.; Hoeschen, C. 3D-visualization of segmented contacts of directional deep brain stimulation electrodes via registration and fusion of CT and FDCT. EJNMMI Rep. 2024, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Beare, R.; Lowekamp, B.; Yaniv, Z. Image Segmentation, Registration and Characterization in R with SimpleITK. J. Stat. Softw. 2018, 86, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Soler-Rico, M.; Peeters, J.B.; Joris, V.; Delavallée, M.; Duprez, T.; Raftopoulos, C. MRI-guided DBS of STN under general anesthesia for Parkinson’s disease: Results and microlesion effect analysis. Acta Neurochir. 2022, 164, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, R.L.; Jarosz, J.M.; Samuel, M.; Clough, C.; Selway, R.P.; Ashkan, K. CT/MR image fusion in the postoperative assessment of electrodes implanted for deep brain stimulation. Stereotact. Funct. Neurosurg. 2009, 87, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Aldosary, G.; Szanto, J.; Holmes, O.; Lavigne, B.; Althobaity, W.; Sheikh, A.; Foottit, C.; Vandervoort, E. Geometric inaccuracy and co-registration errors for CT, DynaCT and MRI images used in robotic stereotactic radiosurgery treatment planning. Phys. Med. 2020, 69, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Geevarghese, R.; O’Gorman Tuura, R.; Lumsden, D.E.; Samuel, M.; Ashkan, K. Registration accuracy of CT/MRI fusion for localisation of deep brain stimulation electrode position: An imaging study and systematic review. Stereotact. Funct. Neurosurg. 2016, 94, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Orth, R.C.; Wallace, M.J.; Kuo, M.D.; Technology Assessment Committee of the Society of Interventional Radiology. C-arm cone-beam CT: General principles and technical considerations for use in interventional radiology. J. Vasc. Interv. Radiol. 2008, 19, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhamari, I.; Helal, R.; Abdelaziz, T.; Waldeck, S.; Paulus, D. Automatic cochlear multimodal 3D image segmentation and analysis using atlas–model-based method. Cochlear Implant. Int. 2024, 25, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Maes, F.; Vandermeulen, D.; Suetens, P. Medical image registration using mutual information. Proc. IEEE Inst. Electr. Electron. Eng. 2003, 91, 1699–1722. [Google Scholar] [CrossRef]
- Chen, M.; Tustison, N.J.; Jena, R.; Gee, J.C. Image Registration: Fundamentals and Recent Advances Based on Deep Learning. In Machine Learning for Brain Disorders; Colliot, O., Ed.; Springer: New York, NY, USA, 2023; pp. 435–458. [Google Scholar] [CrossRef]
- Krüger, M.T.; Naseri, Y.; Cavalloni, F.; Reinacher, P.C.; Kägi, G.; Weber, J.; Brogle, D.; Bozinov, O.; Hägele-Link, S.; Brugger, F. Do directional deep brain stimulation leads rotate after implantation? Acta Neurochir. 2021, 163, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Dembek, T.A.; Asendorf, A.L.; Wirths, J.; Barbe, M.T.; Visser-Vandewalle, V.; Treuer, H. Temporal stability of lead orientation in directional deep brain stimulation. Stereotact. Funct. Neurosurg. 2021, 99, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Al-Jaberi, F.; Fachet, M.; Moeskes, M.; Skalej, M.; Hoeschen, C. Optimization Techniques for Semi-Automated 3D Rigid Registration in Multimodal Image-Guided Deep Brain Stimulation. Curr. Dir. Biomed. Eng. 2023, 9, 355–358. [Google Scholar] [CrossRef]
- Handa, B.; Singh, G.; Kamal, R.; Oinam, A.S.; Kumar, V. Evaluation method for the optimization of 3d rigid image registration on multimodal image datasets. Int. J. Eng. Adv. Technol. 2019, 9, 234. [Google Scholar] [CrossRef]
- Chung, H.T.; Kim, J.H.; Kim, J.W.; Paek, S.H.; Kim, D.G.; Chun, K.J.; Kim, T.H.; Kim, Y.K. Assessment of image co-registration accuracy for frameless gamma knife surgery. PLoS ONE 2018, 13, e0193809. [Google Scholar] [CrossRef] [PubMed]





| Patient | TRE (mm) | SD (mm) | Image Size DynaCT (pixels) | Voxel Size DynaCT (mm) | Image Size T2-w MRI (pixels) | Voxel Size T2-w MRI (mm) | Comment |
|---|---|---|---|---|---|---|---|
| 01 | 2.61 | 0.5 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 02 | 1.24 | 0.35 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 03 | 1.05 | 0.24 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 04 | 1.07 | 0.21 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 05 | 1.94 | 1.21 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | Motion artifacts MRI |
| 06 | 1.86 | 0.68 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | Motion artifacts MRI |
| 07 | 0.72 | 0.28 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 08 | 1.94 | 0.51 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 09 | 1.05 | 0.37 | 512 512 497 | 0.2 0.2 0.2 | 192 192 160 | 1 1 1 | - |
| 10 | 1.3 | 0.27 | 512 512 497 | 0.2 0.2 0.2 | 240 320 80 | 0.8 0.8 2 | 2 mm slice thickness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Jaberi, F.; Moeskes, M.; Skalej, M.; Fachet, M.; Hoeschen, C. Image Fusion of High-Resolution DynaCT and T2-Weighted MRI for Image-Guided Programming of dDBS. Brain Sci. 2025, 15, 521. https://doi.org/10.3390/brainsci15050521
Al-Jaberi F, Moeskes M, Skalej M, Fachet M, Hoeschen C. Image Fusion of High-Resolution DynaCT and T2-Weighted MRI for Image-Guided Programming of dDBS. Brain Sciences. 2025; 15(5):521. https://doi.org/10.3390/brainsci15050521
Chicago/Turabian StyleAl-Jaberi, Fadil, Matthias Moeskes, Martin Skalej, Melanie Fachet, and Christoph Hoeschen. 2025. "Image Fusion of High-Resolution DynaCT and T2-Weighted MRI for Image-Guided Programming of dDBS" Brain Sciences 15, no. 5: 521. https://doi.org/10.3390/brainsci15050521
APA StyleAl-Jaberi, F., Moeskes, M., Skalej, M., Fachet, M., & Hoeschen, C. (2025). Image Fusion of High-Resolution DynaCT and T2-Weighted MRI for Image-Guided Programming of dDBS. Brain Sciences, 15(5), 521. https://doi.org/10.3390/brainsci15050521






