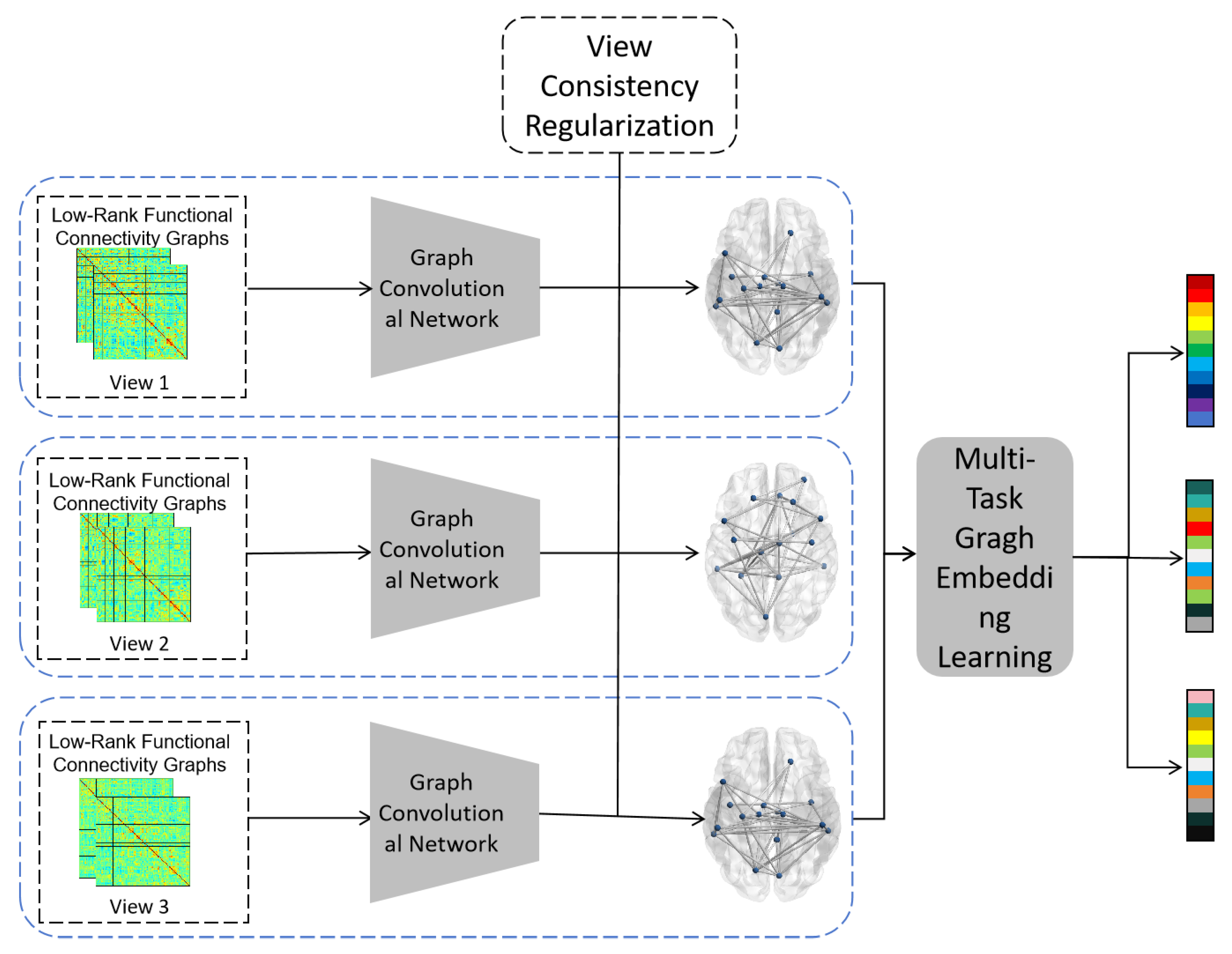
1. Introduction
ASD is a highly heterogeneous neurodevelopmental condition characterized by core symptoms including impaired social interaction and communication, as well as restricted, repetitive patterns of behavior, interests, or activities [
1]. Current evidence suggests that ASD arises from the interaction of genetic predispositions and non-genetic environmental factors, leading to widely varying clinical presentations and significant individual heterogeneity. Currently, clinical diagnosis primarily relies on behavioral observation tools—such as the autism diagnostic observation schedule (ADOS)—and questionnaire-based assessments, which are limited by subjectivity and diagnostic delays. According to the Global Burden of Disease Study 2021, the worldwide prevalence of ASD is estimated to be approximately 0.79%, with a 40% increase in screening positivity among young children over the past five years, underscoring the urgent need for more objective and efficient diagnostic tools. With rapid advances in neuroimaging, rs-fMRI has emerged as a promising technique for aiding ASD diagnosis, owing to its ability to reveal abnormal functional connectivity and potential biomarkers within the brain [
2]. The growing body of rs-fMRI research offers avenues to overcome the subjectivity inherent in conventional diagnostic approaches, thereby improving diagnostic accuracy and facilitating early identification, timely intervention, and effective treatment of autism.
Early neuroimaging-based studies on ASD primarily relied on feature analysis of functional connectivity networks (FCNs). Conventional approaches often employed Pearson’s correlation coefficient, covariance, or time-series information to quantify temporal correlations in functional activation between brain regions, thereby establishing connectivity between spatially defined regions of interest. However, these methods depend on handcrafted features, which are often subjective, limited in interpretability, reliant on prior knowledge, and inadequate for capturing complex nonlinear interactions. With the widespread adoption of deep learning in medical imaging, significant progress has been made in rs-fMRI-based diagnostic methods for ASD. Initial deep learning applications mainly focused on converting brain network data into grid-like structures for processing. For instance, Yin et al. [
3] proposed a semi-supervised autoencoder-based framework for ASD diagnosis, which combines unsupervised autoencoders with supervised classification networks to improve latent feature representation and diagnostic performance compared to traditional hand-engineered FCN features. Heinsfeld et al. [
4] employed convolutional neural networks (CNNs) in an end-to-end manner to automatically extract multi-level features from raw imaging data, improving diagnostic accuracy by approximately 12–15%. The intrinsic nature of functional brain networks is non-Euclidean and graph-structured. Converting such data into grid formats—such as by flattening adjacency matrices—can lead to loss of spatial relationships between brain regions. Consequently, graph convolutional networks (GCNs) and transformer models have been introduced to analyze functional brain connectivity, enabling more discriminative representation learning. GCNs are particularly suited to handling graph-structured data directly [
5]. Ktena et al. [
6] applied GCNs to the ABIDE dataset and directly learning features from functional connectivity graphs for ASD identification, and they have demonstrated significantly better performance compared to traditional machine learning methods. Transformers, known for their ability to model long-range dependencies, have been used to capture dynamic characteristics of rs-fMRI time series. The ST-transformer method proposed by Deng et al. [
7] has shown high effectiveness on the ABIDE dataset in handling class imbalance and capturing spatio-temporal features.
Although deep learning has shown promising potential in ASD diagnosis, conventional approaches often rely on a single brain atlas (e.g., AAL or CC200) to construct FCNs, which may lead to insufficient information utilization. Different atlases are based on distinct brain parcellation principles and prior knowledge, potentially capturing complementary information that a single atlas cannot comprehensively cover. To address this limitation, Wang et al. [
8] proposed a category-consistent and site-independent multi-view hyperedge-aware hypergraph embedding framework that integrates FCNs constructed from multiple atlases. This method uses hypergraph modeling to capture high-order interactions among brain regions and incorporates specifically designed modules to enhance category discrimination and reduce center-related bias, achieving superior performance in ASD identification on the ABIDE dataset compared to other methods. Similarly, Yu et al. [
9] introduced a multi-atlas functional and effective connection attention fusion method, which integrates both functional connectivity and effective connectivity information from multiple atlases using dynamic graph convolutional networks and adaptive self-attention mechanisms, yielding high diagnostic accuracy for ASD. In addition to the limitations of single-atlas approaches, most early studies trained and validated models using single-center data, significantly restricting the generalizability and clinical applicability of these methods. Variations in scanning equipment, acquisition parameters, and participant demographics across imaging centers introduce data heterogeneity, often leading to notable performance degradation when a model trained on one center is applied to another [
10]. To mitigate the impact of multi-center data heterogeneity, some researchers have reframed the multi-center ASD diagnosis task as a domain adaptation problem. For instance, Chu et al. [
11] incorporated domain adaptation techniques into multi-center analysis by introducing mean absolute error and covariance constraints, effectively reducing inter-center distribution discrepancies. Other studies have proposed low-rank representation-based methods for multi-center ASD identification [
12] and low-rank subspace graph convolutional networks [
13]. The former employs label propagation to predict unlabeled samples, while the latter uses graph convolutional networks for feature extraction before classification. Wang et al. [
14] proposed grouping multi-center data by patient and control categories, using similarity-driven multi-center linear reconstruction to learn latent representations, performing clustering within each group, and applying nested singular value decomposition to reduce center-specific heterogeneity.
To further overcome the limitations of single-atlas information insufficiency and multi-center data heterogeneity, this paper proposes a multi-view, low-rank subspace graph structure learning method to integrate multi-atlas and multi-center data for automated ASD diagnosis, termed M3ASD. The proposed framework first constructs functional connectivity matrices from multi-center neuroimaging data using multiple brain atlases. Edge weight filtering is then applied to build multiple brain networks with diverse topological properties, forming several complementary views. Samples from different classes are separately projected into low-rank subspaces within each view to mitigate data heterogeneity. Multi-view consistency regularization is further incorporated to extract more consistent and discriminative features from the low-rank subspaces across views. The proposed method is evaluated on the publicly available ABIDE dataset using multiple data partitioning strategies to demonstrate its effectiveness.
The main contributions of this paper are summarized as follows: (1) We propose a novel multi-view low-rank subspace graph structure learning method, termed M3ASD, to integrate multi-atlas and multi-center rs-fMRI data for automated diagnosis of ASD. (2) Our approach overcomes the limitation of relying on a single brain atlas for constructing functional networks. Instead, it leverages multiple brain atlases to build multi-view brain networks with complementary topological properties. (3) To address the heterogeneity inherent in multi-center data, we employ low-rank subspace projection and consistency regularization techniques, effectively mitigating its adverse effects. (4) The proposed M3ASD framework is extensively evaluated on the publicly available ABIDE dataset under multiple data partitioning strategies. Experimental results demonstrate its effectiveness in enhancing both the accuracy and generalization capability of ASD diagnosis.
The rest of this paper is organized as follows. In the Materials and Methods section, we describe the datasets used in this study and the proposed method. Then, in the Results section, we present the experimental setup, evaluation index, comparison methods, ablation experiment results, and ASD diagnosis outcomes achieved via different methods. In the Discussion section, we investigate the impact of several key components of the proposed M3ASD method. Finally, this paper is concluded in the Conclusions section.
4. Discussion
4.1. Comparison of the Number of Atlases
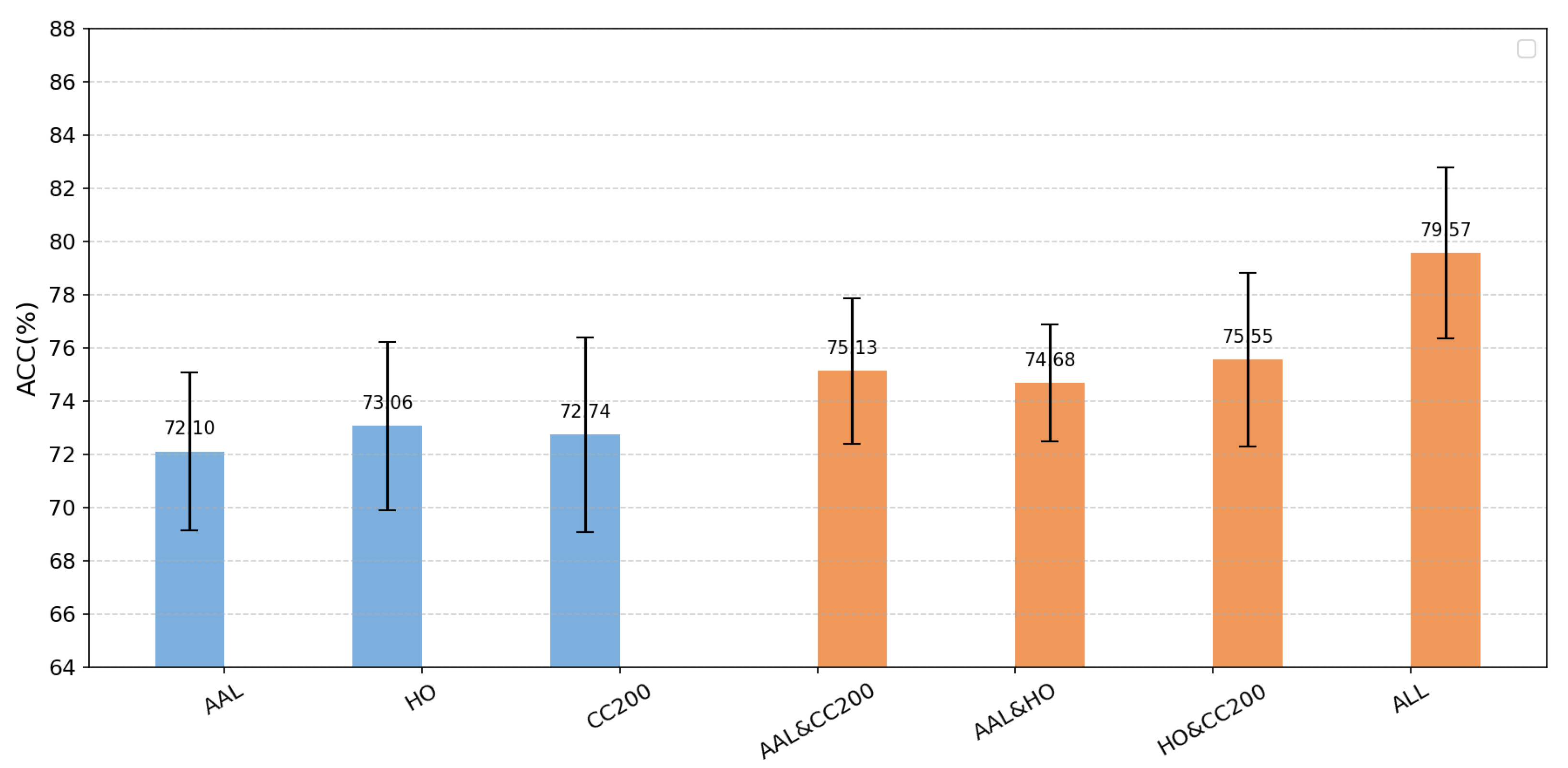
This section aims to investigate the impact of the number of atlases on diagnostic performance. For a systematic comparison, all experiments regarding the number of atlases were conducted under Experimental Setting 1, for which the five centers with the largest sample sizes from the ABIDE dataset were selected. Each time, samples from one center were used as the test set, while those from the remaining centers formed the training set.
We first evaluated the performance using each individual atlas—namely the AAL, CC200, and HO atlases—as input. We then combined the atlases pairwise for comparison and finally integrated all three atlases as input. The corresponding accuracies under these configurations are illustrated in
Figure 5. Evidently, the three-atlas input demonstrates a clear advantage over both dual-atlas and single-atlas approaches. Moreover, the accuracy achieved with any two atlases combined exceeded that of any single-atlas method, indicating that multi-atlas integration effectively enhances the accuracy of autism diagnosis [
30]. Overall, the multi-atlas approach fully leverages information from each atlas and integrates complementary features, leading to improved overall performance, reduced limitations inherent in single-method analyses, and stronger generalization and diagnostic capability [
31].
4.2. The Impact of Multi-View Parameters
To further investigate the impact of the number of views in the multi-view construction module on method performance, the number of views was incrementally increased from 1 to 5. Under Experimental Setting 1, the
ACC values and time efficiency of the M
3ASD method are shown in
Figure 6. When the number of views exceeds 3, the performance improvement becomes marginal, while time efficiency decreases significantly. Therefore, this study sets the number of views to three per sample in the M
3ASD method.
The selection of views also considerably influences method performance. A set of
values,
, was configured, and the accuracy of each single view constructed under different
values was calculated. The results are presented in
Figure 7. It was observed that accuracy does not increase linearly with
; instead, the highest accuracy is achieved when
. The view selection rule in the M
3ASD method prioritizes views in descending order of their individual accuracy [
31]. Thus, when constructing three views, the corresponding
values chosen were 0.6, 0.7, and 0.2.
4.3. Important Brain Functional Connections Affecting Multi-Atlas
This study utilized a multi-center dataset with multiple atlases. After applying low-rank representation, brain network-related weights were learned through a self-attention-based graph convolutional neural network. By analyzing the weights in the functional connectivity weight matrix of each atlas, the top 10 connections with the highest weights were identified, resulting in an important brain functional connectivity graph as shown in
Figure 8. This graph reveals brain regions closely associated with ASD pathology and key functional connections between them. As can be observed from the figure, the connection patterns of the AAL atlas and the HO atlas are similar, indicating that although the atlases differ, the brain regions and connections influencing autism are consistent [
32]. Through the investigation of significant brain functional connectivity graphs, critical regions [
2] potentially strongly linked to autism were identified, including the inferior occipital gyrus, orbital gyrus, insula, superior temporal gyrus [
33], amygdala, temporal lobe, and central operculum [
34].
To strengthen the interpretability of the key functional connections illustrated in
Figure 8, this study conducted systematic statistical validation. Independent samples
t-tests were performed to compare the functional connectivity strength between the autism spectrum disorder (ASD) and healthy control (HC) groups for the salient connections identified through the M
3ASD model. The analysis revealed that all critical connections demonstrated statistically significant group differences (all
, false discovery rate (FDR) corrected). These results confirm that the connectivity patterns visualized in
Figure 8 not only reflect the discriminative basis of the model but also correspond to neuroimaging biomarkers with statistical significance, thereby providing quantitative support for the pathological mechanisms of ASD.
The identified functional connections align well with established models of ASD neurocircuitry. The heightened importance of the connection between the right amygdala and left inferior occipital gyrus [
35] is particularly noteworthy. The amygdala is a core hub for emotional processing and social behavior, and its dysfunction is a hallmark of ASD [
36]. The inferior occipital gyrus is involved in visual processing. Their aberrant connectivity may underlie difficulties in processing emotionally salient visual stimuli, such as facial expressions, a common challenge in ASD [
37]. Furthermore, the consistent identification of the insula and superior temporal gyrus across atlases reinforces their role in ASD. The insula is crucial for interoception and social-emotional awareness, while the superior temporal gyrus is involved in auditory processing and theory of mind. Their disrupted connectivity has been frequently reported in the ASD literature [
38]. These findings not only validate the biological plausibility of M
3ASD but also highlight potential neural pathways for targeted interventions.
4.4. Hyperparameter Analysis
To determine the optimal hyperparameter configuration for the M3ASD model, we adopted a hierarchical optimization strategy: first identifying the optimal number of graph convolutional layers k, followed by a grid search for the regularization parameters and based on the optimal k.
The number of graph convolutional layers k determines the depth of neighborhood information aggregation from the functional connectivity networks. A value of k that is too small (e.g., ) restricts the model’s receptive field, making it difficult to capture long-range dependencies in the brain network, while an excessively large k can easily lead to the over-smoothing phenomenon, causing the node features to lose discriminative power. Therefore, we set the search range as to balance the model capacity and generalization ability. Under Experimental Setting I, with fixed and , we systematically evaluated the impact of different k values on the model’s performance. The experimental results indicate that, as k increased from 2 to 5, the model accuracy improved steadily, with the accuracy being % for k = 2, % for k = 3, % for k = 4, and reaching its peak of % for k = 5. However, when k was increased to 6, the accuracy decreased to %, indicating the occurrence of over-smoothing. This performance trend clearly illustrates the trade-off between network depth and model performance, leading to the determination of k = 5 as the optimal network depth.
The parameter search results, summarized in
Table 7, show that the model achieved the highest classification accuracy of
when
and
. This indicates that a moderate
value effectively filters out noise, while a
value approximately half its strength well preserves the topological structure of the brain network, with the two parameters working synergistically to achieve optimal performance. A further analysis revealed that, with a fixed
, performance initially increased and then decreased as
increased. For instance, with
, accuracy peaked at
, confirming the effectiveness of the graph structural constraint and the importance of balancing its strength. Moreover, in the vicinity of the optimal combination (e.g.,
,
or
,
), the model maintained high performance (accuracy
), demonstrating that M
3ASD is relatively insensitive to small perturbations in hyperparameters and possesses good robustness.
In summary, through systematic hyperparameter analysis, we determined the optimal configuration for the M3ASD model as , , and . This configuration achieves an optimal balance between model expressive power, structure preservation, and noise suppression, providing a reliable parameter foundation for subsequent experiments.
4.5. Limitation and Future Work
4.5.1. Limitation
Despite the encouraging results, this study is subject to several limitations. Firstly, the validation was solely reliant on the ABIDE-I dataset. Although our method is designed to handle multi-center heterogeneity, its generalizability to completely independent cohorts (e.g., ABIDE-II, EU-AIMS) remains to be further verified. The absence of external validation constitutes a limitation of the current work. Secondly, while the multi-center and multi-atlas design is a core strength of this study, it inherently limits the sample size available within each individual center. Consequently, conducting statistically meaningful subgroup analyses (e.g., based on specific gender categories, symptom severity scores, or IQ levels) would result in extremely small and underpowered subgroups per center, potentially leading to unreliable conclusions. Therefore, our current analysis focused on validating the overall framework’s efficacy across centers.
4.5.2. Future Direction
Future research will prioritize the following directions to address these limitations: (1) Applying M3ASD to larger, independent multi-center cohorts to facilitate both robust external validation and meaningful subgroup analyses. (2) Collaborating with multiple institutions to aggregate larger sample sizes within specific demographic or clinical subgroups, enabling the development of more personalized diagnostic models. (3) Exploring the integration of additional neuroimaging modalities to provide a more comprehensive characterization of ASD.
While this study establishes the utility of M3ASD for multi-center, multi-atlas static functional connectivity analysis, several exciting avenues emerge for extending its impact. The inherent temporal dynamics of ASD present a clear next step: integrating models like long short-term memory (LSTM) networks to analyze longitudinal rs-fMRI data could significantly enhance pattern recognition of disease progression.
Beyond temporal analysis, the multi-view architecture of M
3ASD is inherently extensible to other data modalities. Future work could incorporate electrophysiological data (e.g., EEG/EMG) from wearable sensors or leverage advances in photonic sensing technology [
39], moving towards a comprehensive multi-modal diagnostic system. The core methodology of M
3ASD is also not specific to ASD. It holds considerable potential for application to other neurological disorders characterized by aberrant network patterns, such as ADHD or schizophrenia.
The ultimate translational goal is the development of systems for continuous patient monitoring and neurorehabilitation. Achieving this requires addressing challenges such as model lightweighting and integration with portable systems [
40] to enable real-time clinical decision support. To ensure reliability in heterogeneous clinical populations, future iterations must incorporate uncertainty quantification (e.g., via Bayesian deep learning) to provide predictive confidence scores, which is crucial for clinical trust.