Transcriptome-Wide Analysis of Neutrophil-Related Circ_22232 in Neuroinflammation from Ischemic Stroke Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome-Wide Transcriptomic Data Processing
2.2. Screening of Differentially Expressed Genes (DEGs)
2.3. Functional and Pathway Enrichment Analyses
2.4. CircRNA-miRNA-mRNA Coexpression Network
2.5. Animals
2.6. Construction of Circ_22232 Knockdown Mice
2.7. Middle Cerebral Artery Occlusion (MCAO) Model
2.8. Schematic Mode
2.9. Modified Neurological Severity Score (mNSS)
2.10. Triphenyl Tetrazolium Chloride (TTC) Staining
2.11. Brain Water Content Analysis
2.12. Quantitative Real-Time (PCR)
2.13. Enzyme-Linked Immunosorbent Assays (ELISA)
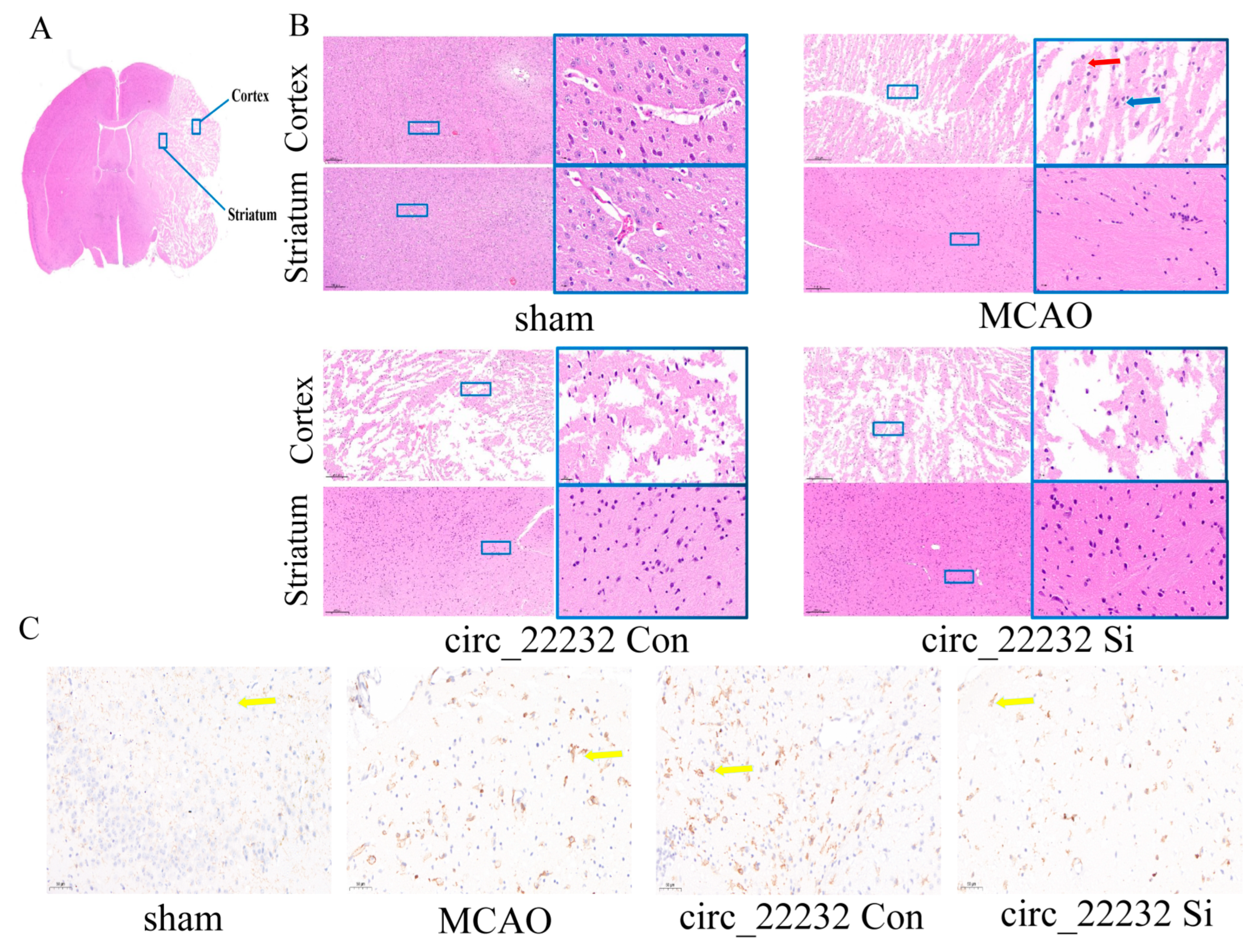
2.14. Hematoxylin-Eosin Staining (H&E Staining)
2.15. Immunohistochemistry
2.16. Statistical Analysis
3. Results
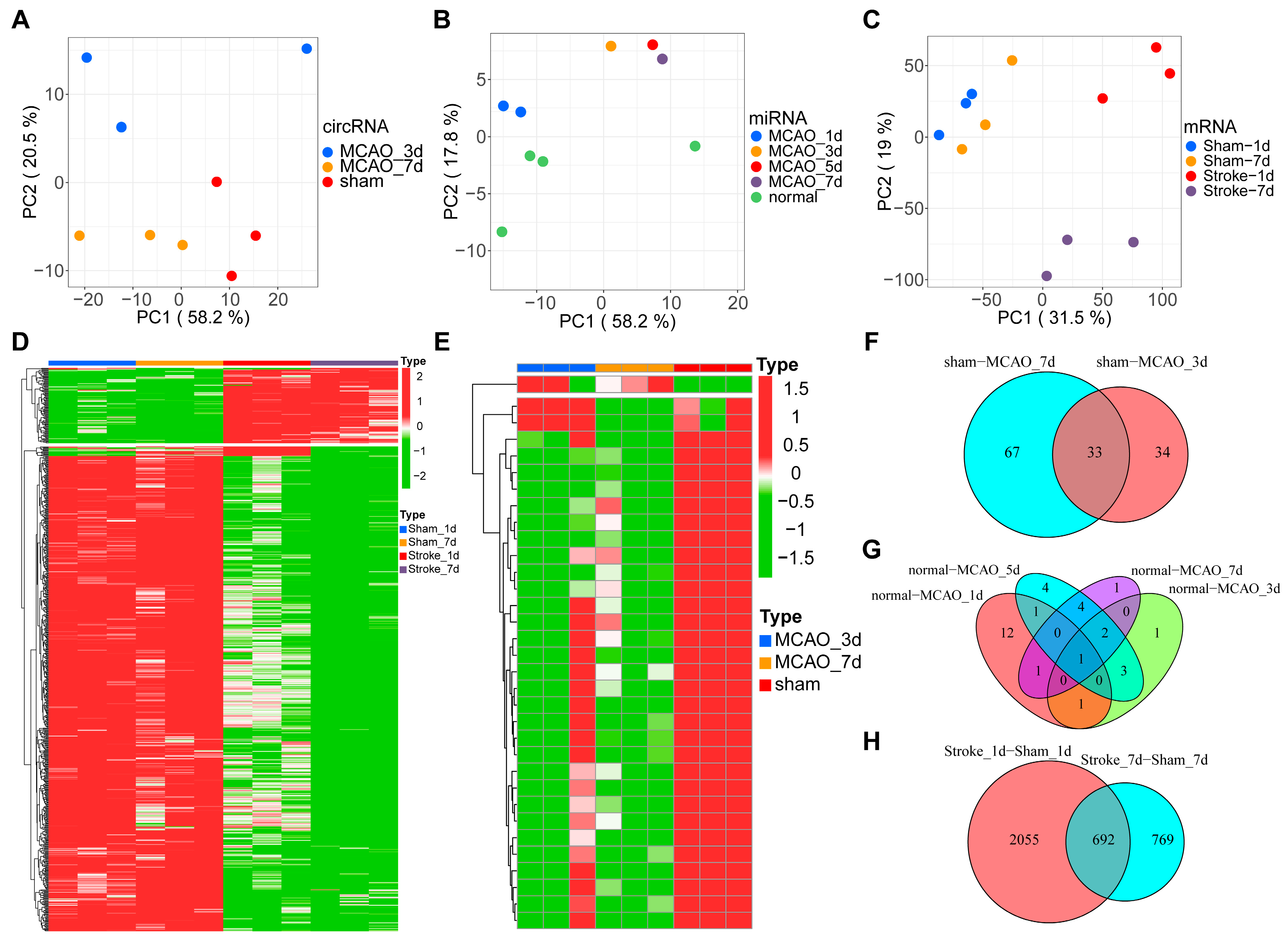
3.1. Sequencing Data Summary and DEG Analysis
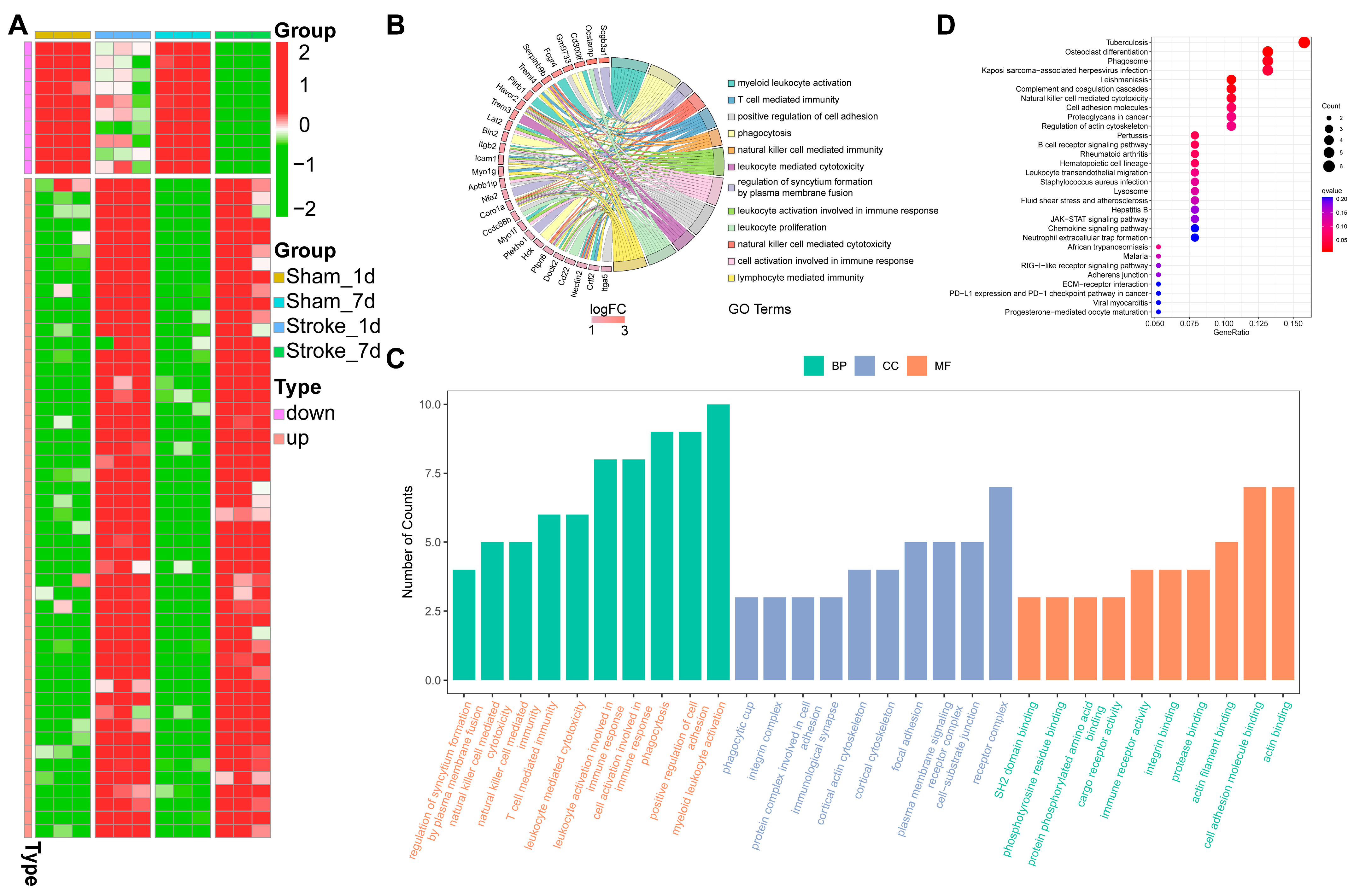
3.2. GO Enrichment and KEGG Pathway Analysis
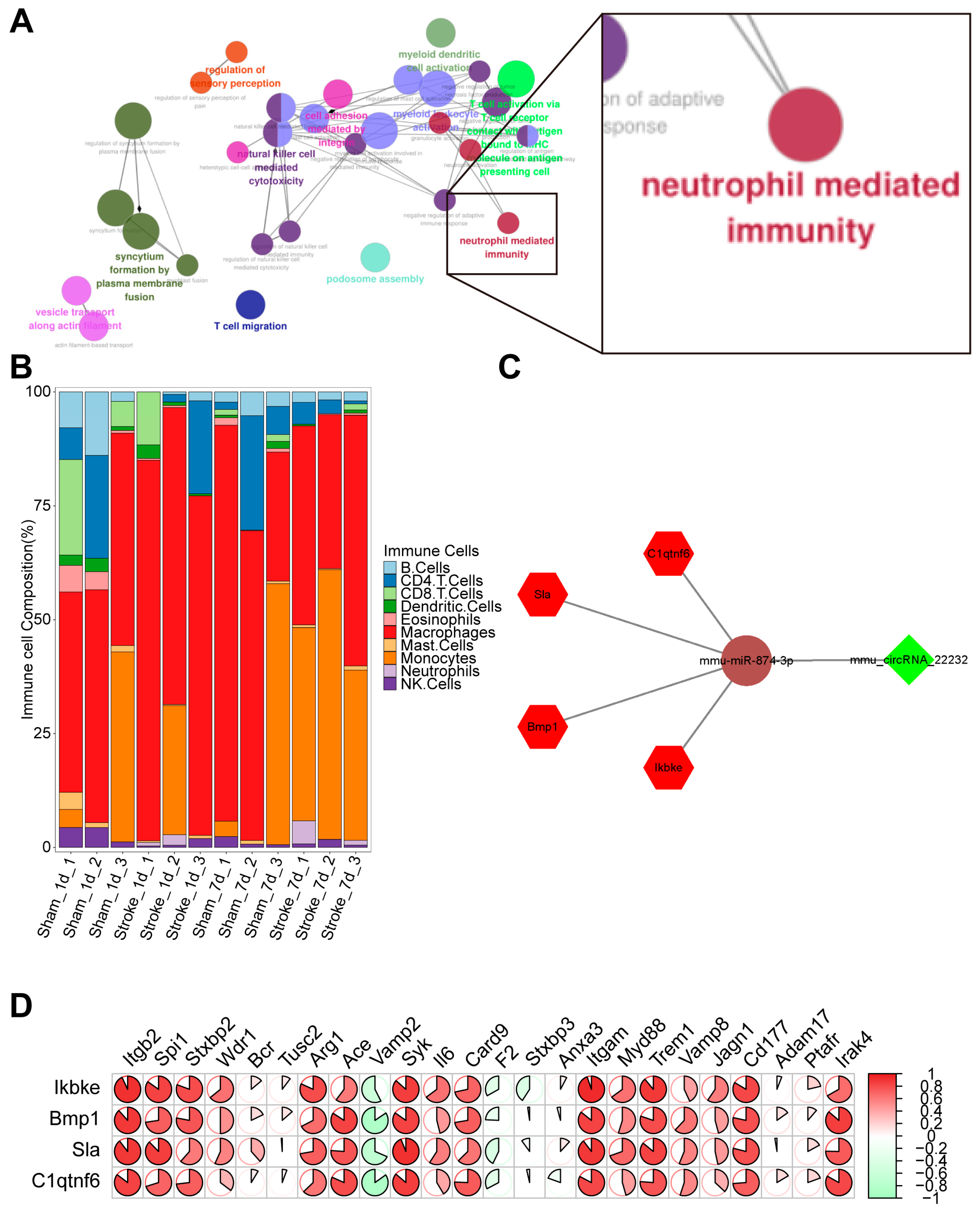
3.3. Coexpression Analysis of CircRNA Related to Inflammation
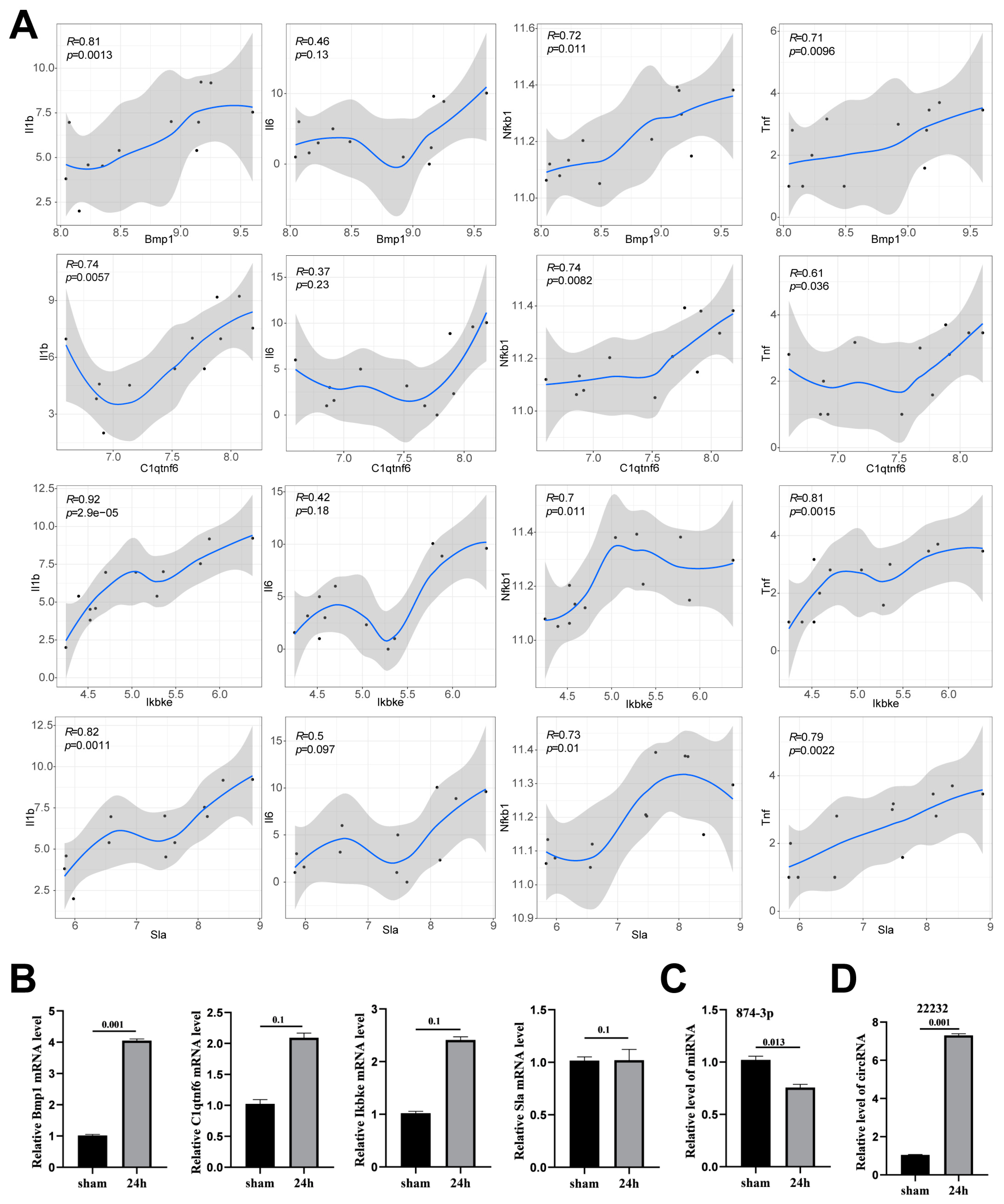
3.4. Verification of Potential circRNA_22232/miR-874-3p/Bmp1 Axis in Neuroinflammation
3.5. Circ_22232 Knockdown Reduced Ischemic Brain Injury and Release Proinflammatory Cytokine
3.6. Circ_22232 Knockdown Reduced Ischemic Brain Injury and Activated Neutrophils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dayon, L.; Turck, N.; Garcí-Berrocoso, T.; Walter, N.; Burkhard, P.R.; Vilalta, A.; Sahuquillo, J.; Montaner, J.; Sanchez, J.C. Brain extracellular fluid protein changes in acute stroke patients. J. Proteome Res. 2011, 10, 1043–1051. [Google Scholar] [CrossRef]
- Gülke, E.; Gelderblom, M.; Magnus, T. Danger signals in stroke and their role on microglia activation after ischemia. Ther. Adv. Neurol. Disord. 2018, 11, 1756286418774254. [Google Scholar] [CrossRef] [PubMed]
- Chamorro, Á.; Dirnagl, U.; Urra, X.; Planas, A.M. Neuroprotection in acute stroke: Targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016, 15, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Land, W.G. Use of DAMPs and SAMPs as Therapeutic Targets or Therapeutics: A Note of Caution. Mol. Diagn. Ther. 2020, 24, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Jha, M.K.; Jo, M.; Kim, J.H.; Suk, K. Microglia-Astrocyte Crosstalk: An Intimate Molecular Conversation. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2019, 25, 227–240. [Google Scholar] [CrossRef]
- Shen, L.; Bai, Y.; Han, B.; Yao, H. Non-coding RNA and neuroinflammation: Implications for the therapy of stroke. Stroke Vasc. Neurol. 2019, 4, 96–98. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Li, X.; Li, Y.; Zhong, Y.; Ling, L. Altered circular RNA expression profiles in the non-ischemic thalamus in focal cortical infarction mice. Aging 2020, 12, 13206–13219. [Google Scholar] [CrossRef]
- Jiang, Q.; Su, D.Y.; Wang, Z.Z.; Liu, C.; Sun, Y.N.; Cheng, H.; Li, X.M.; Yan, B. Retina as neurodegeneration a window to cerebral dysfunction following studies with circRNA signature during. Theranostics 2021, 11, 1814–1827. [Google Scholar] [CrossRef]
- Dong, Z.; Deng, L.; Peng, Q.; Pan, J.; Wang, Y. CircRNA expression profiles and function prediction in peripheral blood mononuclear cells of patients with acute ischemic stroke. J. Cell. Physiol. 2020, 235, 2609–2618. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, L.; Zu, J.; Wang, Z.; Han, B.; Chen, B.; Cheng, M.; Ju, M.; Li, M.; Shu, G.; et al. Circulating Circular RNAs as Biomarkers for the Diagnosis and Prediction of Outcomes in Acute Ischemic Stroke. Stroke 2020, 51, 319–323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Quan, L.; Huang, A.; Zhao, Q.; Yuan, Y.; Yuan, X.; Shen, Q.; Shang, J.; Ben, Y.; Qin, F.X.; et al. seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data. Front. Immunol. 2018, 9, 1286. [Google Scholar] [CrossRef] [PubMed]
- Engel, O.; Kolodziej, S.; Dirnagl, U.; Prinz, V. Modeling stroke in mice—Middle cerebral artery occlusion with the filament model. J. Vis. Exp. JoVE 2011, 47, e2423. [Google Scholar]
- Bieber, M.; Gronewold, J.; Scharf, A.C.; Schuhmann, M.K.; Langhauser, F.; Hopp, S.; Mencl, S.; Geuss, E.; Leinweber, J.; Guthmann, J.; et al. Validity and Reliability of Neurological Scores in Mice Exposed to Middle Cerebral Artery Occlusion. Stroke 2019, 50, 2875–2882. [Google Scholar] [CrossRef]
- Boivin, G.P.; Bottomley, M.A.; Schiml, P.A.; Goss, L.; Grobe, N. Physiologic, Behavioral, and Histologic Responses to Various Euthanasia Methods in C57BL/6NTac Male Mice. J. Am. Assoc. Lab. Anim. Sci. 2017, 56, 69–78. [Google Scholar]
- Shin, T.H.; Lee, D.Y.; Basith, S.; Manavalan, B.; Paik, M.J.; Rybinnik, I.; Mouradian, M.M.; Ahn, J.H.; Lee, G. Metabolome Changes in Cerebral Ischemia. Cells 2020, 9, 1630. [Google Scholar] [CrossRef]
- Kwon, H.S.; Koh, S.H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020, 9, 42. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, H.; Fan, Z.; Li, G.; Ma, Q.; Tao, Z.; Wang, R.; Feng, J.; Luo, Y. Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke 2017, 48, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Ostolaza, A.; Blanco-Luquin, I.; Urdánoz-Casado, A.; Rubio, I.; Labarga, A.; Zandio, B.; Roldán, M.; Martínez-Cascales, J.; Mayor, S.; Herrera, M.; et al. Circular RNA expression profile in blood according to ischemic stroke etiology. Cell Biosci. 2020, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Xie, J.; Liu, Y.; Leng, S.; Zhang, Z.; Yan, F. Down-regulation of circular RNA CDC14A peripherally ameliorates brain injury in acute phase of ischemic stroke. J. Neuroinflamm. 2021, 18, 283. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, Y.; Han, B.; Yang, L.; Chen, X.; Huang, R.; Wu, F.; Chao, J.; Liu, P.; Hu, G.; et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J. Neurosci. 2018, 38, 32–50. [Google Scholar] [CrossRef]
- Yang, L.; Han, B.; Zhang, Z.; Wang, S.; Bai, Y.; Zhang, Y.; Tang, Y.; Du, L.; Xu, L.; Wu, F.; et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 2020, 142, 556–574. [Google Scholar] [CrossRef]
- Peng, X.; Jing, P.; Chen, J.; Xu, L. The role of circular RNA HECTD1 expression in disease risk, disease severity, inflammation, and recurrence of acute ischemic stroke. J. Clin. Lab. Anal. 2019, 33, e22954. [Google Scholar] [CrossRef]
- Hori, H.; Kim, Y. Inflammation and post-traumatic stress disorder. Psychiatry Clin. Neurosci. 2019, 73, 143–153. [Google Scholar] [CrossRef]
- Gesuete, R.; Stevens, S.L.; Stenzel-Poore, M.P. Role of Circulating Immune Cells in Stroke and Preconditioning-Induced Protection. Acta Neurochir. Suppl. 2016, 121, 39–44. [Google Scholar]
- Grønberg, N.V.; Johansen, F.F.; Kristiansen, U.; Hasseldam, H. Leukocyte infiltration in experimental stroke. J. Neuroinflamm. 2013, 10, 115. [Google Scholar] [CrossRef]
- Rosell, A.; Cuadrado, E.; Ortega-Aznar, A.; Hernández-Guillamon, M.; Lo, E.H.; Montaner, J. MMP-9-positive neutrophil infiltration is associated to blood-brain barrier breakdown and basal lamina type IV collagen degradation during hemorrhagic transformation after human ischemic stroke. Stroke 2008, 39, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Mao, L.; Wang, H. Circular RNA in Acute Central Nervous System Injuries: A New Target for Therapeutic Intervention. Front. Mol. Neurosci. 2022, 15, 816182. [Google Scholar] [CrossRef] [PubMed]
- Cuartero, M.I.; Ballesteros, I.; Moraga, A.; Nombela, F.; Vivancos, J.; Hamilton, J.A.; Corbí, Á.L.; Lizasoain, I.; Moro, M.A. N2 neutrophils, novel players in brain inflammation after stroke: Modulation by the PPARγ agonist rosiglitazone. Stroke 2013, 44, 3498–3508. [Google Scholar] [CrossRef] [PubMed]
- Ducroux, C.; Di Meglio, L.; Loyau, S.; Delbosc, S.; Boisseau, W.; Deschildre, C.; Maacha, M.B.; Blanc, R.; Redjem, H.; Ciccio, G.; et al. Thrombus Neutrophil Extracellular Traps Content Impair tPA-Induced Thrombolysis in Acute Ischemic Stroke. Stroke 2018, 49, 754–757. [Google Scholar] [CrossRef]
- Herz, J.; Sabellek, P.; Lane, T.E.; Gunzer, M.; Hermann, D.M.; Doeppner, T.R. Role of Neutrophils in Exacerbation of Brain Injury after Focal Cerebral Ischemia in Hyperlipidemic Mice. Stroke 2015, 46, 2916–2925. [Google Scholar] [CrossRef]
- Zhao, S.C.; Ma, L.S.; Chu, Z.H.; Xu, H.; Wu, W.Q.; Liu, F. Regulation of microglial activation in stroke. Acta Pharmacol. Sin. 2017, 38, 445–458. [Google Scholar] [CrossRef]
- Gordon, S.; Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005, 5, 953–964. [Google Scholar] [CrossRef]
- Cai, W.; Wang, J.; Hu, M.; Chen, X.; Lu, Z.; Bellanti, J.A.; Zheng, S.G. All trans-retinoic acid protects against acute ischemic stroke by modulating neutrophil functions through STAT1 signaling. J. Neuroinflamm. 2019, 16, 175. [Google Scholar] [CrossRef]
- Kuprash, D.V.; Udalova, I.A.; Turetskaya, R.L.; Rice, N.R.; Nedospasov, S.A. Conserved kappa B element located downstream of the tumor necrosis factor alpha gene: Distinct NF-kappa B binding pattern and enhancer activity in LPS activated murine macrophages. Oncogene 1995, 11, 97–106. [Google Scholar]
- Wang, C.; Wan, H.; Wang, Q.; Sun, H.; Sun, Y.; Wang, K.; Zhang, C. Safflor Yellow B Attenuates Ischemic Brain Injury via Downregulation of Long Noncoding AK046177 and Inhibition of MicroRNA-134 Expression in Rats. Oxidative Med. Cell. Longev. 2020, 2020, 4586839. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Li, X.; Zhang, H.; Wang, L.; Wu, X.; Zhang, H.; Luo, Y. Catalytically inactive RIP1 and RIP3 deficiency protect against acute ischemic stroke by inhibiting necroptosis and neuroinflammation. Cell Death Dis. 2020, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Giladi, A.; Amit, I. Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell 2018, 172, 14–21. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Zhou, Y.; Liu, Y.; Luo, R.; Tian, C.; Chen, Q. Transcriptome-Wide Analysis of Neutrophil-Related Circ_22232 in Neuroinflammation from Ischemic Stroke Mice. Brain Sci. 2023, 13, 1283. https://doi.org/10.3390/brainsci13091283
Sun Z, Zhou Y, Liu Y, Luo R, Tian C, Chen Q. Transcriptome-Wide Analysis of Neutrophil-Related Circ_22232 in Neuroinflammation from Ischemic Stroke Mice. Brain Sciences. 2023; 13(9):1283. https://doi.org/10.3390/brainsci13091283
Chicago/Turabian StyleSun, Zheng, Youdong Zhou, Yanting Liu, Ran Luo, Chunlei Tian, and Qianxue Chen. 2023. "Transcriptome-Wide Analysis of Neutrophil-Related Circ_22232 in Neuroinflammation from Ischemic Stroke Mice" Brain Sciences 13, no. 9: 1283. https://doi.org/10.3390/brainsci13091283
APA StyleSun, Z., Zhou, Y., Liu, Y., Luo, R., Tian, C., & Chen, Q. (2023). Transcriptome-Wide Analysis of Neutrophil-Related Circ_22232 in Neuroinflammation from Ischemic Stroke Mice. Brain Sciences, 13(9), 1283. https://doi.org/10.3390/brainsci13091283





