The Potential of Gut Microbiota in Prediction of Stroke-Associated Pneumonia
Abstract
:1. Introduction
2. Methods
2.1. Study Participants
2.2. Data Collection
2.3. DNA Extraction and Sequencing
2.4. Bioinformatics and Statistical Analyses
3. Results
3.1. Baseline Characteristics
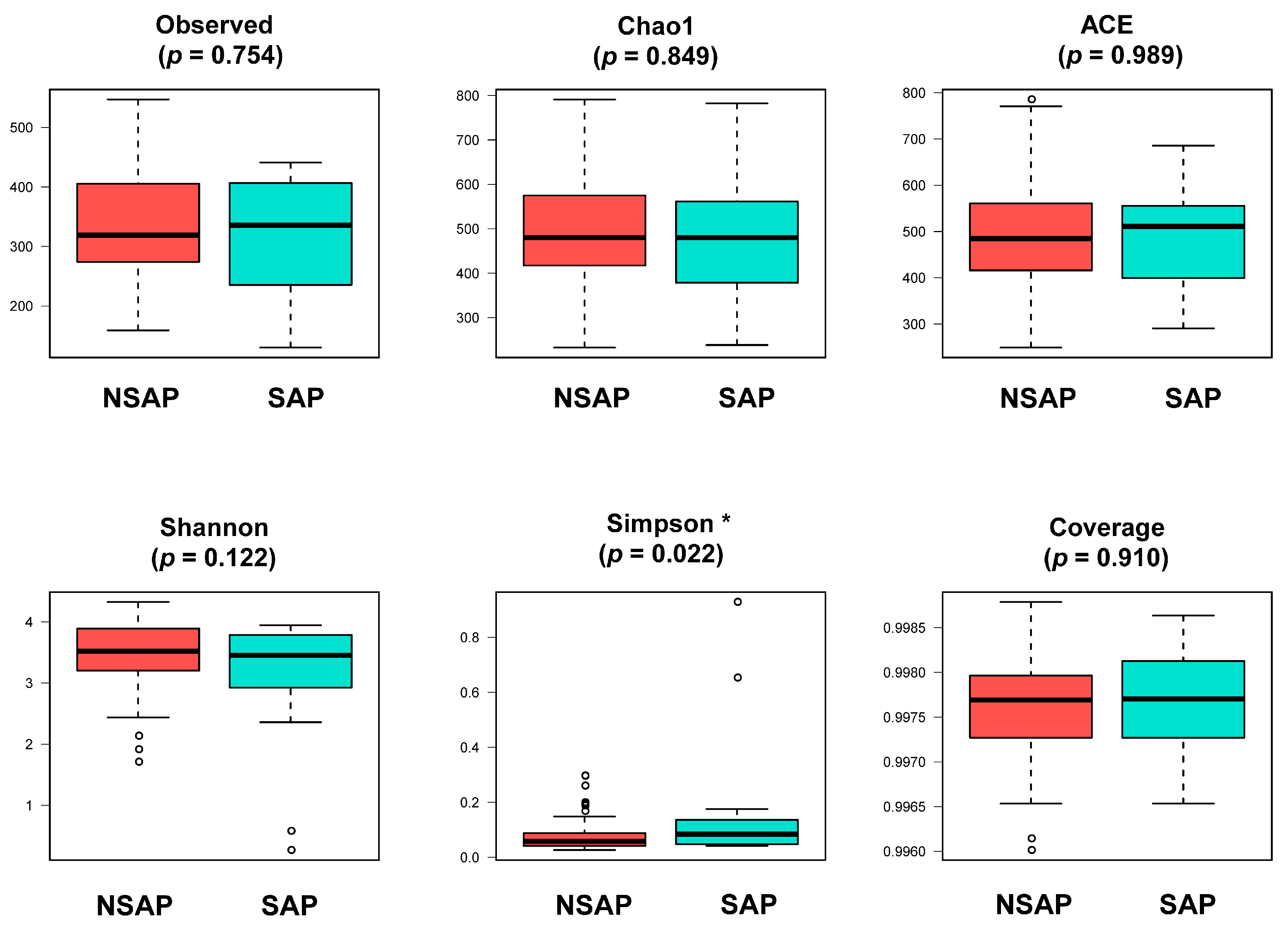
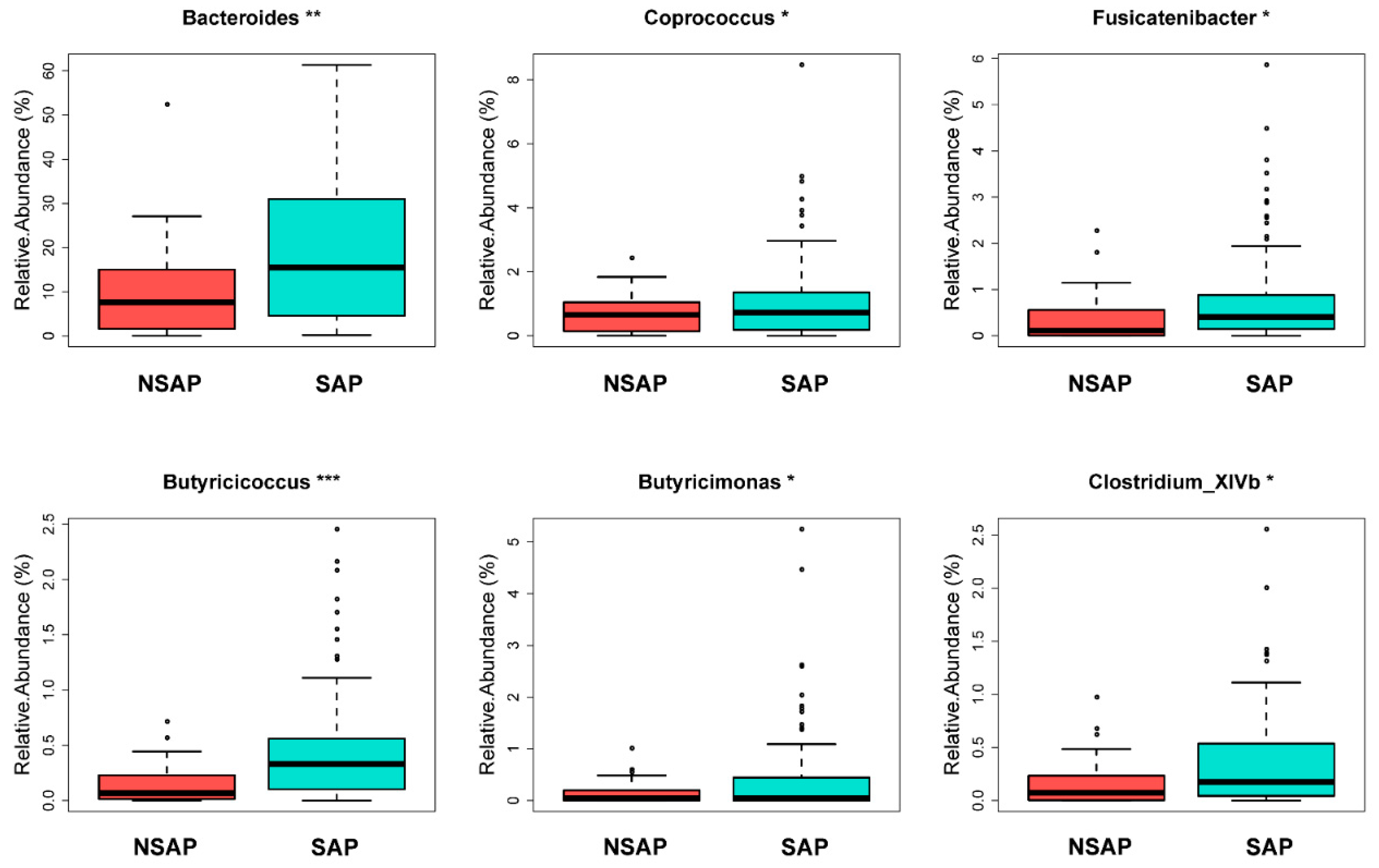
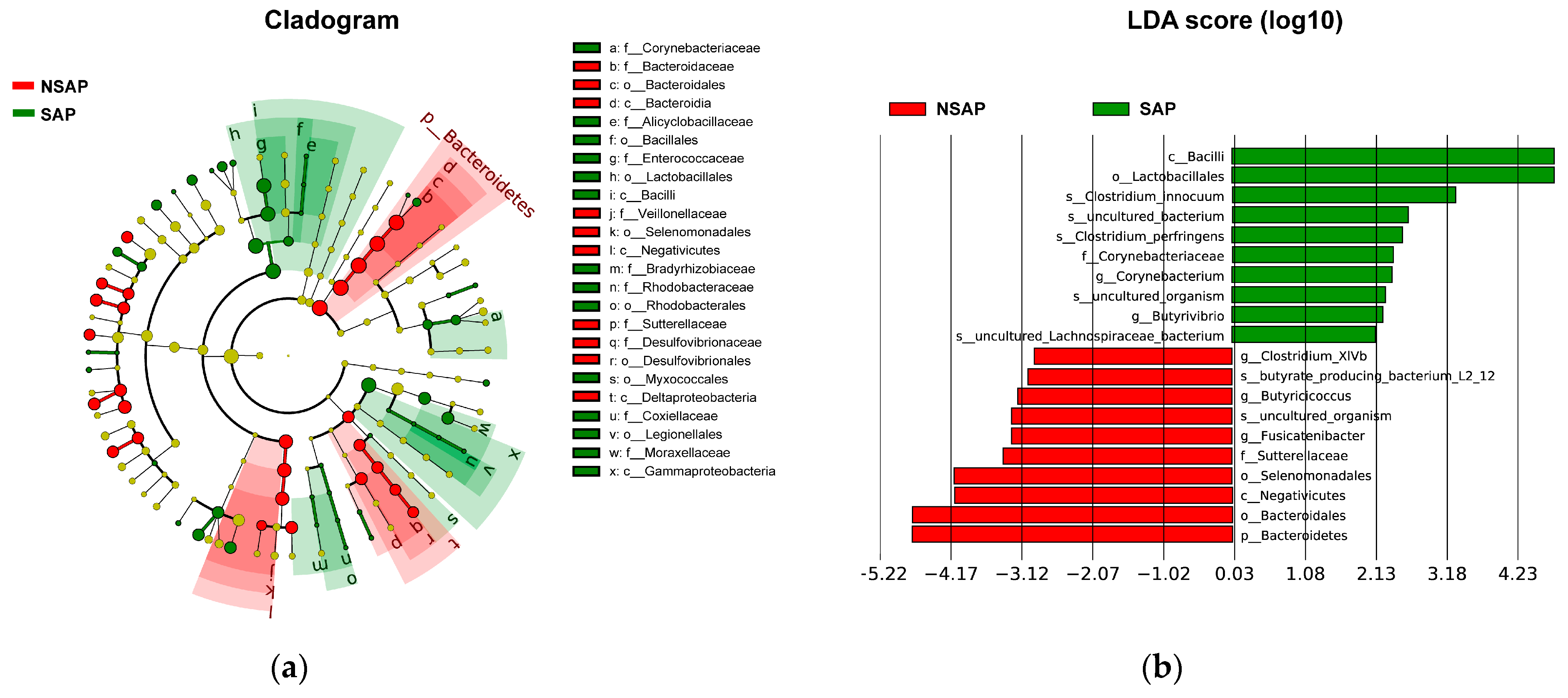
3.2. Altered Gut Microbiota in the SAP Patients
3.3. Gut Microbiota Correlated with SAP Predictive Scores
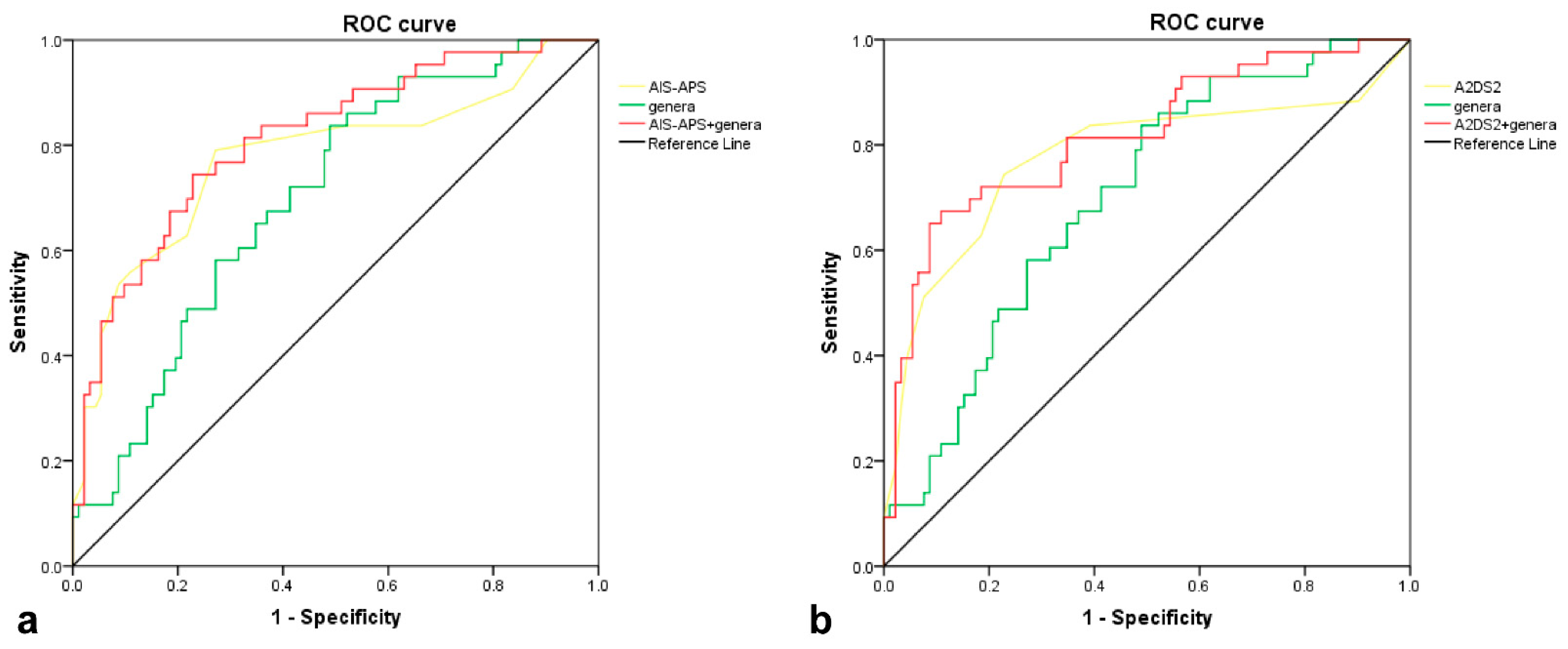
3.4. Predictive Performance of Gut Microbiota for SAP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feigin, V.L.; Norrving, B.; Mensah, G.A. Global Burden of Stroke. Circ. Res. 2017, 120, 439–448. [Google Scholar] [CrossRef]
- Westendorp, W.F.; Dames, C.; Nederkoorn, P.J.; Meisel, A. Immunodepression, Infections, and Functional Outcome in Ischemic Stroke. Stroke 2022, 53, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Faura, J.; Bustamante, A.; Miró-Mur, F.; Montaner, J. Stroke-induced immunosuppression: Implications for the prevention and prediction of post-stroke infections. J. Neuroinflamm. 2021, 18, 127. [Google Scholar] [CrossRef]
- Prass, K.; Meisel, C.; Höflich, C.; Braun, J.; Halle, E.; Wolf, T.; Ruscher, K.; Victorov, I.V.; Priller, J.; Dirnagl, U.; et al. Stroke-induced immunodeficiency promotes spontaneous bacterial infections and is mediated by sympathetic activation reversal by poststroke T helper cell type 1-like immunostimulation. J. Exp. Med. 2003, 198, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Westendorp, W.F.; Nederkoorn, P.J.; Vermeij, J.D.; Dijkgraaf, M.G.; van de Beek, D. Post-stroke infection: A systematic review and meta-analysis. BMC Neurol. 2011, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Vermeij, F.H.; Scholte op Reimer, W.J.; de Man, P.; van Oostenbrugge, R.J.; Franke, C.L.; de Jong, G.; de Kort, P.L.; Dippel, D.W.; Netherlands Stroke Survey, I. Stroke-associated infection is an independent risk factor for poor outcome after acute ischemic stroke: Data from the Netherlands Stroke Survey. Cerebrovasc. Dis. 2009, 27, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.; Hand, P. Infection after acute stroke is associated with poor short-term outcome. Acta Neurol. Scand. 2007, 115, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Hilker, R.; Poetter, C.; Findeisen, N.; Sobesky, J.; Jacobs, A.; Neveling, M.; Heiss, W.D. Nosocomial pneumonia after acute stroke: Implications for neurological intensive care medicine. Stroke 2003, 34, 975–981. [Google Scholar] [CrossRef]
- Katzan, I.L.; Cebul, R.D.; Husak, S.H.; Dawson, N.V.; Baker, D.W.J.N. The effect of pneumonia on mortality among patients hospitalized for acute stroke. Neurology 2003, 60, 620. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, O.; Kapral, M.; Hall, R.; Sllani, E.A.; Selchen, D.; Saposnik, G.J.N. Risk factors, inpatient care, and outcomes of pneumonia after ischemic stroke. Neurology 2011, 77, 1338. [Google Scholar] [CrossRef]
- André, K.; Lev, M.H.; Seyedmehdi, P.; Betensky, R.A.; Jing, Q.; Shihab, M.; Schwamm, L.H.; Jens, M.J.P.O. Hospital Acquired Pneumonia Is Linked to Right Hemispheric Peri-Insular Stroke. PLoS ONE 2013, 8, e71141. [Google Scholar]
- Chumbler, N.R.; Williams, L.S.; Wells, C.K.; Lo, A.; Nadeau, S.; Peixoto, A.J.; Gorman, M.; Boice, J.L.; Concato, J.; Bravata, D.M.J.N. Derivation and Validation of a Clinical System for Predicting Pneumonia in Acute Stroke. Neuroepidemiology 2010, 34, 193–199. [Google Scholar] [CrossRef]
- Ji, R.; Shen, H.; Pan, Y.; Wang, P.; Liu, G.; Wang, Y.; Li, H.; Wang, Y.; China National Stroke Registry, I. Novel risk score to predict pneumonia after acute ischemic stroke. Stroke 2013, 44, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Malzahn, U.; Harms, H.; Koennecke, H.C.; Berger, K.; Kalic, M.; Walter, G.; Meisel, A.; Heuschmann, P.U.; Berlin Stroke, R.; et al. Development of a clinical score (A2DS2) to predict pneumonia in acute ischemic stroke. Stroke 2012, 43, 2617–2623. [Google Scholar] [CrossRef]
- Helmy, T.A.; Abd-Elhady, A.E.; Abdou, M.J.J.o.S.; Association, C.D.t.O.J.o.N.S. Prediction of Ischemic Stroke-Associated Pneumonia: A Comparison between 3 Scores. J. Stroke Cerebrovasc. Dis. 2016, 25, 2756–2761. [Google Scholar] [CrossRef]
- Zapata-Arriaza, E.; Moniche, F.; Blanca, P.G.; Bustamante, A.; Escudero-Martinez, I.; Ucles, O.; Ollero-Ortiz, A.; Sanchez-Garcia, J.A.; Gamero, M.A.; Quesada, A.; et al. External Validation of the ISAN, A2DS2, and AIS-APS Scores for Predicting Stroke-Associated Pneumonia. J. Stroke Cerebrovasc. Dis. 2018, 27, 673–676. [Google Scholar] [CrossRef]
- Ni, J.; Shou, W.; Wu, X.; Sun, J. Prediction of stroke-associated pneumonia by the A2DS2, AIS-APS, and ISAN scores: A systematic review and meta-analysis. Expert. Rev. Respir. Med. 2021, 15, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liao, S.X.; He, Y.; Wang, S.; Xia, G.H.; Liu, F.T.; Zhu, J.J.; You, C.; Chen, Q.; Zhou, L.; et al. Dysbiosis of Gut Microbiota With Reduced Trimethylamine-N-Oxide Level in Patients With Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 2015, 4, e002699. [Google Scholar] [CrossRef]
- Yamashiro, K.; Tanaka, R.; Urabe, T.; Ueno, Y.; Yamashiro, Y.; Nomoto, K.; Takahashi, T.; Tsuji, H.; Asahara, T.; Hattori, N. Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS ONE 2017, 12, e0171521. [Google Scholar] [CrossRef]
- Diaz-Marugan, L.; Gallizioli, M.; Marquez-Kisinousky, L.; Arboleya, S.; Mastrangelo, A.; Ruiz-Jaen, F.; Pedragosa, J.; Casals, C.; Morales, F.J.; Ramos-Romero, S.; et al. Poststroke Lung Infection by Opportunistic Commensal Bacteria Is Not Mediated by Their Expansion in the Gut Microbiota. Stroke 2023, 54, 1875–1887. [Google Scholar] [CrossRef]
- Xu, K.; Gao, X.; Xia, G.; Chen, M.; Yin, J.J.G. Rapid gut dysbiosis induced by stroke exacerbates brain infarction in turn. Gut 2021, 70, 1486–1494. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Mason, L.J.; Mackin, K.E.; Srikhanta, Y.N.; Lyras, D.; Prakash, M.D.; Nurgali, K.; Venegas, A.; Hill, M.D.; Moore, R.J.; et al. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat. Med. 2016, 22, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.H.; Zhang, M.S.; Wu, Q.H.; Wang, H.D.; Zhou, H.W.; He, Y.; Yin, J. Dysbiosis of Gut Microbiota Is an Independent Risk Factor of Stroke-Associated Pneumonia: A Chinese Pilot Study. Front. Cell Infect. Microbiol. 2021, 11, 715475. [Google Scholar] [CrossRef] [PubMed]
- Haak, B.W.; Westendorp, W.F.; van Engelen, T.S.R.; Brands, X.; Brouwer, M.C.; Vermeij, J.D.; Hugenholtz, F.; Verhoeven, A.; Derks, R.J.; Giera, M.; et al. Disruptions of Anaerobic Gut Bacteria Are Associated with Stroke and Post-stroke Infection: A Prospective Case-Control Study. Transl. Stroke Res. 2021, 12, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Kishore, A.K.; Vail, A.; Chamorro, A.; Garau, J.; Hopkins, S.J.; Di Napoli, M.; Kalra, L.; Langhorne, P.; Montaner, J.; et al. Diagnosis of Stroke-Associated Pneumonia: Recommendations From the Pneumonia in Stroke Consensus Group. Stroke 2015, 46, 2335–2340. [Google Scholar] [CrossRef] [PubMed]
- de Andrade, J.B.C.; Mohr, J.P.; Timbó, F.B.; Nepomuceno, C.R.; Moreira, J.; Timbó, I.; Lima, F.O.; Silva, G.S.; Bamford, J. Oxfordshire Community Stroke Project Classification: A proposed automated algorithm. Eur. Stroke J. 2021, 6, 160–167. [Google Scholar] [CrossRef]
- Adams, H.P., Jr.; Bendixen, B.H.; Kappelle, L.J.; Biller, J.; Love, B.B.; Gordon, D.L.; Marsh, E.E., 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993, 24, 35–41. [Google Scholar] [CrossRef]
- Mehta, R.; Chinthapalli, K. Glasgow coma scale explained. BMJ (Clin. Res. Ed.) 2019, 365, l1296. [Google Scholar] [CrossRef]
- Meng, Q.; Ma, M.; Zhang, W.; Bi, Y.; Cheng, P.; Yu, X.; Fu, Y.; Chao, Y.; Ji, T.; Li, J.; et al. The gut microbiota during the progression of atherosclerosis in the perimenopausal period shows specific compositional changes and significant correlations with circulating lipid metabolites. Gut Microbes. 2021, 13, 1880220. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Gu, M.; Li, Z.; Chen, X.; Zhou, J.J.F.i.n. Gut Microbiota Dysbiosis in Acute Ischemic Stroke Associated With 3-Month Unfavorable Outcome. Front. Neurol. 2021, 12, 799222. [Google Scholar] [CrossRef]
- Li, X.J.; You, X.Y.; Wang, C.Y.; Li, X.L.; Sheng, Y.Y.; Zhuang, P.W.; Zhang, Y.J. Bidirectional Brain-gut-microbiota Axis in increased intestinal permeability induced by central nervous system injury. CNS Neurosci. Ther. 2020, 26, 783–790. [Google Scholar] [CrossRef]
- Wassenaar, T.M.; Juncos, V.A.; Zimmermann, K. Interactions between the Gut Microbiome, Lung Conditions, and Coronary Heart Disease and How Probiotics Affect These. Int. J. Mol. Sci. 2021, 22, 9700. [Google Scholar] [CrossRef]
- Chen, L.W.; Chen, P.H.; Hsu, C.M. Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock 2011, 36, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef]
- Brown, R.; Larkinson, M.; Clarke, T. Immunological design of commensal communities to treat intestinal infection and inflammation. PLOS Pathog. 2021, 17, e1009191. [Google Scholar] [CrossRef]
- Chioma, O.S.; Hesse, L.E.; Chapman, A.; Drake, W.P. Role of the Microbiome in Interstitial Lung Diseases. Front Med. 2021, 8, 595522. [Google Scholar] [CrossRef]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Robert, C.; Chassard, C.; Lawson, P.A.; Bernalier-Donadille, A. Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int. J. Syst. Evol. Microbiol. 2007, 57, 1516–1520. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Shen, M.H.; Liu, C.Y.; Pu, C.M.; Hu, J.M.; Huang, C.J. A gut butyrate-producing bacterium Butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol. Lett. 2020, 20, 327. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, G.; Ji, H.; Peng, B.; Huang, Z.; Jin, W.; Chen, X.; Guan, H.; Tang, G.; Zhang, H.; et al. Changes in the gut microbiota during and after commercial helium-oxygen saturation diving in China. Occup. Environ. Med. 2019, 76, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c(-) Patrolling Monocyte Hematopoiesis and CD8(+) T Cell Metabolism. Immunity 2018, 48, 992–1005.e1008. [Google Scholar] [CrossRef] [PubMed]
- Antunes, K.H.; Fachi, J.L.; de Paula, R.; da Silva, E.F.; Pral, L.P.; Dos Santos, A.; Dias, G.B.M.; Vargas, J.E.; Puga, R.; Mayer, F.Q.; et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019, 10, 3273. [Google Scholar] [CrossRef]
- Haak, B.W.; Littmann, E.R.; Chaubard, J.L.; Pickard, A.J.; Fontana, E.; Adhi, F.; Gyaltshen, Y.; Ling, L.; Morjaria, S.M.; Peled, J.U.; et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 2018, 131, 2978–2986. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef]
- Shimizu, K.; Yamada, T.; Ogura, H.; Mohri, T.; Kiguchi, T.; Fujimi, S.; Asahara, T.; Yamada, T.; Ojima, M.; Ikeda, M.; et al. Synbiotics modulate gut microbiota and reduce enteritis and ventilator-associated pneumonia in patients with sepsis: A randomized controlled trial. Crit. Care 2018, 22, 239. [Google Scholar] [CrossRef]
- Benakis, C.; Brea, D.; Caballero, S.; Faraco, G.; Moore, J.; Murphy, M.; Sita, G.; Racchumi, G.; Ling, L.; Pamer, E.G.; et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat. Med. 2016, 22, 516–523. [Google Scholar] [CrossRef]
- Ott, L. Adhesion properties of toxigenic corynebacteria. AIMS Microbiol. 2018, 4, 85–103. [Google Scholar] [CrossRef]
- Chia, J.H.; Wu, T.S.; Wu, T.L.; Chen, C.L.; Chuang, C.H.; Su, L.H.; Chang, H.J.; Lu, C.C.; Kuo, A.J.; Lai, H.C.; et al. Clostridium innocuum is a vancomycin-resistant pathogen that may cause antibiotic-associated diarrhoea. Clin. Microbiol. Infect. 2018, 24, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Highet, A.R.; Berry, A.M.; Bettelheim, K.A.; Goldwater, P.N.J.I.J.o.M.M. Gut microbiome in sudden infant death syndrome (SIDS) differs from that in healthy comparison babies and offers an explanation for the risk factor of prone position. Int. J. Med Microbiol. 2014, 304, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Kuo, Y.C.; Chen, M.C.; Zhang, Y.D.; Chen, C.L.; Le, P.H.; Chiu, C.H. Case-Control Study of Clostridium innocuum Infection, Taiwan. Emerg. Infect. Dis. 2022, 28, 599–607. [Google Scholar] [CrossRef]
- Zanza, C.; Romenskaya, T.; Thangathurai, D.; Ojetti, V.; Saviano, A.; Abenavoli, L.; Robba, C.; Cammarota, G.; Franceschi, F.; Piccioni, A.; et al. Microbiome in Critical Care: An Unconventional and Unknown Ally. Curr. Med. Chem. 2022, 29, 3179–3188. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, K.; Li, Z.; Chai, X.; Fu, X.; Kholodkevich, S.; Kuznetsova, T.; Chen, C.; Ren, N. Effescts of acute diclofenac exposure on intestinal histology, antioxidant defense, and microbiota in freshwater crayfish (Procambarus clarkii). Chemosphere 2021, 263, 128130. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, Y.; Tang, G.; Li, Z.; Yang, X.; Shang, X.; Huang, T.; Huang, G.; Wang, L.; Han, Y.; et al. Gut microbiota composition reflects disease progression, severity and outcome, and dysfunctional immune responses in patients with hypertensive intracerebral hemorrhage. Front. Immunol. 2022, 13, 869846. [Google Scholar] [CrossRef]
- Liu, D.D.; Chu, S.F.; Chen, C.; Yang, P.F.; Chen, N.H.; He, X. Research progress in stroke-induced immunodepression syndrome (SIDS) and stroke-associated pneumonia (SAP). Neurochem. Int. 2018, 114, 42–54. [Google Scholar] [CrossRef]
- Bourassa, M.W.; Alim, I.; Bultman, S.J.; Ratan, R.R.J.N.L. Butyrate, neuroepigenetics and the gut microbiome: Can a high fiber diet improve brain health? Neurosci. Lett. 2016, 625, 56–63. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- CNCB-NGDC. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021, 49, D18–D28. [Google Scholar] [CrossRef] [PubMed]

| Stroke-Associated Pneumonia | p Value | ||
|---|---|---|---|
| Yes (N = 43) | No (N = 92) | ||
| Age, mean (SD), y | 71.5 (10.7) | 66.3 (9.6) | 0.006 |
| Male, n (%) | 22 (51.2) | 69 (75.0) | 0.006 |
| Hypertension, n (%) | 37 (86.0) | 74 (80.4) | 0.427 |
| Diabetes mellitus, n (%) | 15 (34.9) | 35 (38.0) | 0.723 |
| Dyslipidemia, n (%) | 28 (65.1) | 53 (57.6) | 0.407 |
| Atrial fibrillation, n (%) | 19 (44.2) | 14 (15.2) | <0.001 |
| Coronary heart disease, n (%) | 5 (11.6) | 8 (8.7) | 0.591 |
| History of stroke or TIA, n (%) | 11 (25.6) | 16 (17.4) | 0.268 |
| COPD, n (%) | 1 (2.3) | 2 (2.2) | 0.687 |
| Dysphagia, n (%) | 17 (39.5) | 8 (8.7) | <0.001 |
| Speech disorders, n (%) | 37 (86.0) | 61 (66.3) | 0.017 |
| OCSP type, n (%) | <0.001 | ||
| PACI or LACI | 31 (72.1) | 87 (94.6) | |
| TACI or POCI | 12 (27.9) | 5 (5.4) | |
| Fasting Glucose, median (IQR), mmol/L, (n = 130) | 5.8 (5.0–7.9) | 5.1 (4.4–6.6) | 0.004 |
| C-reactive protein, median (IQR), ug/mL, (n = 72) | 9.4 (3.6–34.6) | 2.5 (1.0–5.6) | <0.001 |
| WBC, mean (SD), ×109/L | 9.2 (3.2) | 7.4 (2.6) | 0.001 |
| Neutrophil, median (IQR), ×109/L | 5.6 (5.0–8.8) | 4.4 (3.3–6.5) | <0.001 |
| Lymphocyte, mean (SD), ×109/L | 1.5 (0.9) | 1.7 (0.7) | 0.147 |
| NLR, median (IQR) | 4.8 (3.1–8.4) | 2.7 (2.0–4.2) | <0.001 |
| Baseline NIHSS score, median (IQR) | 11.0 (3.0–16.0) | 3.0 (2.0–4.0) | <0.001 |
| Baseline GCS, median (IQR) | 15 (8–15) | 15 (15–15) | <0.001 |
| Baseline mRS, median (IQR) | 0 (0–0) | 0 (0–0) | 0.216 |
| Thrombolysis, n (%) | 17 (39.5) | 29 (31.5) | 0.360 |
| AIS-APS, median (IQR) | 11.0 (7.0–20.0) | 5.0 (3.0–7.0) | <0.001 |
| A2DS2, median (IQR) | 5.0 (2.0–7.0) | 1.0 (1.0–2.0) | <0.001 |
| NRI (Continuous) | IDI | ||||
|---|---|---|---|---|---|
| Models | Variables | Estimate (95% CI) | p Value | Estimate (95% CI) | p Value |
| AIS-APS | +genera | 0.333 (−0.003–0.700) | 0.052 | 0.038 (0.006–0.070) | 0.018 |
| A2DS2 | +genera | 0.575 (0.245–0.906) | <0.001 | 0.043 (0.012–0.075) | 0.007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Gu, M.; Sun, H.; Chen, X.; Zhou, J.; Zhang, Y. The Potential of Gut Microbiota in Prediction of Stroke-Associated Pneumonia. Brain Sci. 2023, 13, 1217. https://doi.org/10.3390/brainsci13081217
Li Z, Gu M, Sun H, Chen X, Zhou J, Zhang Y. The Potential of Gut Microbiota in Prediction of Stroke-Associated Pneumonia. Brain Sciences. 2023; 13(8):1217. https://doi.org/10.3390/brainsci13081217
Chicago/Turabian StyleLi, Zhongyuan, Mengmeng Gu, Huanhuan Sun, Xiangliang Chen, Junshan Zhou, and Yingdong Zhang. 2023. "The Potential of Gut Microbiota in Prediction of Stroke-Associated Pneumonia" Brain Sciences 13, no. 8: 1217. https://doi.org/10.3390/brainsci13081217
APA StyleLi, Z., Gu, M., Sun, H., Chen, X., Zhou, J., & Zhang, Y. (2023). The Potential of Gut Microbiota in Prediction of Stroke-Associated Pneumonia. Brain Sciences, 13(8), 1217. https://doi.org/10.3390/brainsci13081217






