Resveratrol Promotes Autophagy to Improve neuronal Injury in Parkinson’s Disease by Regulating SNHG1/miR-128-3p/SNCA Axis
Abstract
1. Background
2. Methods
2.1. Cell Culture and Treatments
2.2. Cell Transfection
2.3. Cell Counting Kit-8 (CCK-8) Assay
2.4. Western Blot
2.5. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
- SNHG1 (F): ACGTTGGAACCGAAGAGAGCSNHG1 (R): GCAGCTGAATTCCCCAGGATmiR-128-3p (F): GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAGAGmiR-128-3p (R): CGGTCACAGTGAACCGGTSNCA (F): TGTAGGCTCCAAAACCAAGGSNCA (R): TGTCAGGATCCACAGGCATAβ-actin (F): TGGCACCACACCTTCTACAAβ-actin (R): CCAGAGGCGTACAGGGATAG
2.6. Dual Luciferase Activity Assay
2.7. RNA Immunoprecipitation (RIP) Assay
2.8. Data Analysis
3. Results
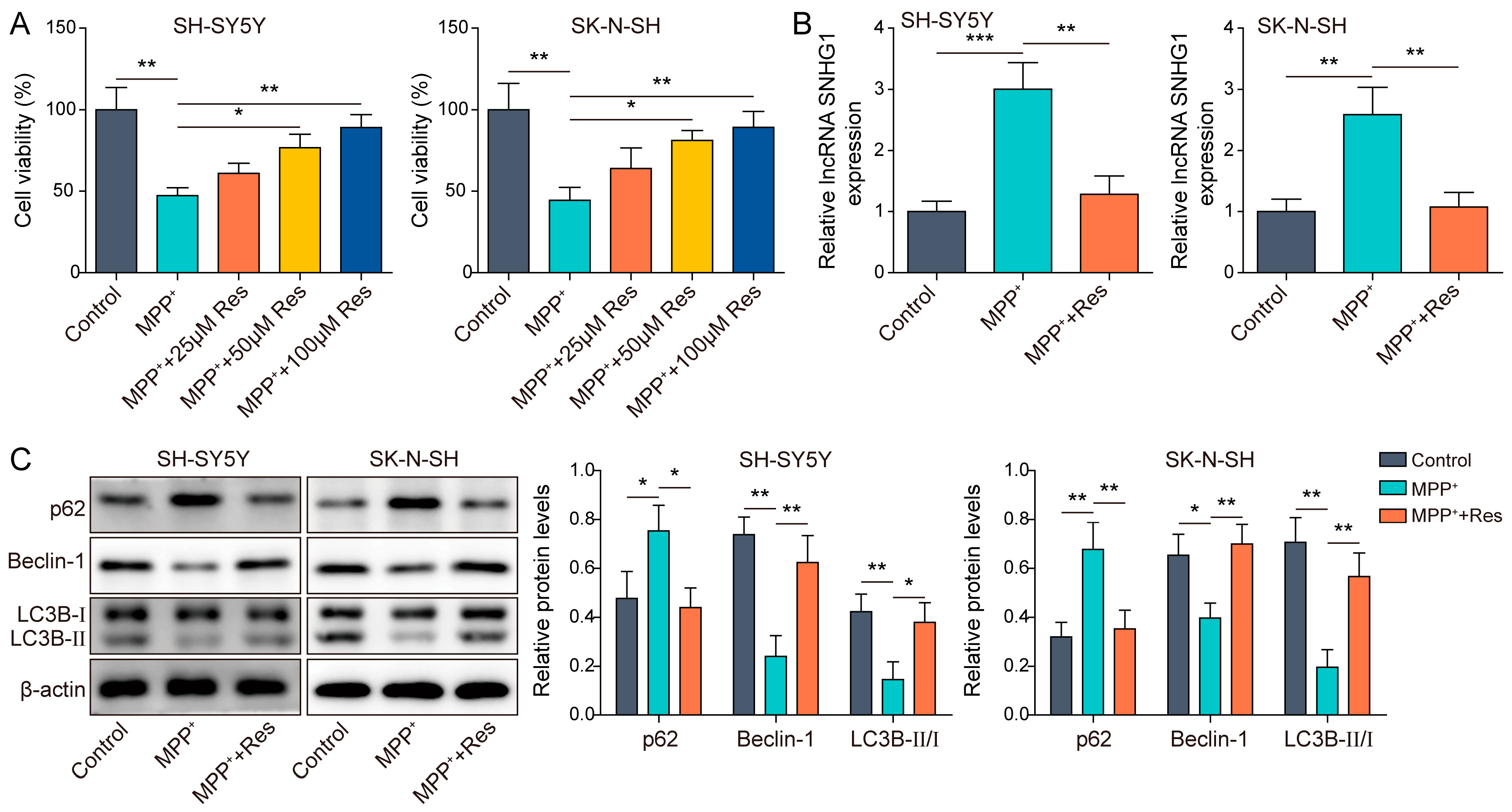
3.1. RES Promoted Autophagy in MPP+-Induced Human Neuroblastoma Cells
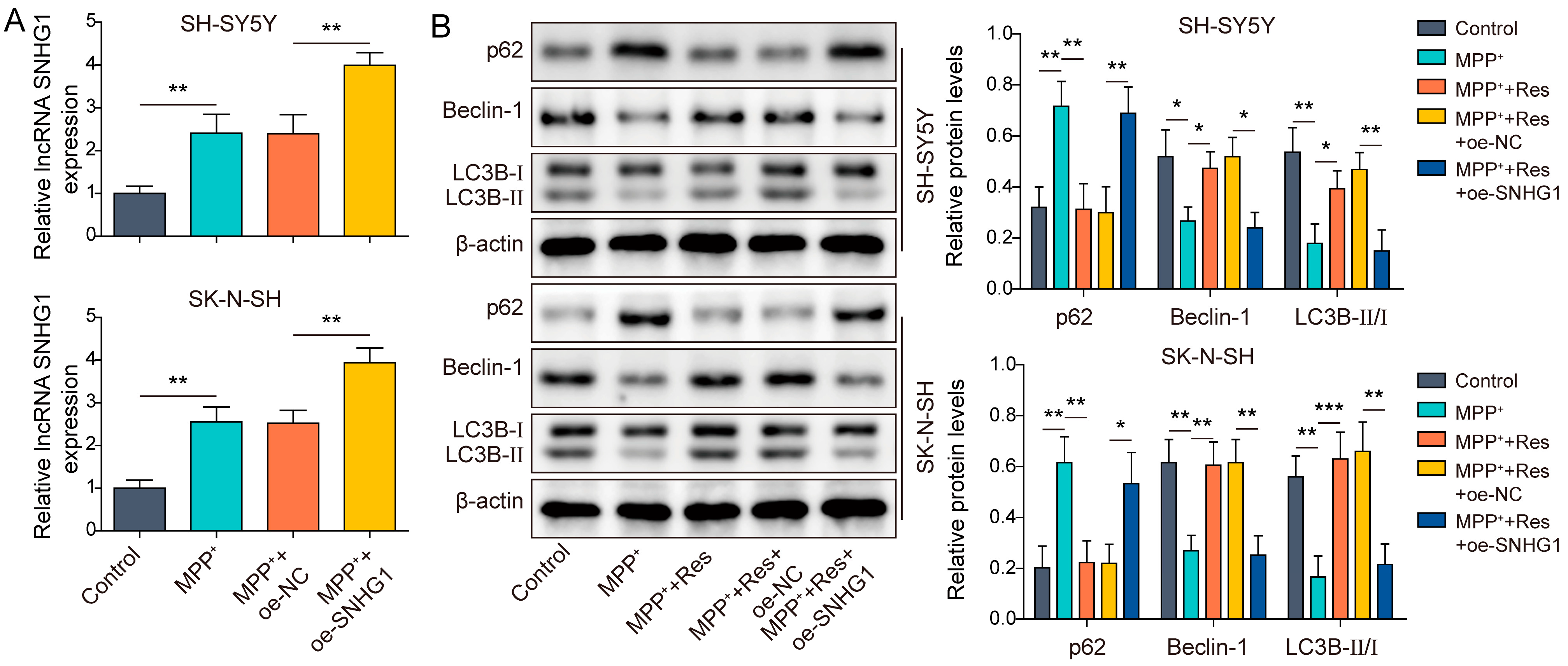
3.2. SNHG1 Overexpression Reversed the Influences of RES on Autophagy in Human Neuroblastoma
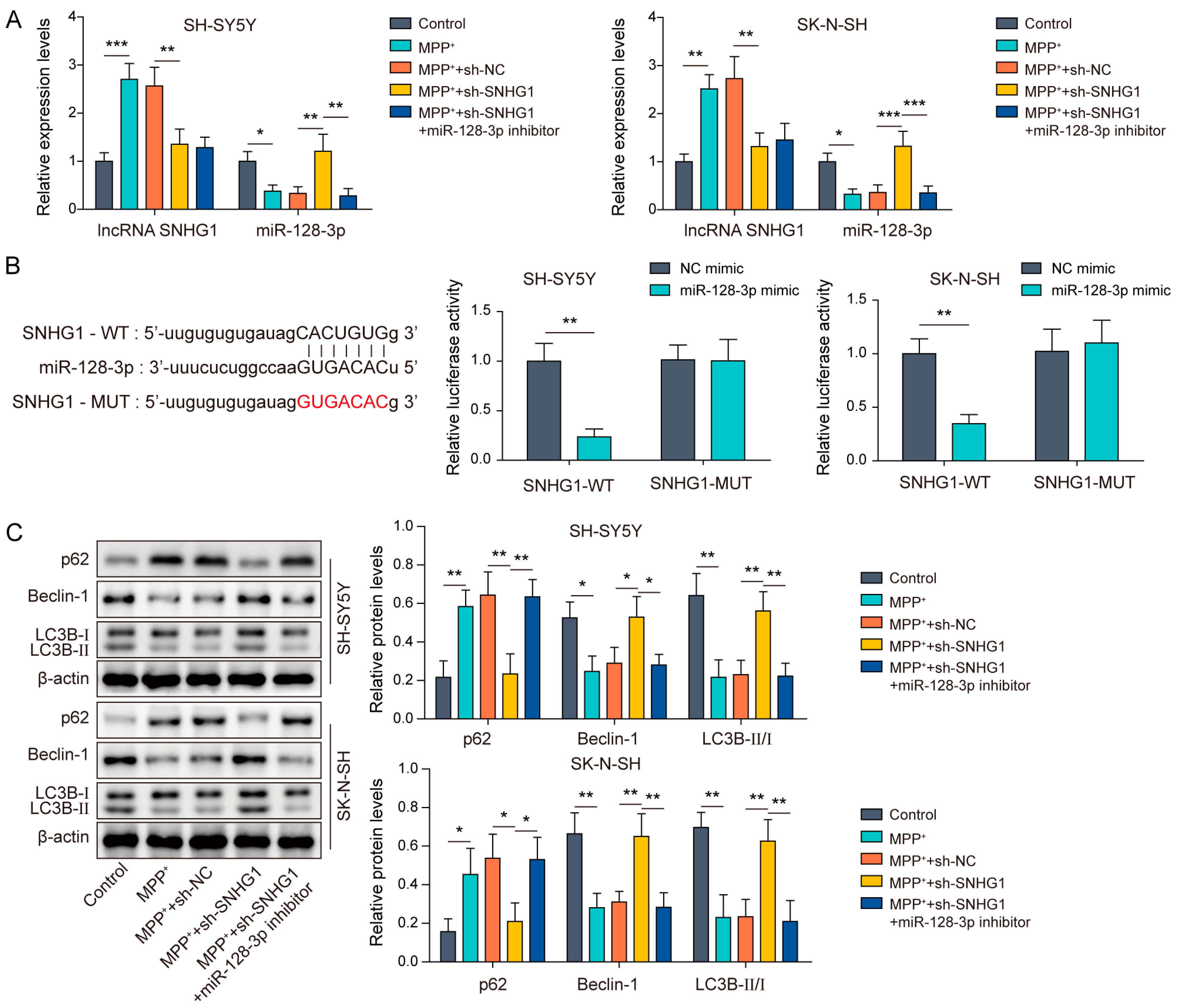
3.3. SNHG1 Knockdown Promoted Autophagy in MPP+-Induced Human Neuroblastoma Cells through Elevating miR-128-3p Expression
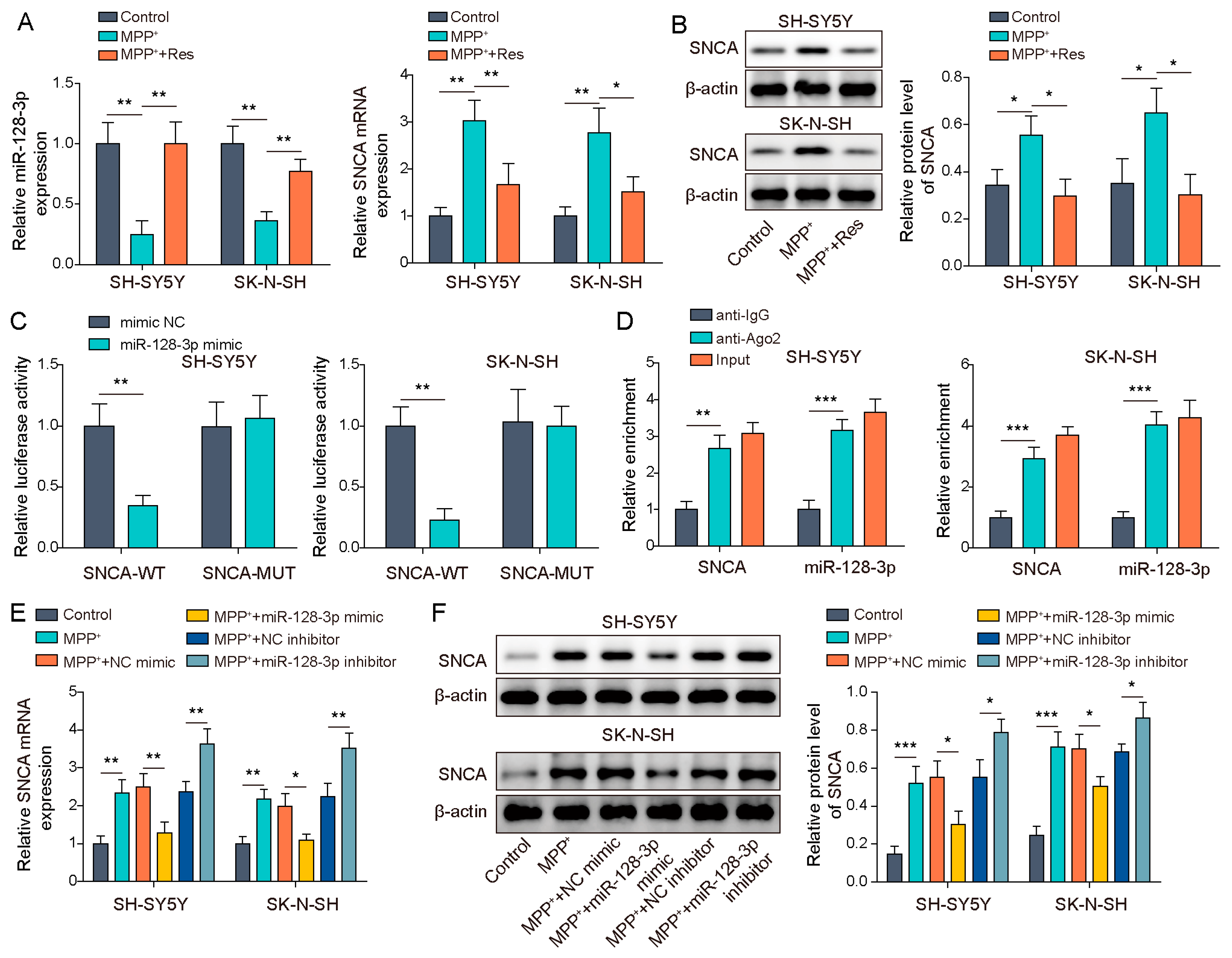
3.4. MiR-128-3p Targeted SNCA
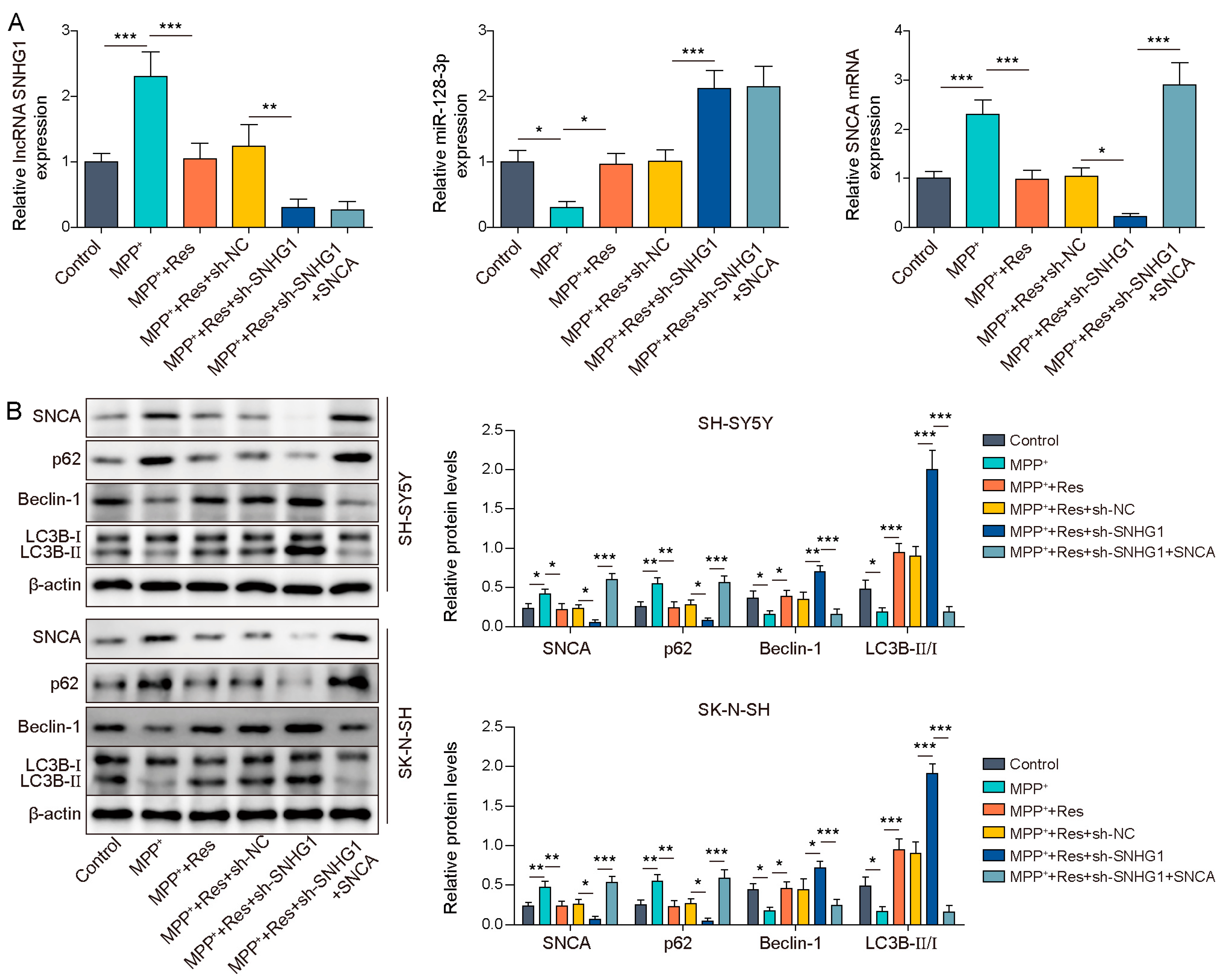
3.5. SNHG1 Knockdown Prompted Cell Autophagy in PD through Reducing SNCA Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silva, R.C.; Domingues, H.S.; Salgado, A.J.; Teixeira, F.G. From regenerative strategies to pharmacological approaches: Can we fine-tune treatment for Parkinson’s disease? Neural Regen. Res. 2022, 17, 933–936. [Google Scholar] [CrossRef]
- Dorsey, E.R.; Elbaz, A.; Nichols, E.; Abbasi, N.; Abd-Allah, F.; Abdelalim, A.; Adsuar, J.C.; Ansha, M.G.; Brayne, C.; Choi, J.Y.; et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Costello, H.; Berry, A.J.; Reeves, S.; Weil, R.S.; Joyce, E.M.; Howard, R.; Roiser, J.P. Disrupted reward processing in Parkinson’s disease and its relationship with dopamine state and neuropsychiatric syndromes: A systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2022, 93, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, M.S. Autophagy—A key player in cellular and body metabolism. Nat. Rev. Endocrinol. 2014, 10, 322–337. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. Embo J. 2021, 40, e108863. [Google Scholar] [CrossRef] [PubMed]
- Lizama, B.N.; Chu, C.T. Neuronal autophagy and mitophagy in Parkinson’s disease. Mol. Asp. Med. 2021, 82, 100972. [Google Scholar] [CrossRef]
- Su, Y.; Li, M.; Wang, Q.; Xu, X.; Qin, P.; Huang, H.; Zhang, Y.; Zhou, Y.; Yan, J. Inhibition of the TLR/NF-κB Signaling Pathway and Improvement of Autophagy Mediates Neuroprotective Effects of Plumbagin in Parkinson’s Disease. Oxidative Med. Cell. Longev. 2022, 2022, 1837278. [Google Scholar] [CrossRef]
- Bonam, S.R.; Tranchant, C.; Muller, S. Autophagy-Lysosomal Pathway as Potential Therapeutic Target in Parkinson’s Disease. Cells 2021, 10, 3547. [Google Scholar] [CrossRef]
- Ren, B.; Kwah, M.X.; Liu, C.; Ma, Z.; Shanmugam, M.K.; Ding, L.; Xiang, X.; Ho, P.C.; Wang, L.; Ong, P.S.; et al. Resveratrol for cancer therapy: Challenges and future perspectives. Cancer Lett. 2021, 515, 63–72. [Google Scholar] [CrossRef]
- Diaz-Gerevini, G.T.; Repossi, G.; Dain, A.; Tarres, M.C.; Das, U.N.; Eynard, A.R. Beneficial action of resveratrol: How and why? Nutrition 2016, 32, 174–178. [Google Scholar] [CrossRef]
- Ko, J.H.; Sethi, G.; Um, J.Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The Role of Resveratrol in Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef]
- Lin, K.L.; Lin, K.J.; Wang, P.W.; Chuang, J.H.; Lin, H.Y.; Chen, S.D.; Chuang, Y.C.; Huang, S.T.; Tiao, M.M.; Chen, J.B.; et al. Resveratrol provides neuroprotective effects through modulation of mitochondrial dynamics and ERK1/2 regulated autophagy. Free Radic. Res. 2018, 52, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.C.; Liu, S.C.; Hsu, Y.F.; Wu, R.M. The role of noncoding RNAs in Parkinson’s disease: Biomarkers and associations with pathogenic pathways. J. Biomed. Sci. 2021, 28, 78. [Google Scholar] [CrossRef]
- Song, Z.; Xie, B. LncRNA OIP5-AS1 reduces α-synuclein aggregation and toxicity by targeting miR-126 to activate PLK2 in human neuroblastoma SH-SY5Y cells. Neurosci. Lett. 2021, 740, 135482. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Ye, Y.; Mao, H.; Yao, L.; Sun, X.; Wang, B.; Zhang, H.; Xie, L.; Zhang, H.; Zhang, Y.; et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222/p27/mTOR pathway in Parkinson’s disease. Exp. Cell Res. 2019, 384, 111614. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, N.; Lv, C.; Li, N.; Li, X.; Li, W. lncRNA SNHG1 Knockdown Alleviates Amyloid-β-Induced Neuronal Injury by Regulating ZNF217 via Sponging miR-361-3p in Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2020, 77, 85–98. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, L.; Liu, J.; Jin, Y.; Lin, Z.; Du, S.; Fu, Z.; Chen, T.; Qin, Y.; Sui, F.; et al. HIF-1α/microRNA-128-3p axis protects hippocampal neurons from apoptosis via the Axin1-mediated Wnt/β-catenin signaling pathway in Parkinson’s disease models. Aging 2020, 12, 4067–4081. [Google Scholar] [CrossRef]
- Rocha, E.M.; De Miranda, B.; Sanders, L.H. Alpha-synuclein: Pathology, mitochondrial dysfunction and neuroinflammation in Parkinson’s disease. Neurobiol. Dis. 2018, 109, 249–257. [Google Scholar] [CrossRef]
- Li, R.; Lu, Y.; Zhang, Q.; Liu, W.; Yang, R.; Jiao, J.; Liu, J.; Gao, G.; Yang, H. Piperine promotes autophagy flux by P2RX4 activation in SNCA/α-synuclein-induced Parkinson disease model. Autophagy 2022, 18, 559–575. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Dehay, B.; Bourdenx, M.; Gorry, P.; Przedborski, S.; Vila, M.; Hunot, S.; Singleton, A.; Olanow, C.W.; Merchant, K.M.; Bezard, E.; et al. Targeting α-synuclein for treatment of Parkinson’s disease: Mechanistic and therapeutic considerations. Lancet Neurol. 2015, 14, 855–866. [Google Scholar] [CrossRef]
- Gao, Q.; Chen, R.; Wu, L.; Huang, Q.; Wang, X.X.; Tian, Y.Y.; Zhang, Y.D. Angiotensin-(1-7) reduces α-synuclein aggregation by enhancing autophagic activity in Parkinson’s disease. Neural Regen. Res. 2022, 17, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Kung, H.C.; Lin, K.J.; Kung, C.T.; Lin, T.K. Oxidative Stress, Mitochondrial Dysfunction, and Neuroprotection of Polyphenols with Respect to Resveratrol in Parkinson’s Disease. Biomedicines 2021, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Moors, T.E.; Hoozemans, J.J.; Ingrassia, A.; Beccari, T.; Parnetti, L.; Chartier-Harlin, M.C.; van de Berg, W.D. Therapeutic potential of autophagy-enhancing agents in Parkinson’s disease. Mol. Neurodegener. 2017, 12, 11. [Google Scholar] [CrossRef]
- Cerri, S.; Blandini, F. Role of Autophagy in Parkinson’s Disease. Curr. Med. Chem. 2019, 26, 3702–3718. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, P.; Yin, H.; Wang, X.; Lv, J.; Yuan, J.; Zhu, J.; Wang, Y. Rapamycin reverses ferroptosis by increasing autophagy in MPTP/MPP(+)-induced models of Parkinson’s disease. Neural Regen. Res. 2023, 18, 2514–2519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Gao, J.Z.; Zhang, X.N.; Dou, K.X.; Shi, W.D.; Xie, A.M. P2X4 receptor participates in autophagy regulation in Parkinson’s disease. Neural Regen. Res. 2021, 16, 2505–2511. [Google Scholar] [CrossRef]
- Breuss, J.M.; Atanasov, A.G.; Uhrin, P. Resveratrol and Its Effects on the Vascular System. Int. J. Mol. Sci. 2019, 20, 1523. [Google Scholar] [CrossRef]
- Nunes, S.; Danesi, F.; Del Rio, D.; Silva, P. Resveratrol and inflammatory bowel disease: The evidence so far. Nutr. Res. Rev. 2018, 31, 85–97. [Google Scholar] [CrossRef]
- Sawda, C.; Moussa, C.; Turner, R.S. Resveratrol for Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2017, 1403, 142–149. [Google Scholar] [CrossRef]
- Lin, T.K.; Chen, S.D.; Chuang, Y.C.; Lin, H.Y.; Huang, C.R.; Chuang, J.H.; Wang, P.W.; Huang, S.T.; Tiao, M.M.; Chen, J.B.; et al. Resveratrol partially prevents rotenone-induced neurotoxicity in dopaminergic SH-SY5Y cells through induction of heme oxygenase-1 dependent autophagy. Int. J. Mol. Sci. 2014, 15, 1625–1646. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Sui, R.; Zhang, Z. Administration of resveratrol improved Parkinson’s disease-like phenotype by suppressing apoptosis of neurons via modulating the MALAT1/miR-129/SNCA signaling pathway. J. Cell. Biochem. 2019, 120, 4942–4951. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Kuo, H.C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 49. [Google Scholar] [CrossRef]
- Vargas-Medrano, J.; Yang, B.; Garza, N.T.; Segura-Ulate, I.; Perez, R.G. Up-regulation of protective neuronal MicroRNAs by FTY720 and novel FTY720-derivatives. Neurosci. Lett. 2019, 690, 178–180. [Google Scholar] [CrossRef]
- Miller, D.B.; O’Callaghan, J.P. Biomarkers of Parkinson’s disease: Present and future. Metab. Clin. Exp. 2015, 64, S40–S46. [Google Scholar] [CrossRef] [PubMed]
- Minakaki, G.; Menges, S.; Kittel, A.; Emmanouilidou, E.; Schaeffner, I.; Barkovits, K.; Bergmann, A.; Rockenstein, E.; Adame, A.; Marxreiter, F.; et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 2018, 14, 98–119. [Google Scholar] [CrossRef]
- Song, J.X.; Lu, J.H.; Liu, L.F.; Chen, L.L.; Durairajan, S.S.; Yue, Z.; Zhang, H.Q.; Li, M. HMGB1 is involved in autophagy inhibition caused by SNCA/α-synuclein overexpression: A process modulated by the natural autophagy inducer corynoxine B. Autophagy 2014, 10, 144–154. [Google Scholar] [CrossRef]
- Nguyen, H.D. Resveratrol, Endocrine Disrupting Chemicals, Neurodegenerative Diseases and Depression: Genes, Transcription Factors, microRNAs, and Sponges Involved. Neurochem. Res. 2023, 48, 604–624. [Google Scholar] [CrossRef]
- Su, J.; Zhang, J.; Bao, R.; Xia, C.; Zhang, Y.; Zhu, Z.; Lv, Q.; Qi, Y.; Xue, J. Mitochondrial dysfunction and apoptosis are attenuated through activation of AMPK/GSK-3β/PP2A pathway in Parkinson’s disease. Eur. J. Pharm. 2021, 907, 174202. [Google Scholar] [CrossRef]
- Li, C.; Wang, Z.; Lei, H.; Zhang, D. Recent progress in nanotechnology-based drug carriers for resveratrol delivery. Drug Deliv. 2023, 30, 2174206. [Google Scholar] [CrossRef]
- Xie, X.; Jin, X.; Huang, J.; Yi, J.; Li, X.; Huang, Z.; Lin, Q.; Guo, B. High resveratrol-loaded microcapsules with trehalose and OSA starch as the wall materials: Fabrication, characterization, and evaluation. Int. J. Biol. Macromol. 2023, 242, 124825. [Google Scholar] [CrossRef] [PubMed]
- González-Sarrías, A.; Iglesias-Aguirre, C.E.; Cortés-Martín, A.; Vallejo, F.; Cattivelli, A.; Del Pozo-Acebo, L.; Del Saz, A.; López de Las Hazas, M.C.; Dávalos, A.; Espín, J.C. Milk-Derived Exosomes as Nanocarriers to Deliver Curcumin and Resveratrol in Breast Tissue and Enhance Their Anticancer Activity. Int. J. Mol. Sci. 2022, 23, 2860. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, D.-F.; Qi, H.-P.; Zhang, W.-N.; Sang, W.-X. Resveratrol Promotes Autophagy to Improve neuronal Injury in Parkinson’s Disease by Regulating SNHG1/miR-128-3p/SNCA Axis. Brain Sci. 2023, 13, 1124. https://doi.org/10.3390/brainsci13081124
Shen D-F, Qi H-P, Zhang W-N, Sang W-X. Resveratrol Promotes Autophagy to Improve neuronal Injury in Parkinson’s Disease by Regulating SNHG1/miR-128-3p/SNCA Axis. Brain Sciences. 2023; 13(8):1124. https://doi.org/10.3390/brainsci13081124
Chicago/Turabian StyleShen, Dong-Fang, Hui-Ping Qi, Wei-Na Zhang, and Wen-Xu Sang. 2023. "Resveratrol Promotes Autophagy to Improve neuronal Injury in Parkinson’s Disease by Regulating SNHG1/miR-128-3p/SNCA Axis" Brain Sciences 13, no. 8: 1124. https://doi.org/10.3390/brainsci13081124
APA StyleShen, D.-F., Qi, H.-P., Zhang, W.-N., & Sang, W.-X. (2023). Resveratrol Promotes Autophagy to Improve neuronal Injury in Parkinson’s Disease by Regulating SNHG1/miR-128-3p/SNCA Axis. Brain Sciences, 13(8), 1124. https://doi.org/10.3390/brainsci13081124






