Correlation of Serum N-Acetylneuraminic Acid with the Risk of Moyamoya Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Data Collection
2.3. Statistical Analyses
3. Results
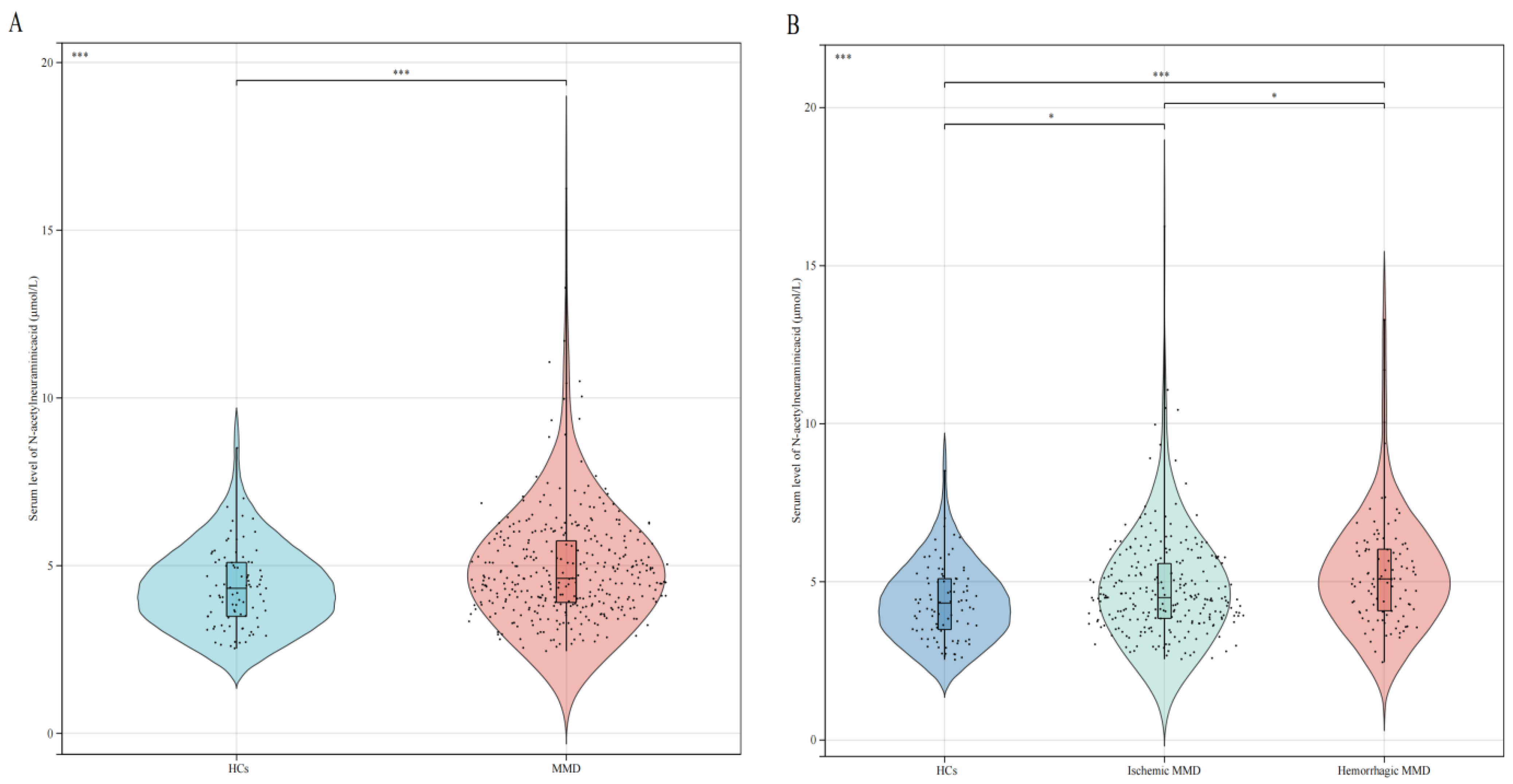
3.1. Clinical and Laboratory Characteristics of the Study Participants
3.2. Characteristics of MMD Patients with Different Neu5Ac Levels
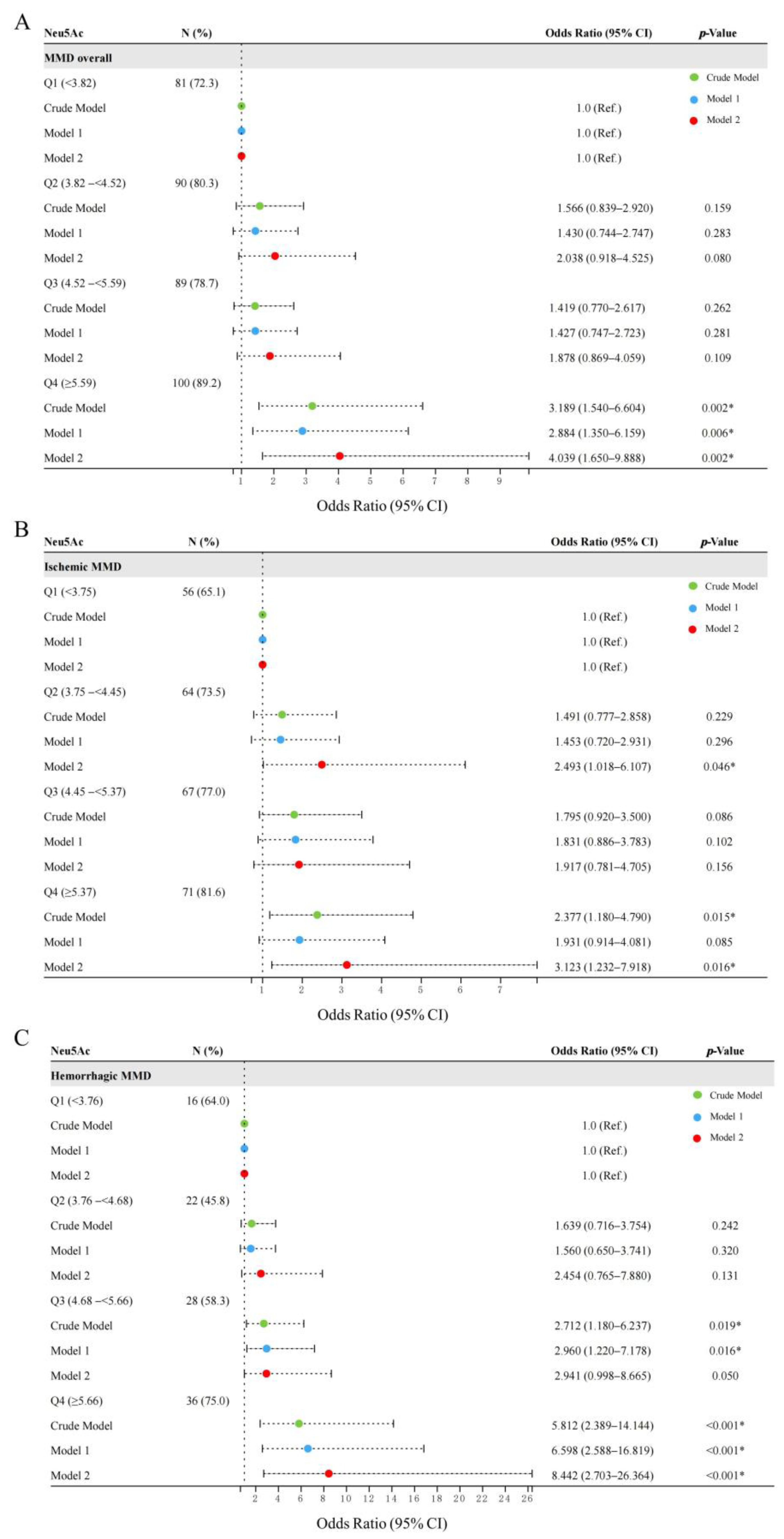
3.3. Association of Neu5Ac Levels with Risk of MMD and MMD Subtypes
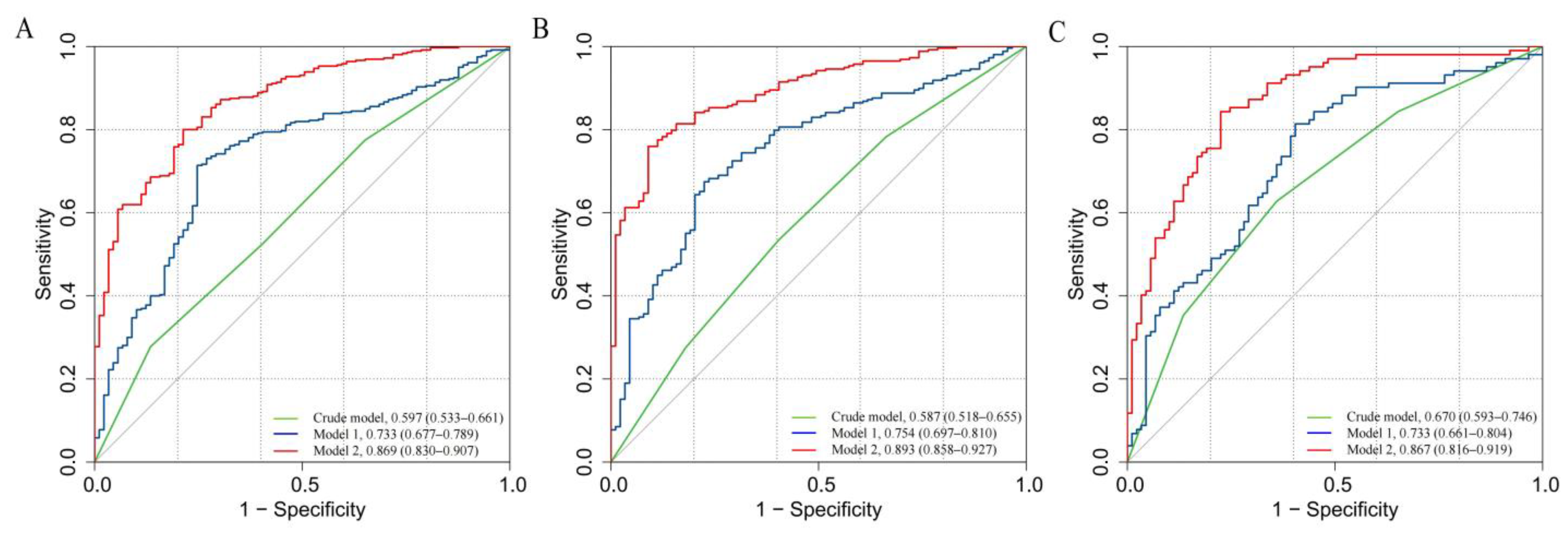
3.4. Predictive Capacity of Neu5Ac in Clinical Risk Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demartini, Z., Jr.; Teixeira, B.C.; Koppe, G.L.; Gatto, L.A.M.; Roman, A.; Munhoz, R.P. Moyamoya disease and syndrome: A review. Radiol. Bras. 2022, 55, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S. Moyamoya Disease: Epidemiology, Clinical Features, and Diagnosis. J. Stroke 2016, 18, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, S.; Houkin, K. Moyamoya disease: Current concepts and future perspectives. Lancet Neurol. 2008, 7, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Oh, C.W.; Bang, J.S.; Kim, J.E.; Cho, W.S. Moyamoya Disease: Treatment and Outcomes. J. Stroke 2016, 18, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Vinjé, S.; Stroes, E.; Nieuwdorp, M.; Hazen, S.L. The gut microbiome as novel cardio-metabolic target: The time has come! Eur. Heart J. 2014, 35, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Kitai, T.; Hazen, S.L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Ishisaka, E.; Watanabe, A.; Murai, Y.; Shirokane, K.; Matano, F.; Tsukiyama, A.; Baba, E.; Nakagawa, S.; Tamaki, T.; Mizunari, T.; et al. Role of RNF213 polymorphism in defining quasi-moyamoya disease and definitive moyamoya disease. Neurosurg. Focus 2021, 51, E2. [Google Scholar] [CrossRef]
- Liu, X.J.; Zhang, D.; Wang, S.; Zhao, Y.L.; Teo, M.; Wang, R.; Cao, Y.; Ye, X.; Kang, S.; Zhao, J.Z. Clinical features and long-term outcomes of moyamoya disease: A single-center experience with 528 cases in China. J. Neurosurg. 2015, 122, 392–399. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.; Yu, L.; Duan, R.; Ma, Y.; Ge, P.; Zhang, D.; Zhang, Y.; Wang, R.; Wang, S.; et al. The Association of the RNF213 p.R4810K Polymorphism with Quasi-Moyamoya Disease and a Review of the Pertinent Literature. World Neurosurg. 2017, 99, 701–708.e701. [Google Scholar] [CrossRef]
- Omori, K.; Katakami, N.; Arakawa, S.; Yamamoto, Y.; Ninomiya, H.; Takahara, M.; Matsuoka, T.A.; Tsugawa, H.; Furuno, M.; Bamba, T.; et al. Identification of Plasma Inositol and Indoxyl Sulfate as Novel Biomarker Candidates for Atherosclerosis in Patients with Type 2 Diabetes. -Findings from Metabolome Analysis Using GC/MS. J. Atheroscler. Thromb. 2020, 27, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Weng, R.; Jiang, Z.; Gu, Y. Noncoding RNA as Diagnostic and Prognostic Biomarkers in Cerebrovascular Disease. Oxid. Med. Cell Longev. 2022, 2022, 8149701. [Google Scholar] [CrossRef] [PubMed]
- Kooner, A.S.; Yu, H.; Chen, X. Synthesis of N-Glycolylneuraminic Acid (Neu5Gc) and Its Glycosides. Front. Immunol. 2019, 10, 2004. [Google Scholar] [CrossRef] [PubMed]
- Perdicchio, M.; Ilarregui, J.M.; Verstege, M.I.; Cornelissen, L.A.; Schetters, S.T.; Engels, S.; Ambrosini, M.; Kalay, H.; Veninga, H.; den Haan, J.M.; et al. Sialic acid-modified antigens impose tolerance via inhibition of T-cell proliferation and de novo induction of regulatory T cells. Proc. Natl. Acad. Sci. USA 2016, 113, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Fougerat, A.; Pan, X.; Smutova, V.; Heveker, N.; Cairo, C.W.; Issad, T.; Larrivée, B.; Medin, J.A.; Pshezhetsky, A.V. Neuraminidase 1 activates insulin receptor and reverses insulin resistance in obese mice. Mol. Metab. 2018, 12, 76–88. [Google Scholar] [CrossRef]
- White, E.J.; Gyulay, G.; Lhoták, Š.; Szewczyk, M.M.; Chong, T.; Fuller, M.T.; Dadoo, O.; Fox-Robichaud, A.E.; Austin, R.C.; Trigatti, B.L.; et al. Sialidase down-regulation reduces non-HDL cholesterol, inhibits leukocyte transmigration, and attenuates atherosclerosis in ApoE knockout mice. J. Biol. Chem. 2018, 293, 14689–14706. [Google Scholar] [CrossRef]
- Kawecki, C.; Bocquet, O.; Schmelzer, C.E.H.; Heinz, A.; Ihling, C.; Wahart, A.; Romier, B.; Bennasroune, A.; Blaise, S.; Terryn, C.; et al. Identification of CD36 as a new interaction partner of membrane NEU1: Potential implication in the pro-atherogenic effects of the elastin receptor complex. Cell. Mol. Life Sci. 2019, 76, 791–807. [Google Scholar] [CrossRef]
- Kawanishi, K.; Dhar, C.; Do, R.; Varki, N.; Gordts, P.; Varki, A. Human species-specific loss of CMP-N-acetylneuraminic acid hydroxylase enhances atherosclerosis via intrinsic and extrinsic mechanisms. Proc. Natl. Acad. Sci. USA 2019, 116, 16036–16045. [Google Scholar] [CrossRef]
- Guidelines for diagnosis and treatment of moyamoya disease (spontaneous occlusion of the circle of Willis). Neurol. Med. Chir. 2012, 52, 245–266. [CrossRef]
- Zeng, C.; Ge, P.; Liu, C.; Yu, X.; Zhai, Y.; Liu, W.; He, Q.; Li, J.; Liu, X.; Wang, J.; et al. Association of circulating branched-chain amino acids with risk of moyamoya disease. Front. Nutr. 2022, 9, 994286. [Google Scholar] [CrossRef]
- Wang, H.; Qing, X.; Wang, H.; Gu, Y. Association between Platelet to Neutrophil Ratio (PNR) and Clinical Outcomes in STEMI Patients after Successful pPCI: A Secondary Analysis Based on a Cohort Study. Cardiovasc. Ther. 2022, 2022, 2022657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, T.T.; Li, Y.; Li, J.; Fan, Y.; Huang, F.Q.; Cai, Y.Y.; Ma, G.; Liu, J.F.; Chen, Q.Q.; et al. Functional Metabolomics Characterizes a Key Role for N-Acetylneuraminic Acid in Coronary Artery Diseases. Circulation 2018, 137, 1374–1390. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Xu, X.; Zhang, M.; Shi, Y.; Gu, G.; Liu, W.; Class, B.; Ciccone, C.; Gahl, W.A.; Huizing, M.; et al. Quantitation of cytidine-5′-monophospho-N-acetylneuraminic acid in human leukocytes using LC-MS/MS: Method development and validation. Biomed. Chromatogr. 2020, 34, e4735. [Google Scholar] [CrossRef] [PubMed]
- Greenland, P. Comments on ‘Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond’ by M.J. Pencina, R.B. D’Agostino Sr, R.B. D’Agostino Jr, R.S. Vasan, Statistics in Medicine (DOI: 10.1002/sim.2929). Stat. Med. 2008, 27, 188–190. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Ganna, A.; Salihovic, S.; Sundström, J.; Broeckling, C.D.; Hedman, A.K.; Magnusson, P.K.; Pedersen, N.L.; Larsson, A.; Siegbahn, A.; Zilmer, M.; et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014, 10, e1004801. [Google Scholar] [CrossRef]
- Rizza, S.; Copetti, M.; Rossi, C.; Cianfarani, M.A.; Zucchelli, M.; Luzi, A.; Pecchioli, C.; Porzio, O.; Di Cola, G.; Urbani, A.; et al. Metabolomics signature improves the prediction of cardiovascular events in elderly subjects. Atherosclerosis 2014, 232, 260–264. [Google Scholar] [CrossRef]
- Fan, Y.; Li, Y.; Chen, Y.; Zhao, Y.J.; Liu, L.W.; Li, J.; Wang, S.L.; Alolga, R.N.; Yin, Y.; Wang, X.M.; et al. Comprehensive Metabolomic Characterization of Coronary Artery Diseases. J. Am. Coll. Cardiol. 2016, 68, 1281–1293. [Google Scholar] [CrossRef]
- Park, S.S. Post-Glycosylation Modification of Sialic Acid and Its Role in Virus Pathogenesis. Vaccines 2019, 7, 171. [Google Scholar] [CrossRef]
- Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009, 19, 507–514. [Google Scholar] [CrossRef]
- Röhrig, C.H.; Choi, S.S.; Baldwin, N. The nutritional role of free sialic acid, a human milk monosaccharide, and its application as a functional food ingredient. Crit. Rev. Food Sci. Nutr. 2017, 57, 1017–1038. [Google Scholar] [CrossRef]
- Hu, W.; Xie, J.; Zhu, T.; Meng, G.; Wang, M.; Zhou, Z.; Guo, F.; Chen, H.; Wang, Z.; Wang, S.; et al. Serum N-Acetylneuraminic Acid Is Associated with Atrial Fibrillation and Left Atrial Enlargement. Cardiol. Res. Pract. 2020, 2020, 1358098. [Google Scholar] [CrossRef]
- Li, C.; Zhao, M.; Xiao, L.; Wei, H.; Wen, Z.; Hu, D.; Yu, B.; Sun, Y.; Gao, J.; Shen, X.; et al. Prognostic Value of Elevated Levels of Plasma N-Acetylneuraminic Acid in Patients With Heart Failure. Circ. Heart Fail. 2021, 14, e008459. [Google Scholar] [CrossRef]
- Li, M.N.; Qian, S.H.; Yao, Z.Y.; Ming, S.P.; Shi, X.J.; Kang, P.F.; Zhang, N.R.; Wang, X.J.; Gao, D.S.; Gao, Q.; et al. Correlation of serum N-Acetylneuraminic acid with the risk and prognosis of acute coronary syndrome: A prospective cohort study. BMC Cardiovasc. Disord. 2020, 20, 404. [Google Scholar] [CrossRef]
- Masuda, J.; Ogata, J.; Yutani, C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke 1993, 24, 1960–1967. [Google Scholar] [CrossRef]
- Altay, M.; Karakoç, M.A.; Çakır, N.; Yılmaz Demirtaş, C.; Cerit, E.T.; Aktürk, M.; Ateş, İ.; Bukan, N.; Arslan, M. Serum Total Sialic Acid Level is Elevated in Hypothyroid Patients as an Atherosclerotic Risk Factor. J. Clin. Lab. Anal. 2017, 31, e22034. [Google Scholar] [CrossRef]
- Rajendiran, K.S.; Ananthanarayanan, R.H.; Satheesh, S.; Rajappa, M. Elevated levels of serum sialic acid and high-sensitivity C-reactive protein: Markers of systemic inflammation in patients with chronic heart failure. Br. J. Biomed. Sci. 2014, 71, 29–32. [Google Scholar] [CrossRef]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef]
- Strubl, S.; Schubert, U.; Kühnle, A.; Rebl, A.; Ahmadvand, N.; Fischer, S.; Preissner, K.T.; Galuska, S.P. Polysialic acid is released by human umbilical vein endothelial cells (HUVEC) in vitro. Cell Biosci. 2018, 8, 64. [Google Scholar] [CrossRef]
- Chiodelli, P.; Urbinati, C.; Paiardi, G.; Monti, E.; Rusnati, M. Sialic acid as a target for the development of novel antiangiogenic strategies. Future Med. Chem. 2018, 10, 2835–2854. [Google Scholar] [CrossRef]
- Rusnati, M.; Urbinati, C.; Tanghetti, E.; Dell’Era, P.; Lortat-Jacob, H.; Presta, M. Cell membrane GM1 ganglioside is a functional coreceptor for fibroblast growth factor 2. Proc. Natl. Acad. Sci. USA 2002, 99, 4367–4372. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; McCarthy, J.; Ladisch, S. Membrane ganglioside enrichment lowers the threshold for vascular endothelial cell angiogenic signaling. Cancer Res. 2006, 66, 10408–10414. [Google Scholar] [CrossRef] [PubMed]
- Chandler, K.B.; Leon, D.R.; Meyer, R.D.; Rahimi, N.; Costello, C.E. Site-Specific N-Glycosylation of Endothelial Cell Receptor Tyrosine Kinase VEGFR-2. J. Proteome Res. 2017, 16, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Chiodelli, P.; Rezzola, S.; Urbinati, C.; Federici Signori, F.; Monti, E.; Ronca, R.; Presta, M.; Rusnati, M. Contribution of vascular endothelial growth factor receptor-2 sialylation to the process of angiogenesis. Oncogene 2017, 36, 6531–6541. [Google Scholar] [CrossRef] [PubMed]
- Imamaki, R.; Ogawa, K.; Kizuka, Y.; Komi, Y.; Kojima, S.; Kotani, N.; Honke, K.; Honda, T.; Taniguchi, N.; Kitazume, S. Glycosylation controls cooperative PECAM-VEGFR2-β3 integrin functions at the endothelial surface for tumor angiogenesis. Oncogene 2018, 37, 4287–4299. [Google Scholar] [CrossRef] [PubMed]



| Variables | Healthy Controls (n = 89) | MMD Patients (n = 360) | p-Value |
|---|---|---|---|
| Age (years), mean ± SD | 39.81 ± 11.57 | 41.54 ± 10.32 | 0.200 |
| Gender (female), n (%) | 52 (58.4) | 210 (58.3) | 0.987 |
| Clinical features, mean ± SD | |||
| Heart rate, bpm | 77.79 ± 9.73 | 78.54 ± 6.42 | 0.486 |
| SBP, mmHg | 123.64 ± 11.77 | 132.34 ± 12.83 | <0.001 * |
| DBP, mmHg | 78.46 ± 8.35 | 81.84 ± 9.32 | 0.002 * |
| BMI, kg/m2 | 23.96 ± 3.39 | 25.45 ± 4.52 | 0.004 * |
| History of risk factors, n (%) | |||
| Current smoking | 2 (2.2) | 71 (19.7) | <0.001 * |
| Current alcohol abuse | 0 (0.0) | 42 (11.7) | 0.001 * |
| Diabetes mellitus | 0 (0.0) | 59 (16.4) | <0.001 * |
| Hypertension | 0 (0.0) | 131 (36.4) | <0.001 * |
| Hyperlipidemia | 0 (0.0) | 54 (15.0) | <0.001 * |
| Laboratory results, median (IQR) | |||
| WBC count, 109/L | 6.03 (1.88) | 6.81 (2.48) | <0.001 * |
| LY count, 109/L | 1.91 (0.71) | 1.92 (0.87) | 0.247 |
| NEUT count, 109/L | 3.44 (1.62) | 4.21 (1.88) | <0.001 * |
| MONO count, 109/L | 0.35 (0.14) | 0.35 (0.17) | 0.295 |
| PLT count, 109/L | 233.00 (87.00) | 248.00 (75.75) | 0.290 |
| ALT, U/L | 18.40 (10.05) | 21.05 (17.58) | 0.045 * |
| ALP, U/L | 57.00 (21.50) | 69.30 (26.88) | <0.001 * |
| Glu, mmol/L | 5.04 (0.62) | 5.11 (1.07) | 0.237 |
| Urea, mmol/L | 4.70 (1.85) | 4.55 (1.80) | 0.208 |
| Cr, μmol/L | 57.7 (19.20) | 54.65 (20.65) | 0.146 |
| TG, mmol/L | 0.87 (0.62) | 1.20 (0.80) | <0.001 * |
| TC, mmol/L | 4.62 (0.98) | 4.23 (1.28) | <0.001 * |
| HDL-C, mmol/L | 1.53 (0.41) | 1.30 (0.37) | <0.001 * |
| LDL-C, mmol/L | 2.69 (0.87) | 2.40 (1.13) | <0.001 * |
| ApoA, g/L | 1.39 (0.28) | 1.29 (0.30) | <0.001 * |
| ApoB, g/L | 0.77 (0.27) | 0.82 (0.27) | 0.318 |
| Hcy, μmol/L | 10.62 (3.97) | 12.00 (5.78) | 0.001 * |
| PNR | 67.42 (37.22) | 60.17 (31.13) | 0.001 * |
| SII | 414.33 (289.40) | 536.13 (383.84) | <0.001 * |
| MHR | 0.23 (0.11) | 0.28 (0.18) | <0.001 * |
| Neu5Ac, μmol/L | 4.33 (1.63) | 4.62 (1.85) | 0.001 * |
| Variables | Health Controls (n = 89) | Ischemic MMD (n = 258) | p-Value | Hemorrhagic MMD (n = 102) | p-Value |
|---|---|---|---|---|---|
| Age (years) mean ± SD | 39.81 ± 11.57 | 41.53 ± 10.14 | 0.214 | 41.55 ± 10.82 | 0.284 |
| Gender (female), n (%) | 52 (58.4) | 142 (55.0) | 0.621 | 68 (66.7) | 0.294 |
| Clinical features, mean ± SD | |||||
| Heart rate, bpm | 77.79 ± 9.73 | 78.25 ± 6.60 | 0.678 | 79.29 ± 5.89 | 0.205 |
| SBP, mmHg | 123.64 ± 11.77 | 133.48 ± 12.78 | <0.001 * | 129.45 ± 12.54 | 0.001 * |
| DBP, mmHg | 78.46 ± 8.35 | 82.39 ± 9.39 | 0.001 * | 80.46 ± 9.03 | 0.116 |
| BMI, kg/m2 | 23.96 ± 3.39 | 25.93 ± 4.60 | <0.001 * | 24.27 ± 4.12 | 0.581 |
| History of risk factors, n (%) | |||||
| Current smoking | 2 (2.2) | 54 (20.9) | <0.001 * | 17 (16.7) | 0.001 * |
| Current alcohol abuse | 0 (0.0) | 34 (13.2) | <0.001 * | 8 (7.8) | 0.008 * |
| Diabetes mellitus | 0 (0.0) | 55 (21.3) | <0.001 * | 4 (3.9) | 0.125 |
| Hypertension | 0 (0.0) | 101 (39.1) | <0.001 * | 30 (29.4) | <0.001 * |
| Hyperlipidemia | 0 (0.0) | 45 (17.4) | <0.001 * | 9 (8.8) | 0.004 * |
| Laboratory results, median (IQR) | |||||
| WBC count, 109/L | 6.03 (1.88) | 7.00 (2.42) | <0.001 * | 6.43 (2.48) | 0.021 * |
| LY count, 109/L | 1.91 (0.71) | 2.04 (0.87) | 0.021 * | 1.70 (0.83) | 0.106 |
| NEUT count, 109/L | 3.44 (1.62) | 4.31 (1.83) | 0.158 | 3.88 (1.85) | 0.004 * |
| MONO count, 109/L | 0.35 (0.14) | 0.36 (0.17) | <0.001 * | 0.35 (0.17) | 0.991 |
| PLT count, 109/L | 233.00 (87.00) | 249.50 (71.75) | 0.156 | 243.50 (75.75) | 0.993 |
| ALT, U/L | 18.40 (10.05) | 21.90 (19.90) | 0.014 * | 18.80 (15.20) | 0.611 |
| ALP, U/L | 57.00 (21.50) | 68.80 (26.75) | <0.001 * | 69.45 (26.35) | <0.001 * |
| Glu, mmol/L | 5.04 (0.62) | 5.17 (1.14) | 0.015 * | 4.91 (0.68) | 0.068 |
| Urea, mmol/L | 4.70 (1.85) | 4.50 (1.80) | 0.177 | 4.60 (2.03) | 0.465 |
| Cr, μmol/L | 57.7 (19.20) | 55.65 (21.05) | 0.279 | 52.95 (21.63) | 0.063 |
| TG, mmol/L | 0.87 (0.62) | 1.22 (0.79) | <0.001 * | 1.13 (0.82) | 0.012 * |
| TC, mmol/L | 4.62 (0.98) | 4.10 (1.33) | <0.001 * | 4.35 (1.16) | 0.170 |
| HDL-C, mmol/L | 1.53 (0.41) | 1.27 (0.38) | <0.001 * | 1.34 (0.35) | 0.001 * |
| LDL-C, mmol/L | 2.69 (0.87) | 2.24 (1.14) | <0.001 * | 2.55 (1.06) | 0.716 |
| ApoA, g/L | 1.39 (0.28) | 1.29 (0.31) | <0.001 * | 1.30 (0.28) | 0.004 * |
| ApoB, g/L | 0.77 (0.27) | 0.82 (0.27) | 0.692 | 0.83 (0.30) | 0.043 * |
| Hcy, μmol/L | 10.62 (3.97) | 12.03 (6.33) | 0.001 * | 11.95 (4.94) | 0.005 * |
| PNR | 67.42 (37.22) | 60.17 (28.09) | <0.001 * | 60.29 (39.24) | 0.016 * |
| SII | 414.33 (289.40) | 532.68 (350.76) | 0.001 * | 541.80 (444.31) | 0.003 * |
| MHR | 0.23 (0.11) | 0.29 (0.18) | <0.001 * | 0.24 (0.16) | 0.088 |
| Neu5Ac, μmol/L | 4.33 (1.63) | 4.50 (1.76) | 0.019 * | 5.09 (1.97) | <0.001 * |
| Variables | Low Neu5Ac (2.45–4.62, n = 180) | High Neu5Ac (4.62–16.25, n = 180) | p-Value |
|---|---|---|---|
| Age (years), mean ± SD | 41.38 ± 10.35 | 41.69 ± 10.32 | 0.771 |
| Gender (female), n (%) | 113 (62.8) | 97 (53.9) | 0.109 |
| Clinical features, mean ± SD | |||
| Heart rate, bpm | 77.8 ± 5.68 | 79.29 ± 7.02 | 0.028 * |
| SBP, mmHg | 131.77 ± 13.27 | 132.9 ± 12.38 | 0.405 |
| DBP, mmHg | 80.98 ± 9.22 | 82.71 ± 9.36 | 0.078 |
| BMI, kg/m2 | 25.27 ± 4.77 | 25.64 ± 4.27 | 0.431 |
| RNF213 p.R4810K, n (%) | |||
| Wild type | 145 (81.9) | 141 (81.5) | 0.919 |
| Mutant | 32 (18.1) | 32 (18.5) | |
| History of risk factors, n (%) | |||
| Current smoking | 30 (16.7) | 41 (22.8) | 0.185 |
| Current alcohol abuse | 15 (8.3) | 27 (15.0) | 0.070 |
| Diabetes mellitus | 29 (16.1) | 30 (16.7) | 0.887 |
| Hypertension | 62 (34.4) | 69 (38.3) | 0.511 |
| Hyperlipidemia | 29 (16.1) | 25 (13.9) | 0.658 |
| Laboratory results, median (IQR) | |||
| WBC count, 109/L | 6.85 (2.54) | 6.81 (2.43) | 0.917 |
| LY count, 109/L | 1.88 (0.88) | 1.99 (0.85) | 0.061 |
| NEUT count, 109/L | 4.29 (1.80) | 4.19 (1.94) | 0.468 |
| MONO count, 109/L | 0.34 (0.16) | 0.37 (0.17) | 0.237 |
| PLT count, 109/L | 247.50 (74.25) | 248.50 (85.25) | 0.433 |
| ALT, U/L | 21.15 (18.08) | 21.00 (18.03) | 0.687 |
| ALP, U/L | 68.30 (28.38) | 69.95 (24.72) | 0.721 |
| Glu, mmol/L | 5.13 (1.14) | 5.06 (0.98) | 0.979 |
| Urea, mmol/L | 4.45 (1.88) | 4.60 (1.95) | 0.425 |
| Cr, μmol/L | 53.55 (18.80) | 56.75 (22.08) | 0.047 * |
| TG, mmol/L | 1.20 (0.74) | 1.19 (0.83) | 0.936 |
| TC, mmol/L | 4.03 (1.30) | 4.26 (1.22) | 0.040 * |
| HDL-C, mmol/L | 1.31 (0.39) | 1.29 (0.33) | 0.495 |
| LDL-C, mmol/L | 2.24 (1.11) | 2.43 (1.23) | 0.018 * |
| ApoA, g/L | 1.30 (0.30) | 1.28 (0.31) | 0.308 |
| ApoB, g/L | 0.79 (0.27) | 0.86 (0.28) | 0.017 * |
| Hcy, μmol/L | 12.05 (6.32) | 12.00 (5.38) | 0.919 |
| PNR | 58.90 (28.77) | 60.80 (36.26) | 0.213 |
| SII | 544.14 (434.77) | 525.64 (344.73) | 0.219 |
| MHR | 0.26 (0.18) | 0.29 (0.18) | 0.376 |
| Neu5Ac, μmol/L | 3.90 (0.83) | 5.74 (1.27) | <0.001 * |
| Clinical type, n (%) | 0.002 * | ||
| Ischemic type | 142 (78.9) | 116 (64.4) | |
| Hemorrhagic type | 38 (21.1) | 64 (35.6) | |
| Admission mRS, n (%) | 0.859 | ||
| 0–2 | 163 (90.6) | 162 (90.0) | |
| 3–5 | 17 (9.4) | 18 (10.0) | |
| Suzuki stage, n (%) | 0.754 | ||
| 1–2 | 48 (26.7) | 52 (28.9) | |
| 3–4 | 95 (52.8) | 88 (48.9) | |
| 5–6 | 37 (20.6) | 40 (22.2) |
| Variables | Neu5Ac | p-Value | Kendall’s Tau-b Coefficient | p-Value | |||
|---|---|---|---|---|---|---|---|
| Q1 (2.45–3.89, n = 90) | Q2 (3.89–4.62, n = 90) | Q3 (4.62–5.75, n = 90) | Q4 (5.75–16.25, n = 90) | ||||
| Age (years), mean ± SD | 40.32 ± 9.94 | 42.43 ± 10.70 | 41.82 ± 11.24 | 41.57 ± 9.37 | 0.522 | 0.022 | 0.579 |
| Gender (female), n (%) | 55 (61.1) | 58 (64.4) | 49 (54.4) | 48 (53.3) | 0.379 | 0.069 | 0.152 |
| Clinical features, mean ± SD | |||||||
| Heart rate, bpm | 77.51 ± 5.75 | 78.09 ± 5.63 | 79.64 ± 7.56 | 78.93 ± 6.45 | 0.054 | 0.070 | 0.090 |
| SBP, mmHg | 130.78 ± 12.29 | 132.77 ± 14.17 | 133.43 ± 12.41 | 132.37 ± 12.40 | 0.370 | 0.044 | 0.277 |
| DBP, mmHg | 80.78 ± 8.97 | 81.18 ± 9.50 | 82.47 ± 9.60 | 82.96 ± 9.16 | 0.076 | 0.081 | 0.046 * |
| BMI, kg/m2 | 24.55 ± 4.18 | 25.98 ± 5.21 | 25.38 ± 4.41 | 25.91 ± 4.14 | 0.104 | 0.076 | 0.055 |
| RNF213 p.R4810K, n (%) | 0.477 | −0.020 | 0.682 | ||||
| Wild type | 68 (77.30) | 77 (86.5) | 71 (81.6) | 70 (81.4) | |||
| Mutant | 20 (22.7) | 12 (13.5) | 16 (18.4) | 16 (18.6) | |||
| History of risk factors, n (%) | |||||||
| Current smoking | 18 (20.0) | 12 (13.3) | 19 (21.1) | 22 (24.4) | 0.305 | 0.054 | 0.261 |
| Current alcohol abuse | 8 (8.9) | 7 (7.8) | 12 (13.3) | 15 (16.7) | 0.252 | 0.092 | 0.057 |
| Diabetes mellitus | 13 (14.4) | 16 (17.8) | 12 (13.3) | 18 (20.0) | 0.623 | 0.034 | 0.484 |
| Hypertension | 26 (28.9) | 36 (40.0) | 37 (41.1) | 32 (35.6) | 0.323 | 0.045 | 0.353 |
| Hyperlipidemia | 14 (15.6) | 15 (16.7) | 15 (16.7) | 10 (11.1) | 0.740 | −0.038 | 0.429 |
| Laboratory results, median (IQR) | |||||||
| WBC count, 109/L | 6.92 (2.46) | 6.82 (2.52) | 6.65 (2.24) | 6.90 (2.57) | 0.953 | 0.016 | 0.685 |
| LY count, 109/L | 1.87 (0.91) | 1.88 (0.84) | 1.98 (0.76) | 2.03 (0.93) | 0.010 * | 0.083 | 0.036 * |
| NEUT count, 109/L | 4.29 (1.72) | 4.28 (1.84) | 4.18 (1.90) | 4.22 (2.10) | 0.393 | −0.018 | 0.654 |
| MONO count, 109/L | 0.35 (0.20) | 0.33 (0.14) | 0.37 (0.17) | 0.36 (0.14) | 0.509 | 0.013 | 0.752 |
| PLT count, 109/L | 241.00 (74.50) | 251.00 (66.50) | 245.50 (73.50) | 254.50 (91.00) | 0.081 | 0.061 | 0.124 |
| ALT, U/L | 22.10 (21.60) | 20.80 (14.80) | 19.25 (18.80) | 21.55 (17.18) | 0.669 | −0.009 | 0.816 |
| ALP, U/L | 65.70 (27.30) | 69.70 (28.80) | 70.30 (31.93) | 68.85 (23.15) | 0.577 | 0.010 | 0.791 |
| Glu, mmol/L | 5.06 (0.90) | 5.19 (1.19) | 4.95 (0.85) | 5.17 (1.08) | 0.511 | 0.055 | 0.161 |
| Urea, mmol/L | 4.40 (2.05) | 4.60 (1.48) | 4.60 (1.85) | 4.60 (2.10) | 0.330 | 0.027 | 0.492 |
| Cr, μmol/L | 54.25 (19.15) | 53.20 (18.73) | 54.15 (19.90) | 60.50 (24.25) | 0.007 * | 0.092 | 0.019 * |
| TG, mmol/L | 1.16 (0.77) | 1.22 (0.76) | 1.22 (1.02) | 1.16 (0.74) | 0.292 | 0.003 | 0.935 |
| TC, mmol/L | 3.92 (1.12) | 4.13 (1.42) | 4.23 (1.38) | 4.26 (0.95) | 0.021 * | 0.092 | 0.019 * |
| HDL-C, mmol/L | 1.29 (0.41) | 1.34 (0.36) | 1.33 (0.37) | 1.26 (0.29) | 0.642 | −0.033 | 0.407 |
| LDL-C, mmol/L | 2.18 (1.11) | 2.33 (1.14) | 2.40 (1.35) | 2.51 (0.99) | 0.016 * | 0.108 | 0.006 * |
| ApoA, g/L | 1.28 (0.27) | 1.32 (0.36) | 1.30 (0.31) | 1.23 (0.27) | 0.409 | −0.036 | 0.359 |
| ApoB, g/L | 0.76 (0.27) | 0.82 (0.27) | 0.85 (0.29) | 0.87 (0.27) | 0.007 * | 0.118 | 0.003 * |
| Hcy, μmol/L | 12.00 (5.79) | 12.05 (6.21) | 11.65 (5.57) | 12.26 (5.44) | 0.877 | 0.028 | 0.480 |
| PNR | 56.90 (27.30) | 59.49 (30.08) | 60.45 (32.95) | 61.38 (38.77) | 0.136 | 0.055 | 0.167 |
| SII | 526.35 (457.77) | 573.51 (426.91) | 507.85 (303.91) | 532.61 (382.66) | 0.309 | −0.039 | 0.324 |
| MHR | 0.29 (0.19) | 0.25 (0.15) | 0.28 (0.18) | 0.29 (0.17) | 0.699 | 0.011 | 0.771 |
| Neu5Ac, μmol/L | 3.43 (0.64) | 4.26 (0.39) | 5.09 (0.49) | 6.35 (1.08) | <0.001 * | 0.867 | <0.001 * |
| Clinical type, n (%) | 0.024 * | 0.131 | 0.007 * | ||||
| Ischemic type | 70 (77.8) | 72 (80) | 59 (65.6) | 57 (63.3) | |||
| Hemorrhagic type | 20 (22.2) | 18 (20.0) | 31 (34.4) | 33 (36.7) | |||
| Admission mRS, n (%) | 0.160 | 0.034 | 0.475 | ||||
| 0–2 | 86 (95.6) | 77 (85.6) | 80 (88.9) | 82 (91.1) | |||
| 3–5 | 4 (4.4) | 13 (14.4) | 10 (11.1) | 8 (8.9) | |||
| Suzuki stage, n (%) | 0.666 | −0.006 | 0.898 | ||||
| 1–2 | 20 (22.2) | 28 (31.1) | 30 (33.3) | 22 (24.4) | |||
| 3–4 | 50 (55.6) | 45 (50.0) | 41 (45.6) | 47 (52.2) | |||
| 5–6 | 20 (22.2) | 17 (18.9) | 19 (21.1) | 21 (23.3) | |||
| Neu5Ac | No. of Events (%) | Crude Model | Model 1 | Model 2 | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | OR (95% CI) | p-Value | ||
| MMD overall | |||||||
| Continuous | 360 (80.2) | 1.395 (1.141–1.706) | 0.001 * | 1.380 (1.125–1.694) | 0.002 * | 1.528 (1.178–1.983) | 0.001 * |
| Neu5Ac level | |||||||
| Low (2.45–4.52) | 171 (76.3) | 1.0 (Ref.) | 1.0 (Ref.) | 1.0 (Ref.) | |||
| High (4.52–16.25) | 189 (84.0) | 1.627 (1.016–2.606) | 0.043 * | 1.628 (0.988–2.681) | 0.056 | 1.882 (1.037–3.413) | 0.037 * |
| Ischemic MMD | |||||||
| Continuous | 258 (74.3) | 1.299 (1.059–1.593) | 0.012 * | 1.265 (1.025–1.561) | 0.029 * | 1.414 (1.077–1.857) | 0.013 * |
| Neu5Ac level | |||||||
| Low (2.53–4.45) | 120 (69.3) | 1.0 (Ref.) | 1.0 (Ref.) | 1.0 (Ref.) | |||
| High (4.45–16.25) | 138 (79.3) | 0.591 (0.362–0.963) | 0.035 * | 0.636 (0.374–1.080) | 0.094 | 0.611 (0.316–1.182) | 0.144 |
| Hemorrhagic MMD | |||||||
| Continuous | 102 (53.4) | 1.666 (1.294–2.144) | <0.001 * | 1.758 (1.343–2.302) | <0.001 * | 1.806 (1.306–2.498) | <0.001 * |
| Neu5Ac level | |||||||
| Low (2.45–4.68) | 38 (40.0) | 1.0 (Ref.) | 1.0 (Ref.) | 1.0 (Ref.) | |||
| High (4.68–13.29) | 64 (66.6) | 3.000 (1.662–5.414) | <0.001 * | 3.420 (1.823–6.416) | <0.001 * | 3.154 (1.472–6.757) | 0.003 * |
| AUC (95% CI) | Categorical NRI (95% CI) | p-Value | IDI (95% CI) | p-Value | |
|---|---|---|---|---|---|
| MMD overall | |||||
| Basic model | 0.858 (0.817–0.900) | 1.0 (Ref.) | 1.0 (Ref.) | ||
| Basic model + Neu5Ac binary | 0.869 (0.830–0.907) | 0.070 (0.001–0.140) | 0.047 * | 0.010 (−0.002–0.023) | 0.120 |
| Basic model + Neu5Ac quartiles | 0.863 (0.824–0.903) | 0.062 (−0.007–0.131) | 0.081 | 0.020 (0.003–0.037) | 0.024 * |
| Basic model + Neu5Ac continuous | 0.873 (0.836–0.911) | 0.124 (0.033–0.214) | 0.007 * | 0.028 (0.008–0.047) | 0.005 * |
| Ischemic MMD | |||||
| Basic model | 0.887 (0.851–0.924) | 1.0 (Ref.) | 1.0 (Ref.) | ||
| Basic model + Neu5Ac binary | 0.893 (0.858–0.927) | −0.015 (−0.088–0.057) | 0.677 | 0.004 (−0.005–0.014) | 0.387 |
| Basic model + Neu5Ac quartiles | 0.890 (0.854–0.926) | 0.048 (−0.038–0.135) | 0.273 | 0.011 (−0.004–0.027) | 0.154 |
| Basic model + Neu5Ac continuous | 0.896 (0.862–0.930) | 0.067 (−0.013–0.147) | 0.101 | 0.018 (0.001–0.035) | 0.042 * |
| Hemorrhagic MMD | |||||
| Basic model | 0.849 (0.793–0.905) | 1.0 (Ref.) | 1.0 (Ref.) | ||
| Basic model + Neu5Ac binary | 0.867 (0.816–0.919) | 0.152 (0.029–0.274) | 0.015 * | 0.034 (0.007–0.061) | 0.014 * |
| Basic model + Neu5Ac quartiles | 0.858 (0.804–0.911) | 0.166 (0.033–0.298) | 0.014 * | 0.056 (0.022–0.089) | 0.001 * |
| Basic model + Neu5Ac continuous | 0.872 (0.821–0.923) | 0.199 (0.074–0.324) | 0.002 * | 0.065 (0.030–0.100) | <0.001 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Ge, P.; Zeng, C.; Yu, X.; Zhai, Y.; Liu, W.; He, Q.; Li, J.; Liu, X.; Wang, J.; et al. Correlation of Serum N-Acetylneuraminic Acid with the Risk of Moyamoya Disease. Brain Sci. 2023, 13, 913. https://doi.org/10.3390/brainsci13060913
Liu C, Ge P, Zeng C, Yu X, Zhai Y, Liu W, He Q, Li J, Liu X, Wang J, et al. Correlation of Serum N-Acetylneuraminic Acid with the Risk of Moyamoya Disease. Brain Sciences. 2023; 13(6):913. https://doi.org/10.3390/brainsci13060913
Chicago/Turabian StyleLiu, Chenglong, Peicong Ge, Chaofan Zeng, Xiaofan Yu, Yuanren Zhai, Wei Liu, Qiheng He, Junsheng Li, Xingju Liu, Jia Wang, and et al. 2023. "Correlation of Serum N-Acetylneuraminic Acid with the Risk of Moyamoya Disease" Brain Sciences 13, no. 6: 913. https://doi.org/10.3390/brainsci13060913
APA StyleLiu, C., Ge, P., Zeng, C., Yu, X., Zhai, Y., Liu, W., He, Q., Li, J., Liu, X., Wang, J., Ye, X., Zhang, Q., Wang, R., Zhang, Y., Zhao, J., & Zhang, D. (2023). Correlation of Serum N-Acetylneuraminic Acid with the Risk of Moyamoya Disease. Brain Sciences, 13(6), 913. https://doi.org/10.3390/brainsci13060913





