mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources for Exposure
2.2. Data Sources for the Outcome
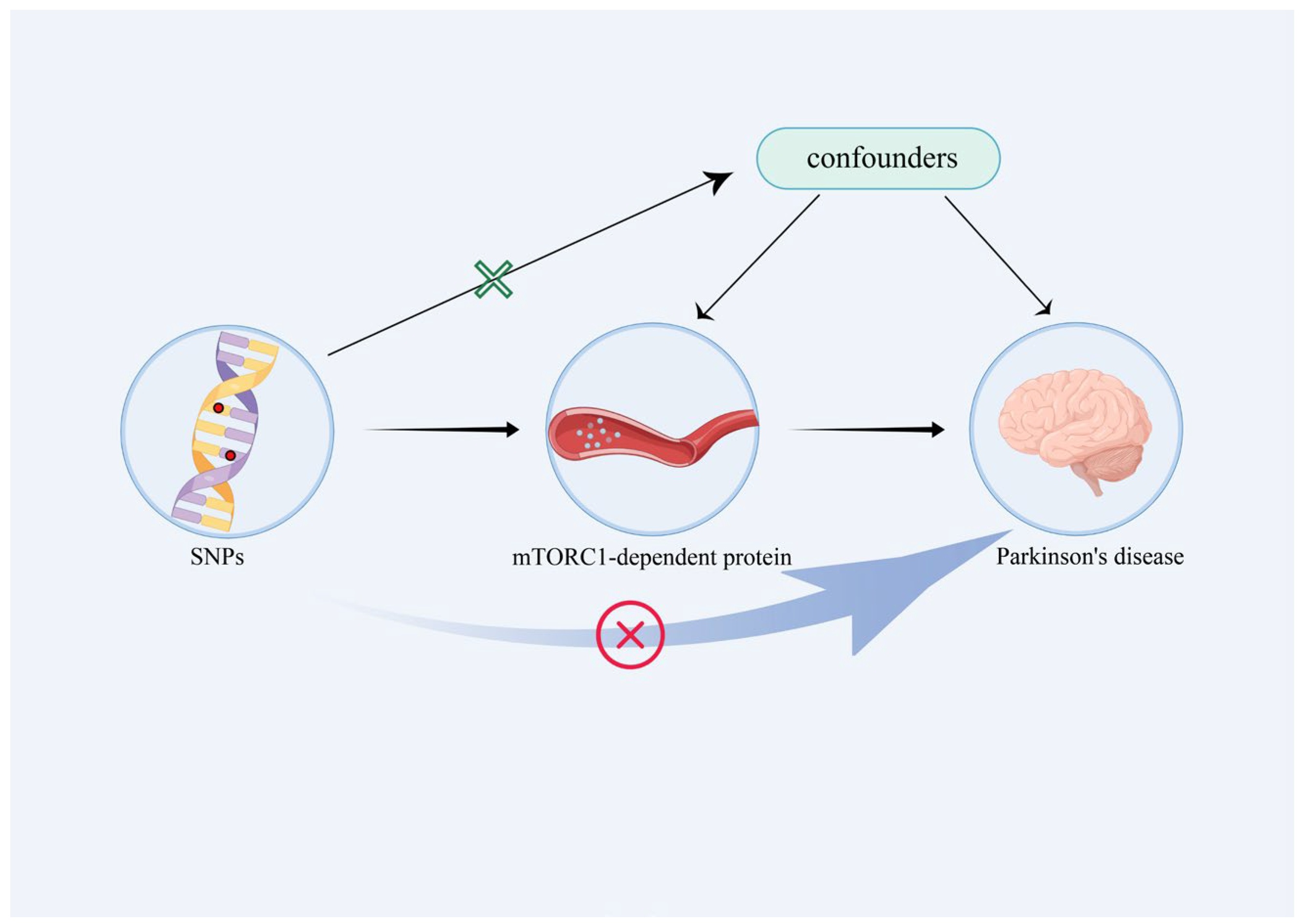
2.3. Instrumental Variable
2.4. Mendelian Randomization Analyses
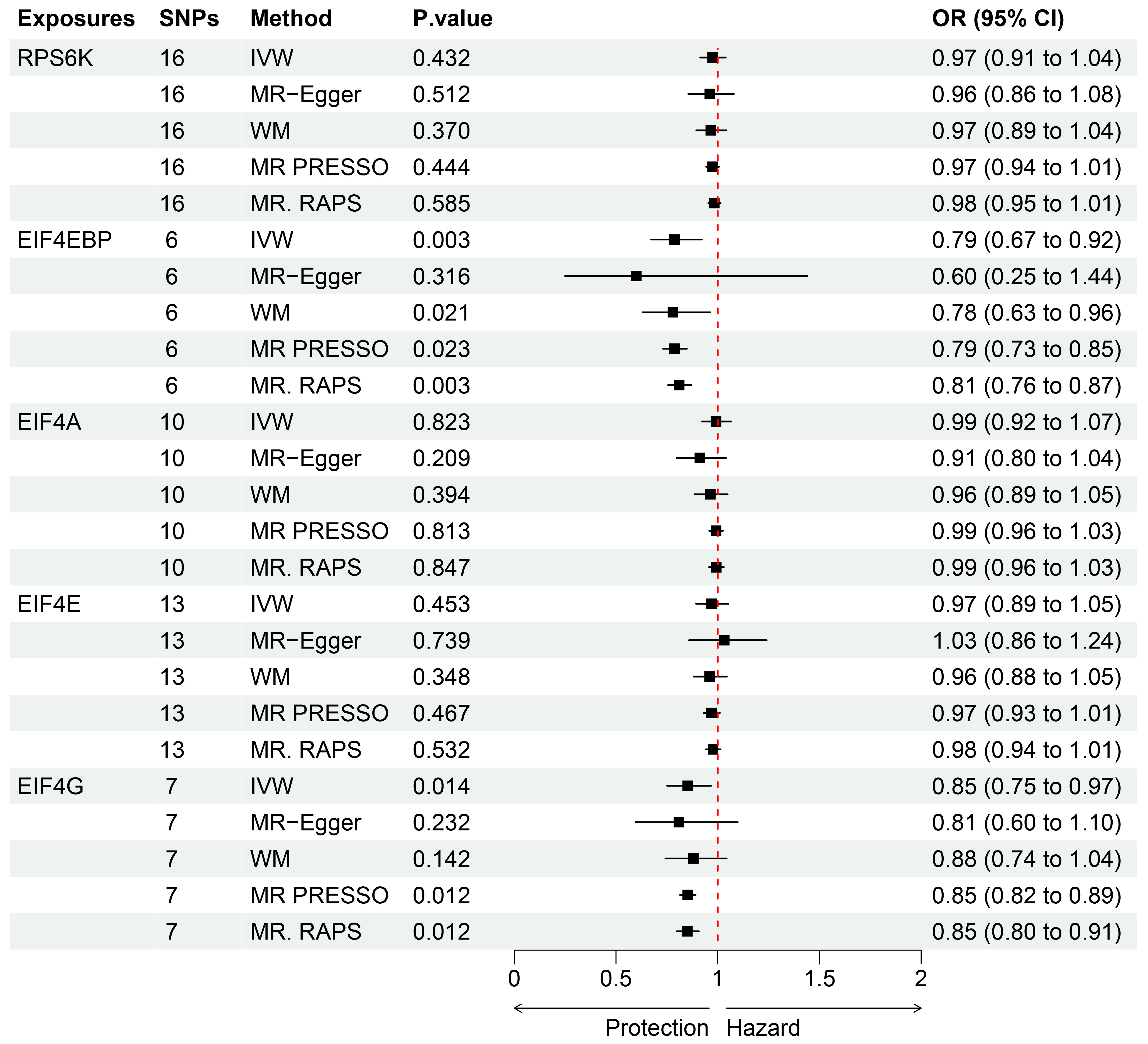
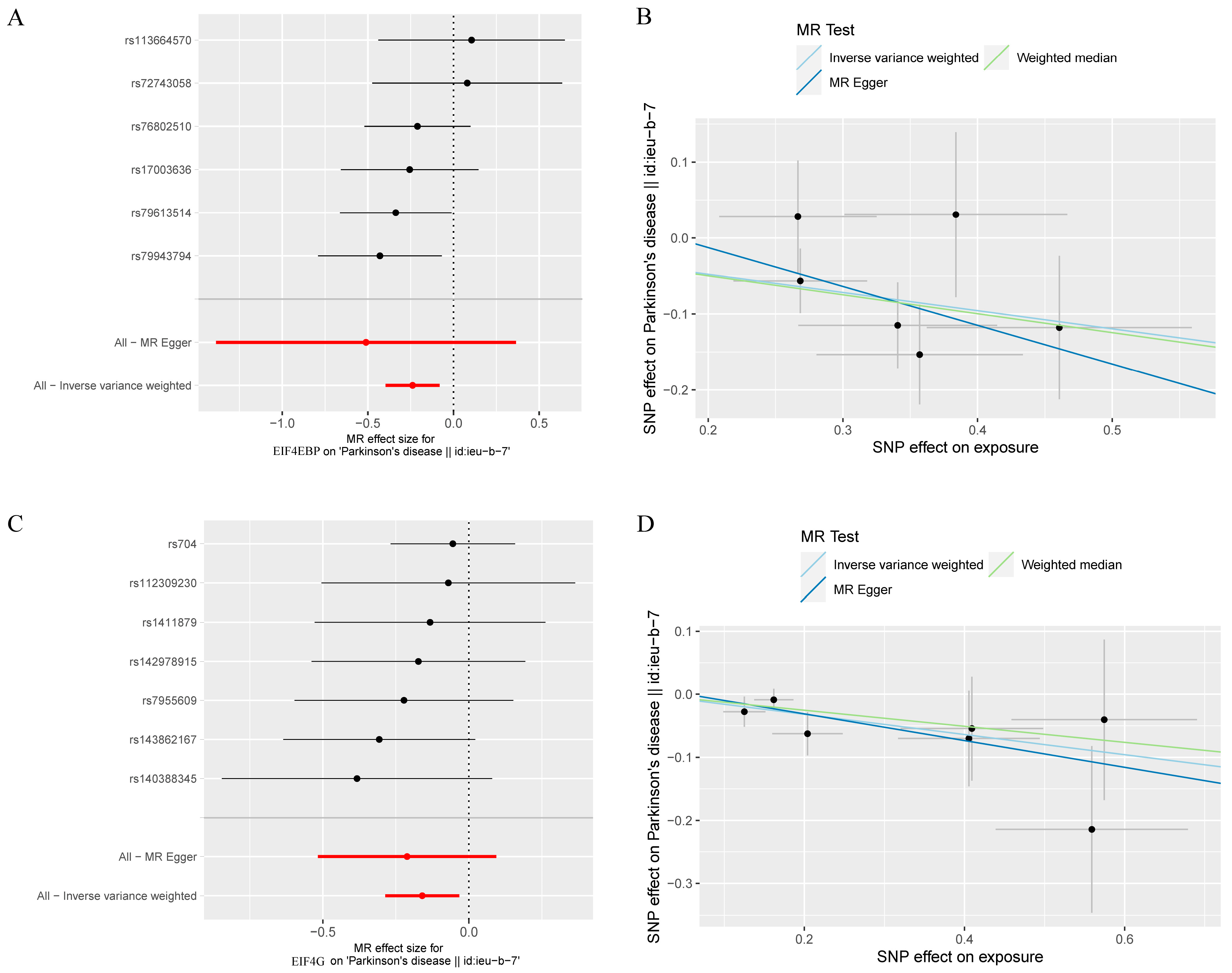
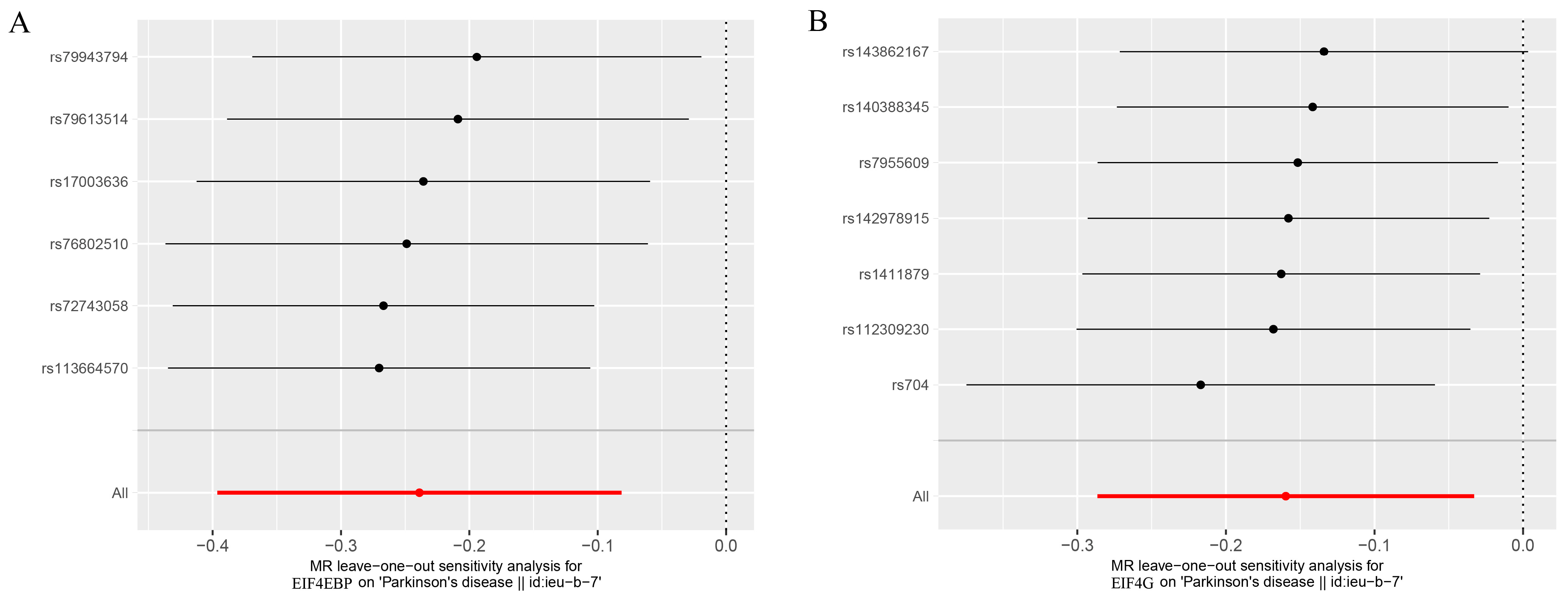
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rocca, W.A. The Burden of Parkinson’s Disease: A Worldwide Perspective. Lancet Neurol. 2018, 17, 928–929. [Google Scholar] [CrossRef] [PubMed]
- Gelb, D.J.; Oliver, E.; Gilman, S. Diagnostic Criteria for Parkinson Disease. Arch. Neurol. 1999, 56, 33. [Google Scholar] [CrossRef] [PubMed]
- Dexter, D.T.; Jenner, P. Parkinson Disease: From Pathology to Molecular Disease Mechanisms. Free. Radic. Biol. Med. 2013, 62, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, J.; Sheng, M. The Growing Role of MTOR in Neuronal Development and Plasticity. Mol. Neurobiol. 2006, 34, 205–220. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. MTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Maiese, K.; Chong, Z.Z.; Shang, Y.C.; Wang, S. MTOR: On Target for Novel Therapeutic Strategies in the Nervous System. Trends Mol. Med. 2013, 19, 51–60. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR Complexes, Only One of Which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Chiarini, F.; Evangelisti, C.; McCubrey, J.A.; Martelli, A.M. Current Treatment Strategies for Inhibiting MTOR in Cancer. Trends Pharmacol. Sci. 2015, 36, 124–135. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.-H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a Novel Binding Partner of MTOR, Defines a Rapamycin-Insensitive and Raptor-Independent Pathway That Regulates the Cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef]
- Ma, X.M.; Blenis, J. Molecular Mechanisms of MTOR-Mediated Translational Control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. MTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A Unifying Model for MTORC1-Mediated Regulation of MRNA Translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef]
- Tang, Z.; Ioja, E.; Bereczki, E.; Hultenby, K.; Li, C.; Guan, Z.; Winblad, B.; Pei, J.-J. MTor Mediates Tau Localization and Secretion: Implication for Alzheimer’s Disease. Biochim. Et Biophys. Acta 2015, 1853, 1646–1657. [Google Scholar] [CrossRef]
- Lan, A.; Chen, J.; Zhao, Y.; Chai, Z.; Hu, Y. MTOR Signaling in Parkinson’s Disease. Neuromol. Med. 2017, 19, 1–10. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925. [Google Scholar] [CrossRef]
- Davey Smith, G.; Hemani, G. Mendelian Randomization: Genetic Anchors for Causal Inference in Epidemiological Studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic Atlas of the Human Plasma Proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Di Angelantonio, E.; Thompson, S.G.; Kaptoge, S.; Moore, C.; Walker, M.; Armitage, J.; Ouwehand, W.H.; Roberts, D.J.; Danesh, J. INTERVAL Trial Group Efficiency and Safety of Varying the Frequency of Whole Blood Donation (INTERVAL): A Randomised Trial of 45 000 Donors. Lancet 2017, 390, 2360–2371. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of Novel Risk Loci, Causal Insights, and Heritable Risk for Parkinson’s Disease: A Meta-Analysis of Genome-Wide Association Studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining Information on Multiple Instrumental Variables in Mendelian Randomization: Comparison of Allele Score and Summarized Data Methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Del Greco, F.M.; Minelli, C.; Smith, G.D.; Sheehan, N.A.; Thompson, J.R. Assessing the Suitability of Summary Data for Two-Sample Mendelian Randomization Analyses Using MR-Egger Regression: The Role of the I-2 Statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, Y.; Wang, J.; Small, D.S. Powerful Three-Sample Genome-Wide Design and Robust Statistical Inference in Summary-Data Mendelian Randomization. Int. J. Epidemiol. 2019, 48, 1478–1492. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting Findings from Mendelian Randomization Using the MR-Egger Method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Ong, J.-S.; MacGregor, S. Implementing MR-PRESSO and GCTA-GSMR for Pleiotropy Assessment in Mendelian Randomization Studies from a Practitioner’s Perspective. Genet. Epidemiol. 2019, 43, 609–616. [Google Scholar] [CrossRef]
- Querfurth, H.; Lee, H.-K. Mammalian/Mechanistic Target of Rapamycin (MTOR) Complexes in Neurodegeneration. Mol. Neurodegener. 2021, 16, 44. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 Complexes Mediate MTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell. 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 Induces Autophagy by Phosphorylating Beclin-1 and Activating VPS34 Lipid Kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef]
- Webb, J.L.; Ravikumar, B.; Atkins, J.; Skepper, J.N.; Rubinsztein, D.C. Alpha-Synuclein Is Degraded by Both Autophagy and the Proteasome. J. Biol. Chem. 2003, 278, 25009–25013. [Google Scholar] [CrossRef]
- Gingras, A.C.; Raught, B.; Sonenberg, N. Regulation of Translation Initiation by FRAP/MTOR. Genes Dev. 2001, 15, 807–826. [Google Scholar] [CrossRef]
- Gingras, A.C.; Raught, B.; Gygi, S.P.; Niedzwiecka, A.; Miron, M.; Burley, S.K.; Polakiewicz, R.D.; Wyslouch-Cieszynska, A.; Aebersold, R.; Sonenberg, N. Hierarchical Phosphorylation of the Translation Inhibitor 4E-BP1. Genes Dev. 2001, 15, 2852–2864. [Google Scholar] [CrossRef]
- Mythri, R.B.; Venkateshappa, C.; Harish, G.; Mahadevan, A.; Muthane, U.B.; Yasha, T.C.; Bharath, M.M.S.; Shankar, S.K. Evaluation of Markers of Oxidative Stress, Antioxidant Function and Astrocytic Proliferation in the Striatum and Frontal Cortex of Parkinson’s Disease Brains. Neurochem. Res. 2011, 36, 1452–1463. [Google Scholar] [CrossRef]
- Teleman, A.A.; Chen, Y.W.; Cohen, S.M. 4E-BP Functions as a Metabolic Brake Used under Stress Conditions but Not during Normal Growth. Genes Dev. 2005, 19, 1844–1848. [Google Scholar] [CrossRef]
- Tettweiler, G.; Miron, M.; Jenkins, M.; Sonenberg, N.; Lasko, P.F. Starvation and Oxidative Stress Resistance in Drosophila Are Mediated through the EIF4E-Binding Protein, D4E-BP. Genes Dev. 2005, 19, 1840–1843. [Google Scholar] [CrossRef]
- Clemens, M.J. Translational Regulation in Cell Stress and Apoptosis. Roles of the EIF4E Binding Proteins. J. Cell. Mol. Med. 2001, 5, 221–239. [Google Scholar] [CrossRef]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 Mutations in Autosomal Dominant Late-Onset Parkinson’s Disease: A Genome-Wide Linkage and Sequencing Study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Ogaki, K.; Koga, S.; Heckman, M.G.; Fiesel, F.C.; Ando, M.; Labbé, C.; Lorenzo-Betancor, O.; Moussaud-Lamodière, E.L.; Soto-Ortolaza, A.I.; Walton, R.L.; et al. Mitochondrial Targeting Sequence Variants of the CHCHD2 Gene Are a Risk for Lewy Body Disorders. Neurology 2015, 85, 2016–2025. [Google Scholar] [CrossRef]
- Meng, H.; Yamashita, C.; Shiba-Fukushima, K.; Inoshita, T.; Funayama, M.; Sato, S.; Hatta, T.; Natsume, T.; Umitsu, M.; Takagi, J.; et al. Loss of Parkinson’s Disease-Associated Protein CHCHD2 Affects Mitochondrial Crista Structure and Destabilizes Cytochrome c. Nat. Commun. 2017, 8, 15500. [Google Scholar] [CrossRef]
- Tain, L.S.; Mortiboys, H.; Tao, R.N.; Ziviani, E.; Bandmann, O.; Whitworth, A.J. Rapamycin Activation of 4E-BP Prevents Parkinsonian Dopaminergic Neuron Loss. Nat. Neurosci. 2009, 12, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Lubbe, S.; Morris, H.R. Recent Advances in Parkinson’s Disease Genetics. J. Neurol. 2014, 261, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Kim, J.W.; Dawson, V.L.; Dawson, T.M. LRRK2 Pathobiology in Parkinson’s Disease. J. Neurochem. 2014, 131, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Gehrke, S.; Wang, H.-Q.; Takahashi, R.; Hasegawa, K.; Oota, E.; Lu, B. Phosphorylation of 4E-BP by LRRK2 Affects the Maintenance of Dopaminergic Neurons in Drosophila. EMBO J. 2008, 27, 2432–2443. [Google Scholar] [CrossRef]
- Landis, G.N.; Abdueva, D.; Skvortsov, D.; Yang, J.; Rabin, B.E.; Carrick, J.; Tavaré, S.; Tower, J. Similar Gene Expression Patterns Characterize Aging and Oxidative Stress in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2004, 101, 7663–7668. [Google Scholar] [CrossRef]
| Exposure | Cochran’s Q Test | MR–Egger | MR-PRESSO | |||
|---|---|---|---|---|---|---|
| Q Value | p Value | Egger Intercept | p Value | Global Test | p Value | |
| RPS6K | 15.53 | 0.41 | 0.0049 | 0.78 | 16.60 | 0.51 |
| EIF4EBP | 4.27 | 0.51 | 0.0895 | 0.57 | 5.63 | 0.60 |
| EIF4A | 7.64 | 0.57 | 0.0226 | 0.17 | 12.74 | 0.50 |
| EIF4E | 15.31 | 0.23 | −0.0228 | 0.47 | 16.61 | 0.32 |
| EIF4G | 2.88 | 0.82 | 0.0111 | 0.73 | 4.68 | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, C.; Ai, J.; Zhu, Y. mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study. Brain Sci. 2023, 13, 536. https://doi.org/10.3390/brainsci13040536
Tan C, Ai J, Zhu Y. mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study. Brain Sciences. 2023; 13(4):536. https://doi.org/10.3390/brainsci13040536
Chicago/Turabian StyleTan, Cheng, Jianzhong Ai, and Ye Zhu. 2023. "mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study" Brain Sciences 13, no. 4: 536. https://doi.org/10.3390/brainsci13040536
APA StyleTan, C., Ai, J., & Zhu, Y. (2023). mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study. Brain Sciences, 13(4), 536. https://doi.org/10.3390/brainsci13040536






