Get a New Perspective on EEG: Convolutional Neural Network Encoders for Parametric t-SNE
Abstract
1. Introduction
2. Materials and Methods
2.1. EEG Data
2.1.1. TUH EEG Corpus
2.1.2. Ethical Considerations
2.1.3. Extracted Data
2.1.4. Preprocessing
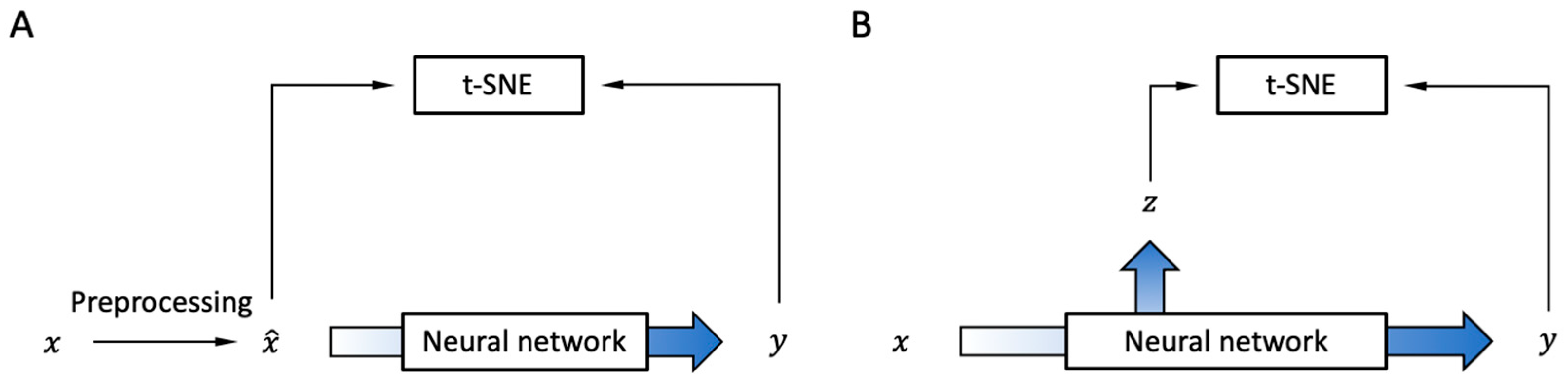
2.2. Encoders
2.2.1. Modification of t-SNE
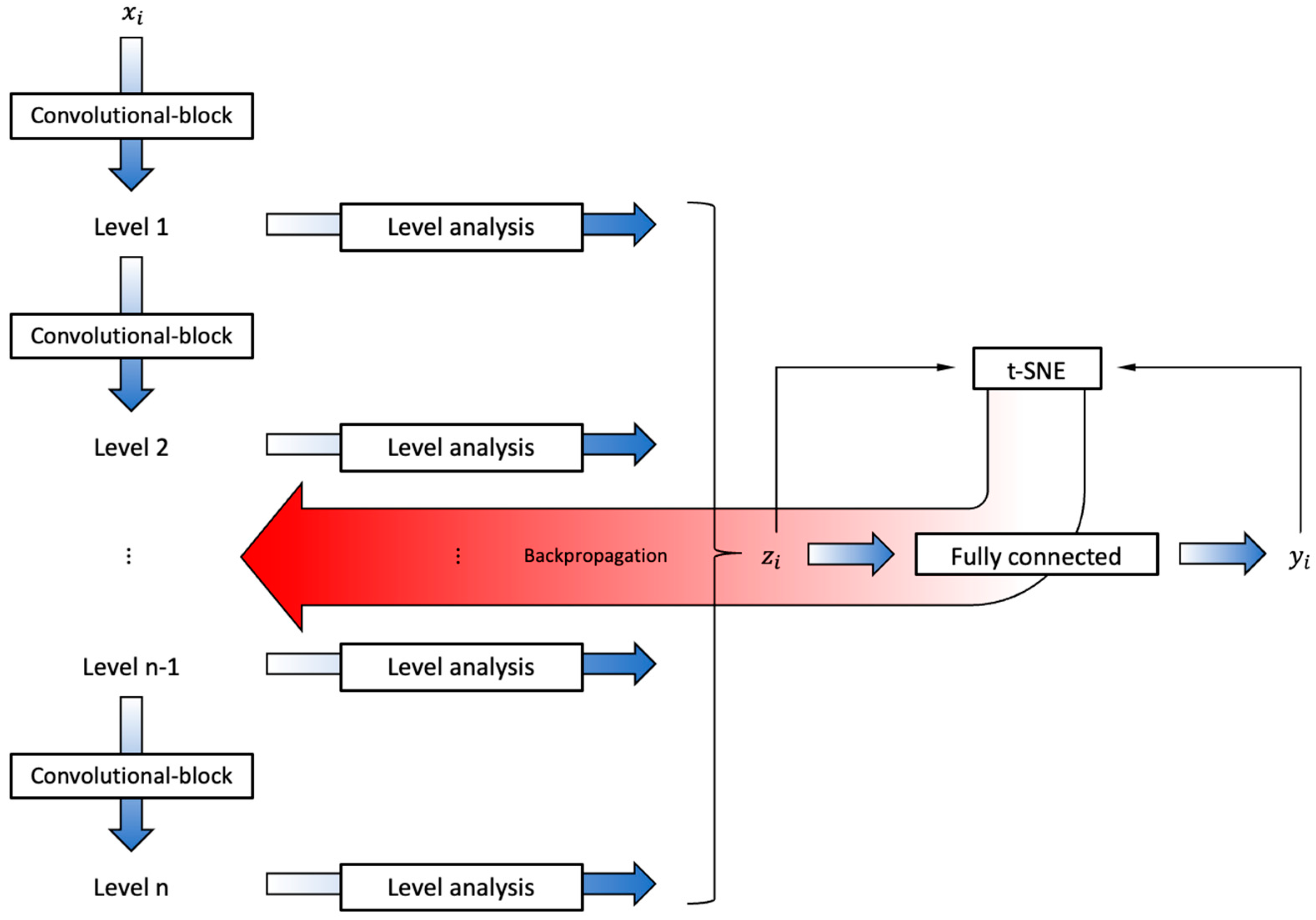
2.2.2. CNN Encoder Architecture
2.3. Evaluation
2.3.1. Comparative Methods
2.3.2. Quantitative Measures
2.4. Software and Hardware
3. Results
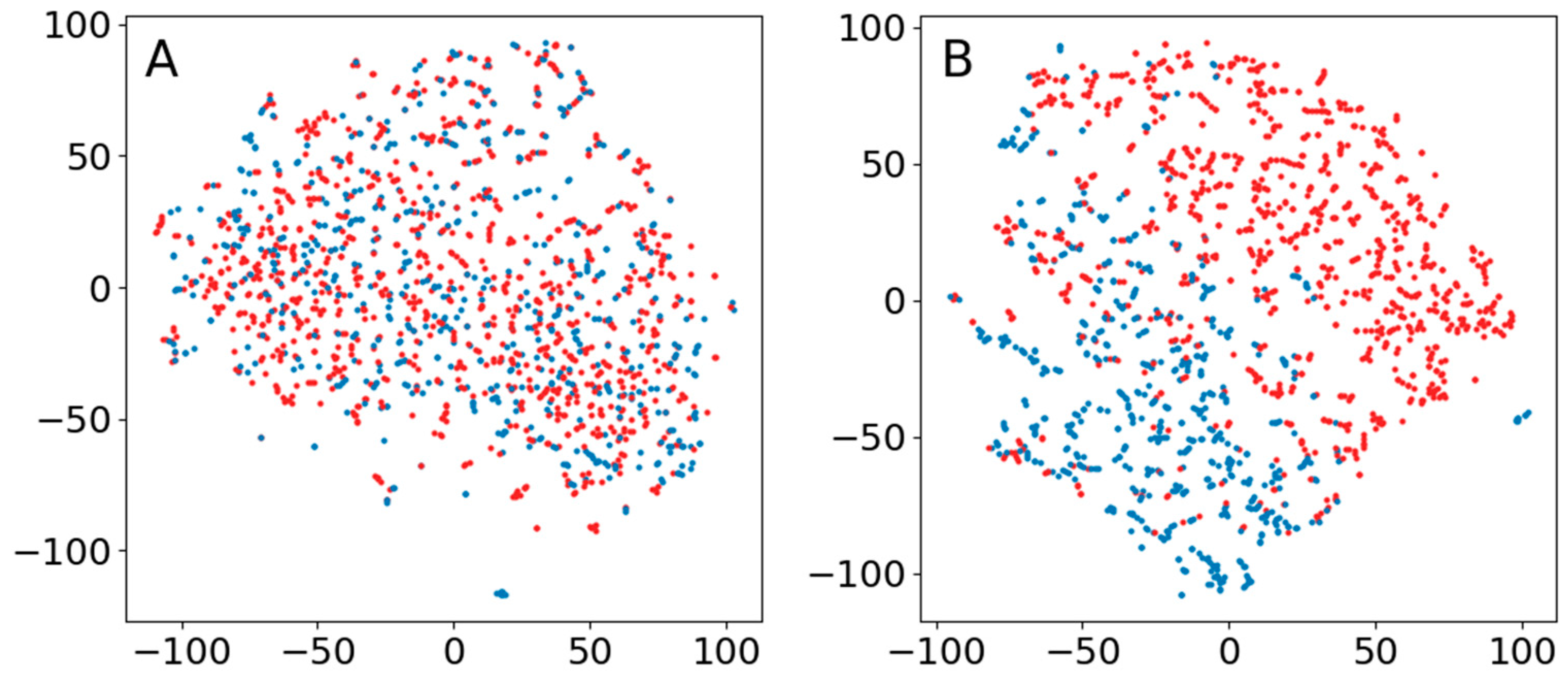
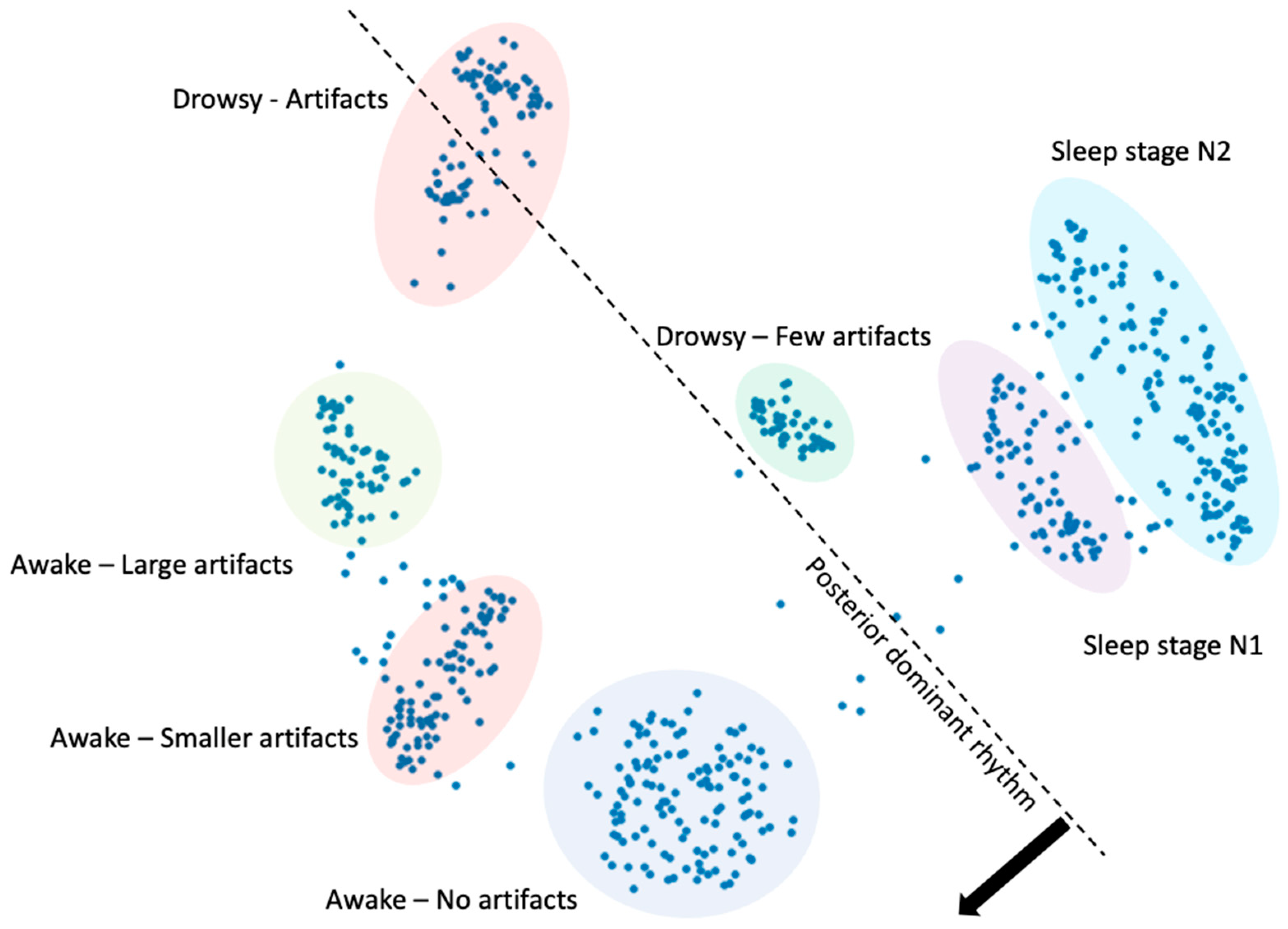
3.1. Sleep–Wake
3.2. IEDs
3.3. Seizure Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Data Selection and Annotation
| Train 0 | Train 1 | Test 0 | Test 1 | |
|---|---|---|---|---|
| Sleep–wake | 17,386 | 10,990 | 7261 | 3460 |
| IEDs | 5932 | 3246 | 2012 | 1138 |
| Seizures | 5592 | 1949 | 1875 | 600 |
Appendix A.2. Example Duration
Appendix A.3. Overlapping vs. Non-Overlapping Examples
Appendix B
Appendix B.1. The Original t-SNE
Appendix B.2. Modified t-SNE Using Custom Probability Distributions

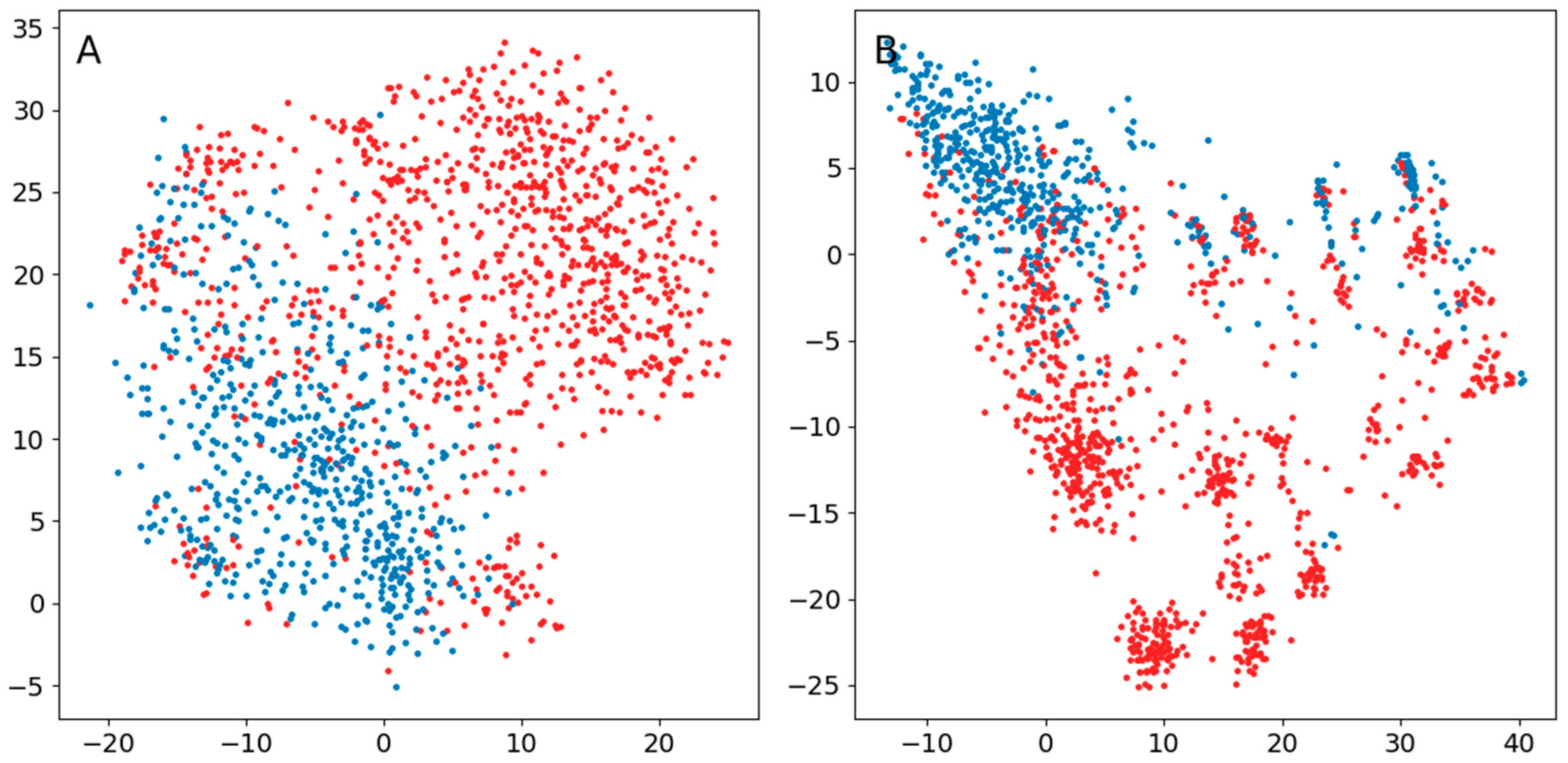
Appendix B.3. Comparison of the Use of a Normal Distribution or a Distribution Based on Ranked Distances in the Original t-SNE
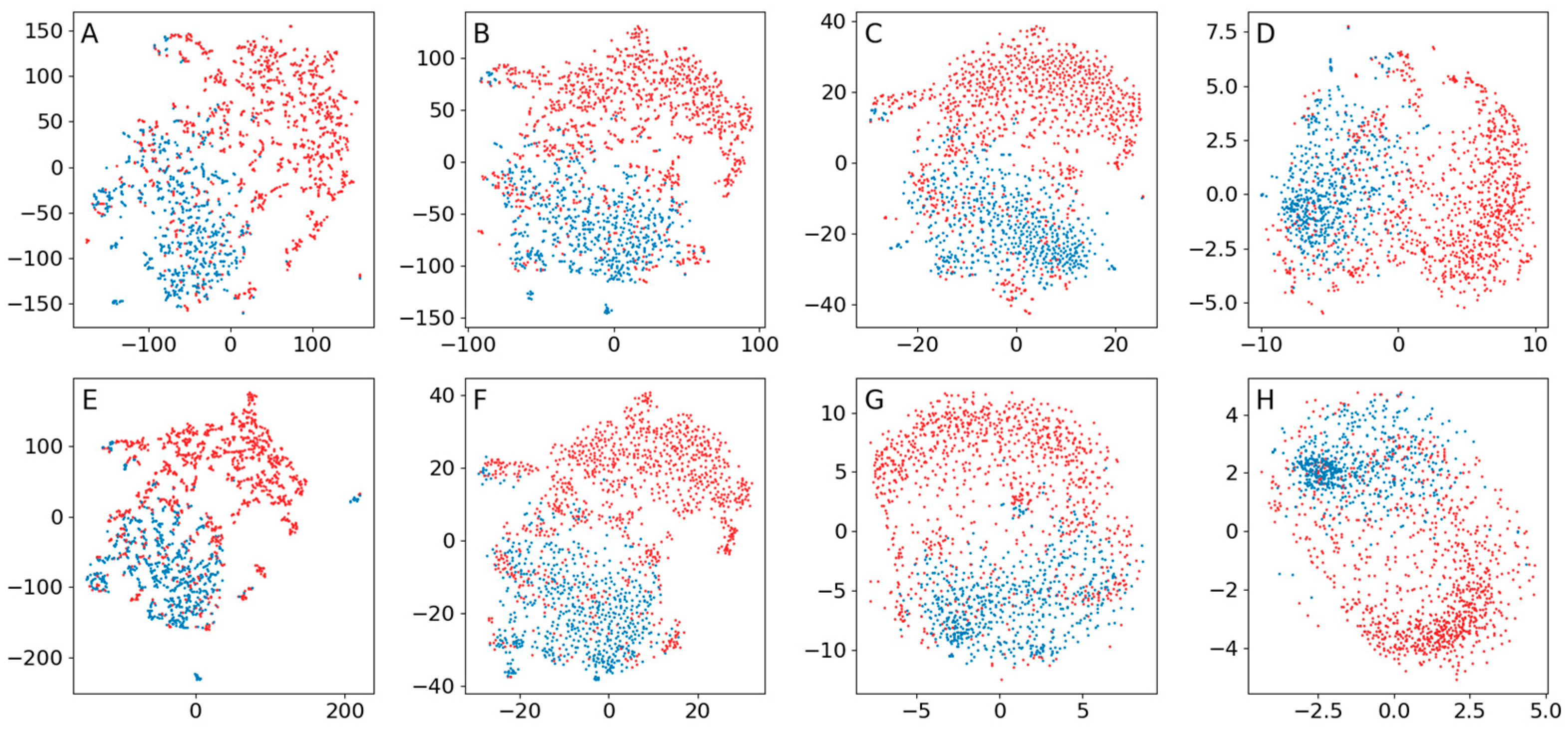
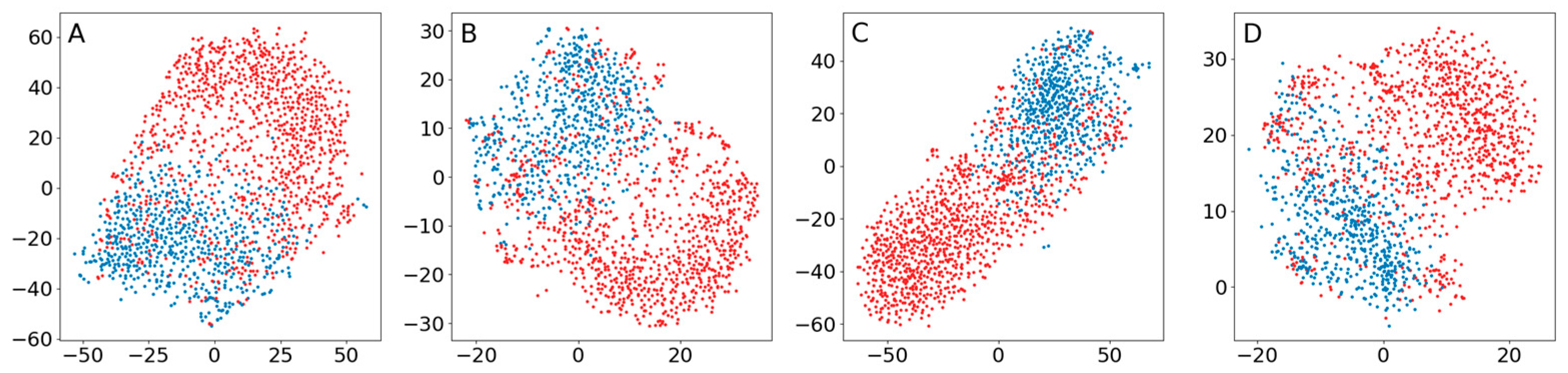
Appendix B.4. Comparison of the Use of a Normal Distribution or a Distribution Based on Ranked Distances When Training Encoders
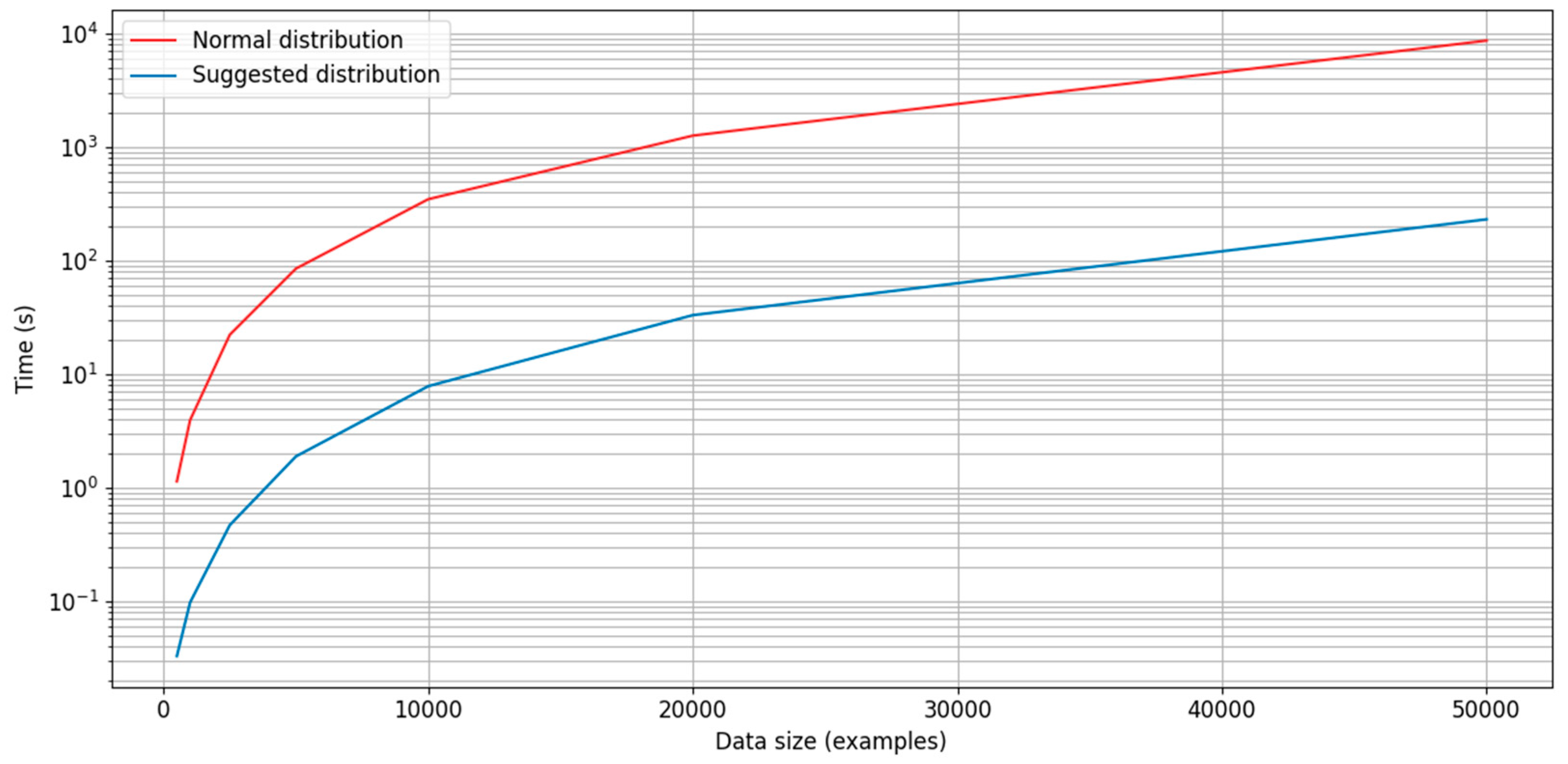
Appendix B.5. Time Consumption for a Normal Distribution and a Distribution Based on Ranked Distances
Appendix C
Appendix C.1. Encoder Architecture

Appendix C.1.1. Convolutional Blocks
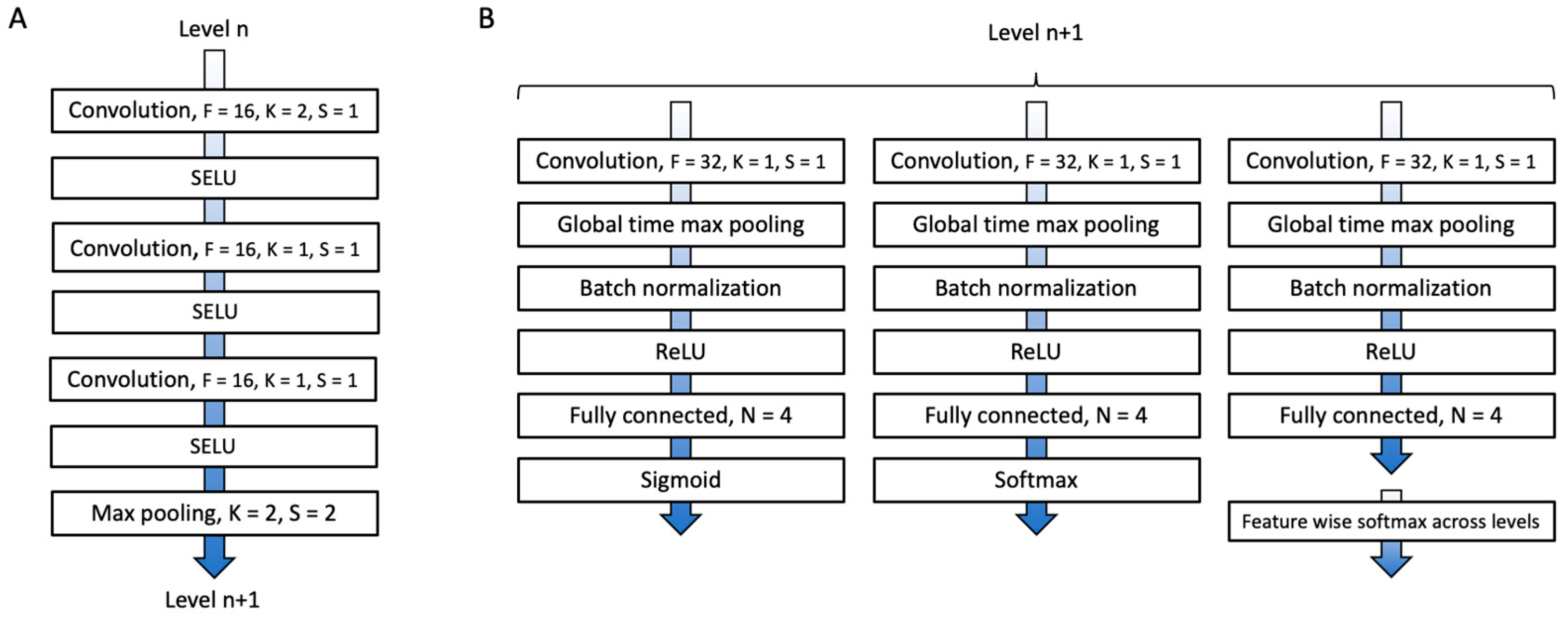
Appendix C.1.2. Level Analysis

Appendix C.1.3. Fully Connected Block

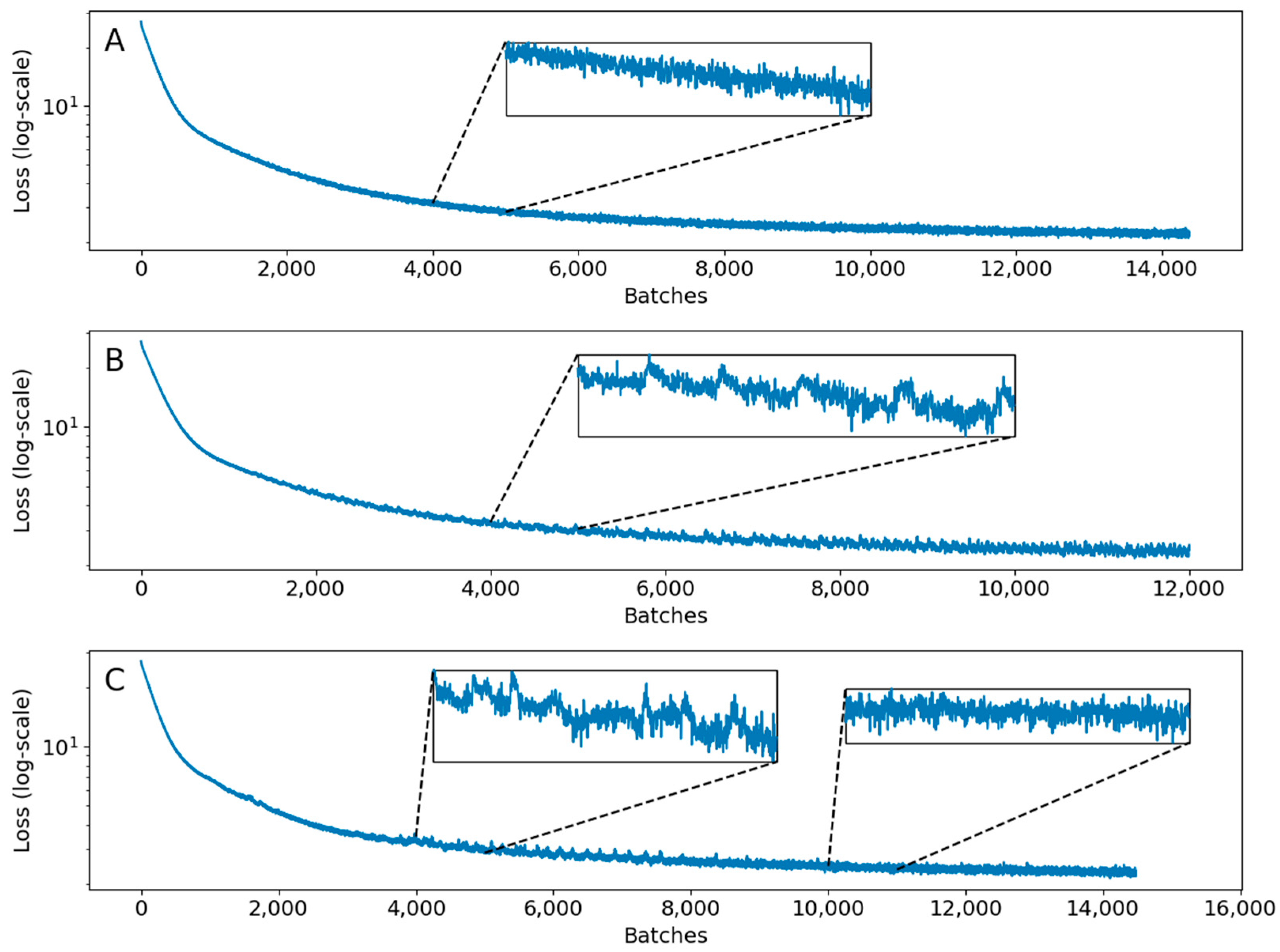
Appendix C.2. Loss Function

Appendix C.3. Hyperparameters
Appendix D
Appendix D.1. Parametric t-SNE

Appendix D.2. Features
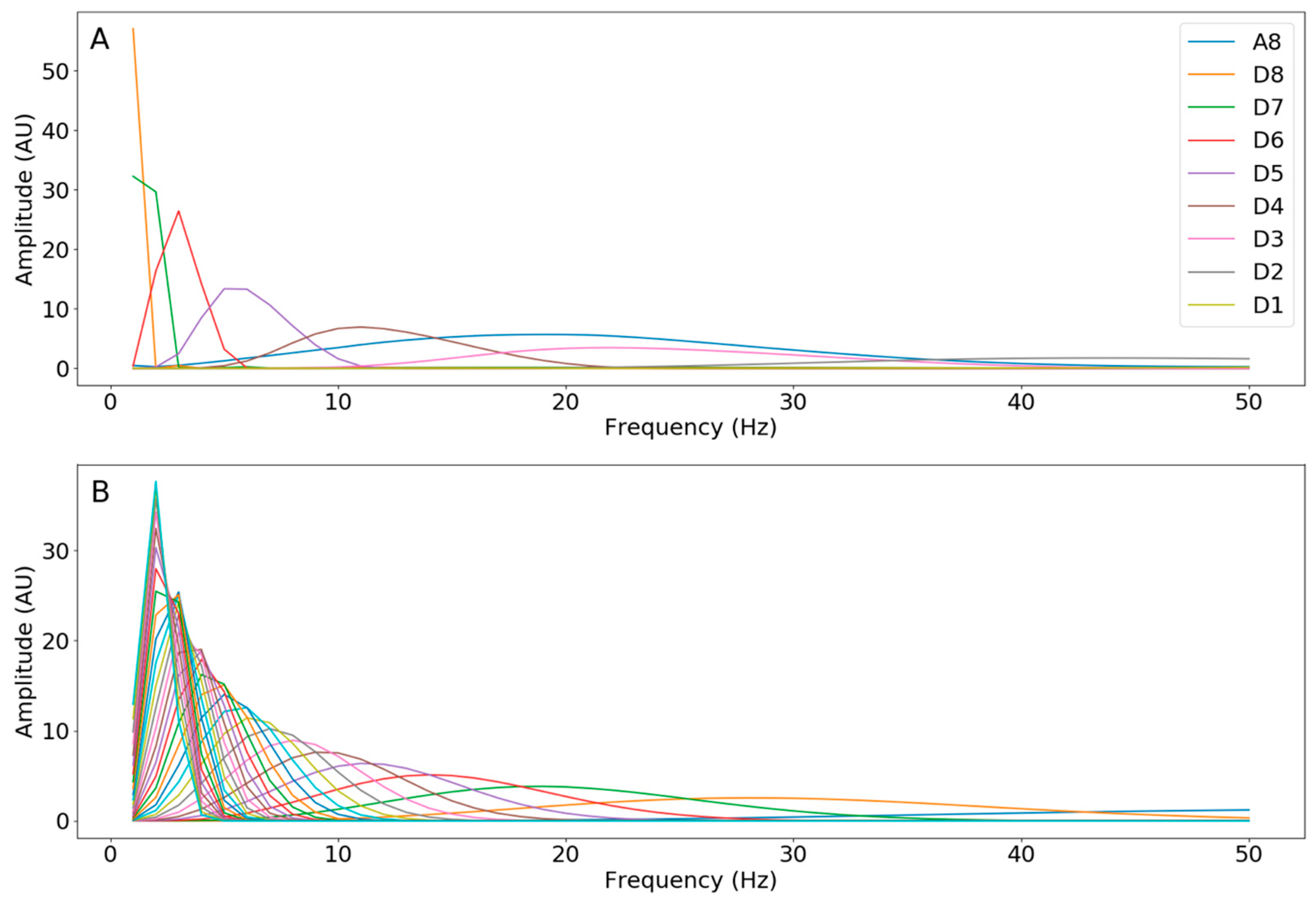
Appendix D.2.1. STFTs
Appendix D.2.2. CWTs
Appendix E
Appendix E.1. Data
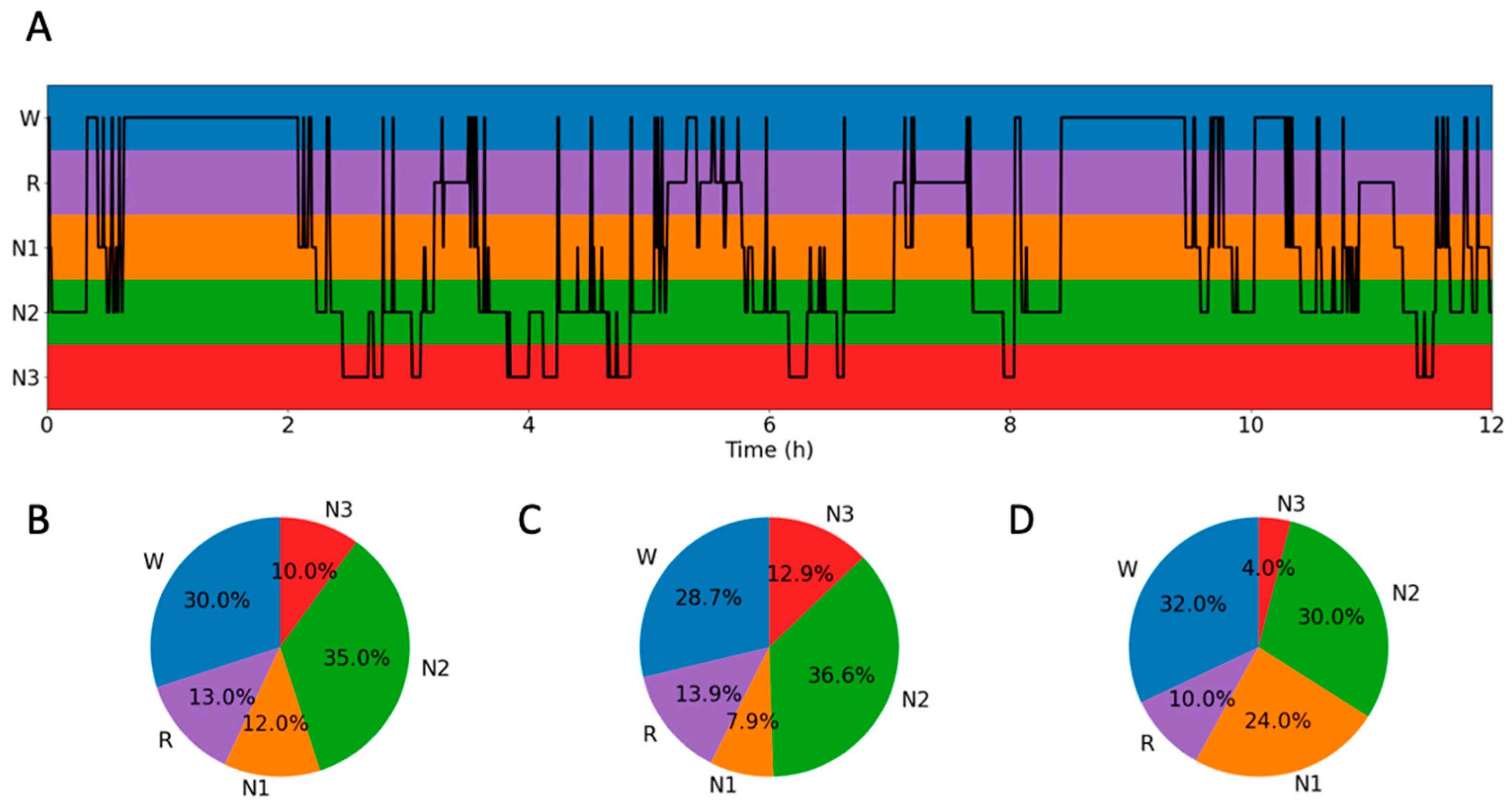
Appendix E.2. Training Encoders
Appendix E.3. Results

| -Train | -Test | -Train | -Test | -Train | -Test | |
|---|---|---|---|---|---|---|
| CNN | 0.55 | 0.35 | 0.85 | 0.19 | 12 | 2 |
| STFT | 0.77 | 0.51 | 0.96 | 0.33 | 2 | 6 |
| CWT | 0.36 | 0.30 | 0.92 | 0.30 | 2 | 2 |
| CNN * | 0.63 | 0.53 | 0.79 | 0.52 | 3 | 2 |
References
- van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Jing, J.; d’Angremont, E.; Zafar, S.; Rosenthal, E.S.; Tabaeizadeh, M.; Ebrahim, S.; Dauwels, J.; Westover, M.B. Rapid Annotation of Seizures and Interictalictal Continuum EEG Patterns. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 3394–3397. [Google Scholar] [CrossRef]
- Kinney-Lang, E.; Spyrou, L.; Ebied, A.; Chin, R.F.M.; Escudero, J. Tensor-driven extraction of developmental features from varying paediatric EEG datasets. J. Neural Eng. 2018, 15, 046024. [Google Scholar] [CrossRef]
- Al-Fahad, R.; Yeasin, M.; Bidelman, G.M. Decoding of single-trial EEG reveals unique states of functional brain connectivity that drive rapid speech categorization decisions. J. Neural Eng. 2020, 17, 016045. [Google Scholar] [CrossRef]
- Ravi, A.; Beni, N.H.; Manuel, J.; Jiang, N. Comparing user-dependent and user-independent training of CNN for SSVEP BCI. J. Neural Eng. 2020, 17, 026028. [Google Scholar] [CrossRef]
- Suetani, H.; Kitajo, K. A manifold learning approach to mapping individuality of human brain oscillations through beta-divergence. Neurosci. Res. 2020, 156, 188–196. [Google Scholar] [CrossRef]
- Idowu, O.P.; Ilesanmi, A.E.; Li, X.; Samuel, O.W.; Fang, P.; Li, G. An integrated deep learning model for motor intention recognition of multi-class EEG Signals in upper limb amputees. Comput. Methods Programs Biomed. 2021, 206, 106121. [Google Scholar] [CrossRef]
- Jeon, E.; Ko, W.; Yoon, J.S.; Suk, H.I. Mutual Information-Driven Subject-Invariant and Class-Relevant Deep Representation Learning in BCI. IEEE Trans. Neural Netw. Learn. Syst. 2021, 34, 739–749. [Google Scholar] [CrossRef]
- Kottlarz, I.; Berg, S.; Toscano-Tejeida, D.; Steinmann, I.; Bähr, M.; Luther, S.; Wilke, M.; Parlitz, U.; Schlemmer, A. Extracting Robust Biomarkers from Multichannel EEG Time Series Using Nonlinear Dimensionality Reduction Applied to Ordinal Pattern Statistics and Spectral Quantities. Front. Physiol. 2021, 11, 614565. [Google Scholar] [CrossRef]
- Malafeev, A.; Hertig-Godeschalk, A.; Schreier, D.R.; Skorucak, J.; Mathis, J.; Achermann, P. Automatic Detection of Microsleep Episodes with Deep Learning. Front. Neurosci. 2021, 15, 564098. [Google Scholar] [CrossRef]
- George, O.; Smith, R.; Madiraju, P.; Yahyasoltani, N.; Ahamed, S.I. Data augmentation strategies for EEG-based motor imagery decoding. Heliyon 2022, 8, e10240. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, M.; Hu, L.; Zhang, L.; Hua, Z. A Novel Recurrent Neural Network to Classify EEG Signals for Customers’ Decision-Making Behavior Prediction in Brand Extension Scenario. Front. Hum. Neurosci. 2021, 15, 610890. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhao, Q.; Li, S.; Li, K.; Liu, C.; Wang, J. Decoding Digital Visual Stimulation from Neural Manifold with Fuzzy Leaning on Cortical Oscillatory Dynamics. Front. Comput. Neurosci. 2022, 16, 852281. [Google Scholar] [CrossRef]
- van der Maaten, L. Learning a Parametric Embedding by Preserving Local Structure. In Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics 2009, PMLR 5, Clearwater Beach, FL, USA, 16–18 April 2009; pp. 384–391. [Google Scholar]
- Li, M.; Luo, X.; Yang, J. Extracting the nonlinear features of motor imagery EEG using parametric t-SNE. Neurocomputing 2016, 218, 317–381. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, H.; Wang, J.; Li, D.; Fang, X. Recognition of EEG Signal Motor Imagery Intention Based on Deep Multi-View Feature Learning. Sensors 2020, 20, 3496. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Aljalbout, E.; Golkov, V.; Siddiqui, Y.; Strobel, M.; Cremers, D. Clustering with deep learning: Taxonomy and new methods. arXiv 2018. [Google Scholar] [CrossRef]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A Survey of Clustering with Deep Learning: From the Perspective of Network Architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- Ren, Y.; Pu, J.; Yang, Z.; Xu, J.; Li, G.; Pu, X.; Yu, P.S.; He, L. Deep Clustering: A Comprehensive Survey. arXiv 2022. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, H.; Zheng, Z.; Chen, J.; Li, Z.; Bu, J.; Wu, J.; Wang, X.; Zhu, W.; Ester, M. A Comprehensive Survey on Deep Clustering: Taxonomy, Challenges, and Future Directions. arXiv 2022. [Google Scholar] [CrossRef]
- Obeid, I.; Picone, J. The Temple University Hospital EEG data corpus. Front. Neurosci. 2016, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Jasper, H.H. The ten twenty electrode system of the international federation. Electroencephalogr. Clin. Neurophysiol. 1958, 10, 367–380. [Google Scholar]
- Daubechies, I. Orthonormal bases of compactly supported wavelets, Comm. Pure Appl. Math. 1988, 41, 909–996. [Google Scholar] [CrossRef]
- Mallat, S. A Wavelet Tour of Signal Processing; Academic Press: Cambridge, MA, USA; Elsevier Inc.: Amsterdam, The Netherlands, 2009; pp. 180–181. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65, ISSN 0377-0427. [Google Scholar] [CrossRef]
- Nahrstaedt, H.; Lee-Messer, C. 2019. Holgern/Pyedflib. 2017. Available online: https://github.com/holgern/pyedflib (accessed on 21 September 2019).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- Sejdić, E.; Djurović, I.; Jiang, J. Time–frequency feature representation using energy concentration: An overview of recent advances. Digit. Signal Process. 2009, 19, 153–183. [Google Scholar] [CrossRef]
- Stanković, L.; Ivanović, V.; Petrović, Z. Unified approach to noise analysis in the Wigner distribution and spectrogram. Ann. Télécommun. 1996, 51, 585–594. [Google Scholar] [CrossRef]
- Dakovic, M.; Ivanovic, V.; Stankovic, L. On the S-method based instantaneous frequency estimation. In Proceedings of the Seventh International Symposium on Signal Processing and Its Applications 2003, Paris, France, 4 June 2003; Volume 1, pp. 605–608. [Google Scholar] [CrossRef]
- van der Maaten, L. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 2014, 15, 3221–3245. [Google Scholar]
- Linderman, G.C.; Rachh, M.; Hoskins, J.G.; Steinerberger, S.; Kluger, Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat. Methods 2019, 16, 243–245. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.; Quan, S. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, 1st ed.; American Academy of Sleep Medicine: Westchester, IL, USA, 2007. [Google Scholar]
- Kane, N.; Acharya, J.; Benickzy, S.; Caboclo, L.; Finnigan, S.; Kaplan, P.W.; Shibasaki, H.; Pressler, R.; van Putten, M.J.A.M. A revised glossary of terms most commonly used by clinical electroencephalographers and updated proposal for the report format of the EEG findings. Clin. Neurophysiol. Pract. 2017, 2, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, L.J.; Fong, M.W.K.; Leitinger, M.; LaRoche, S.M.; Beniczky, S.; Abend, N.S.; Lee, J.W.; Wusthoff, C.J.; Hahn, C.D.; Westover, M.B.; et al. American Clinical Neurophysiology Society’s Standardized Critical Care EEG Terminology: 2021 Version. J. Clin. Neurophysiol. 2021, 38, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Kobak, D.; Berens, P. The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 2019, 10, 5416. [Google Scholar] [CrossRef] [PubMed]
- Klambauer, G.; Unterthiner, T.; Mayr, A.; Hochreiter, S. Self-normalizing neural networks. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 972–981. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Lee, G.R.; Gommers, R.; Wasilewski, F.; Wohlfahrt, K.; O’Leary, A. PyWavelets: A Python package for wavelet analysis. J. Open Source Softw. 2019, 4, 1237. [Google Scholar] [CrossRef]
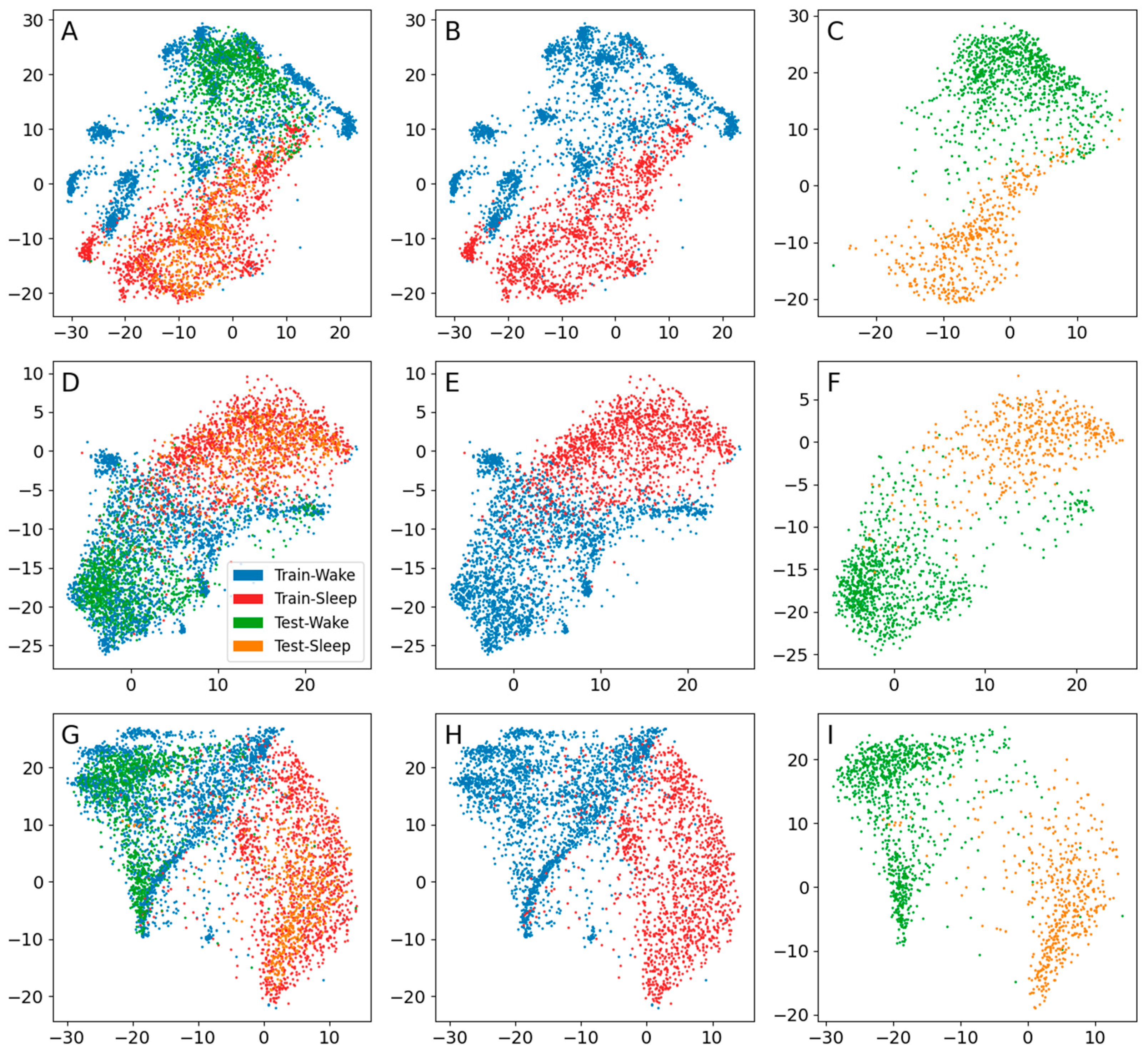
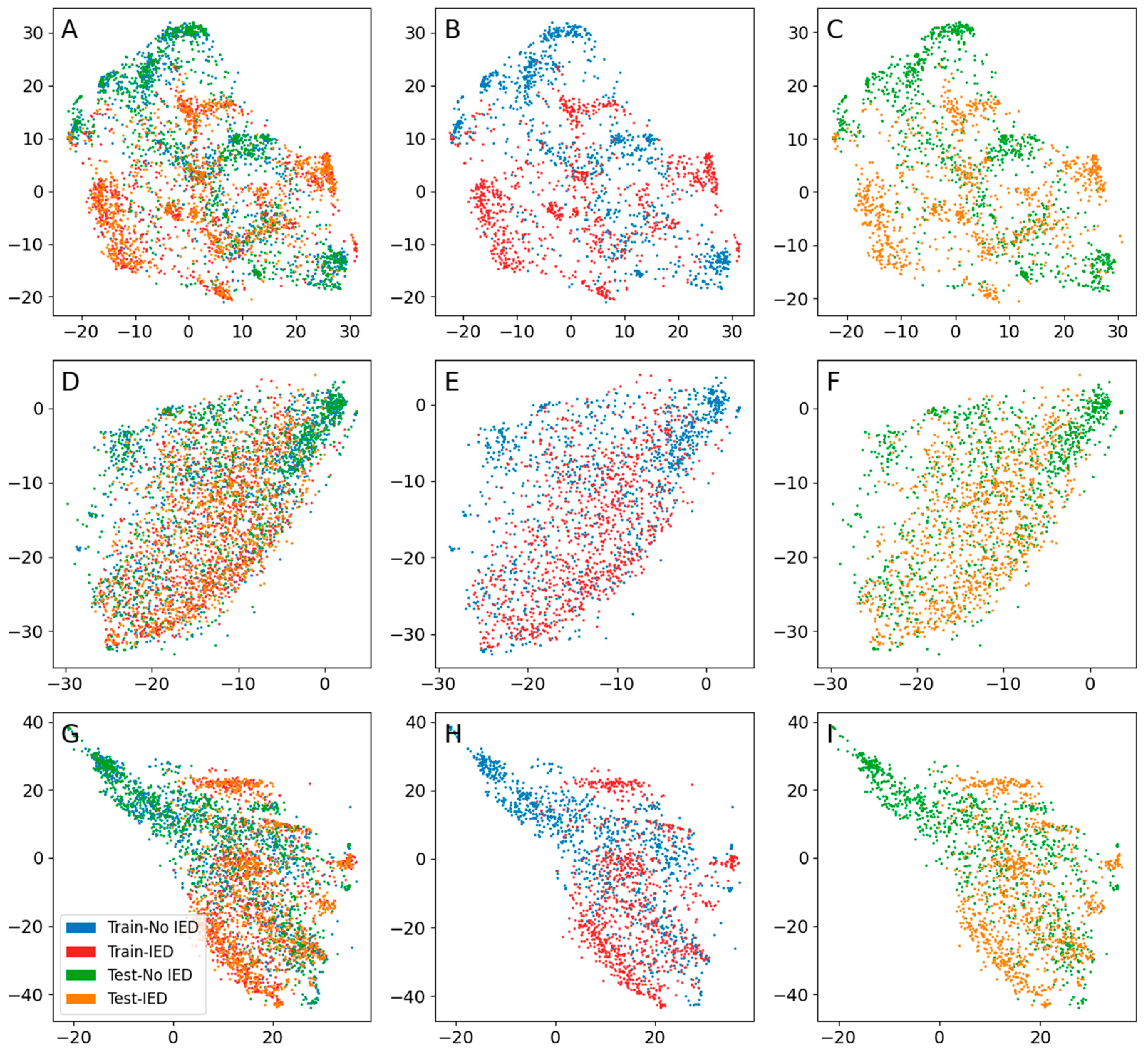
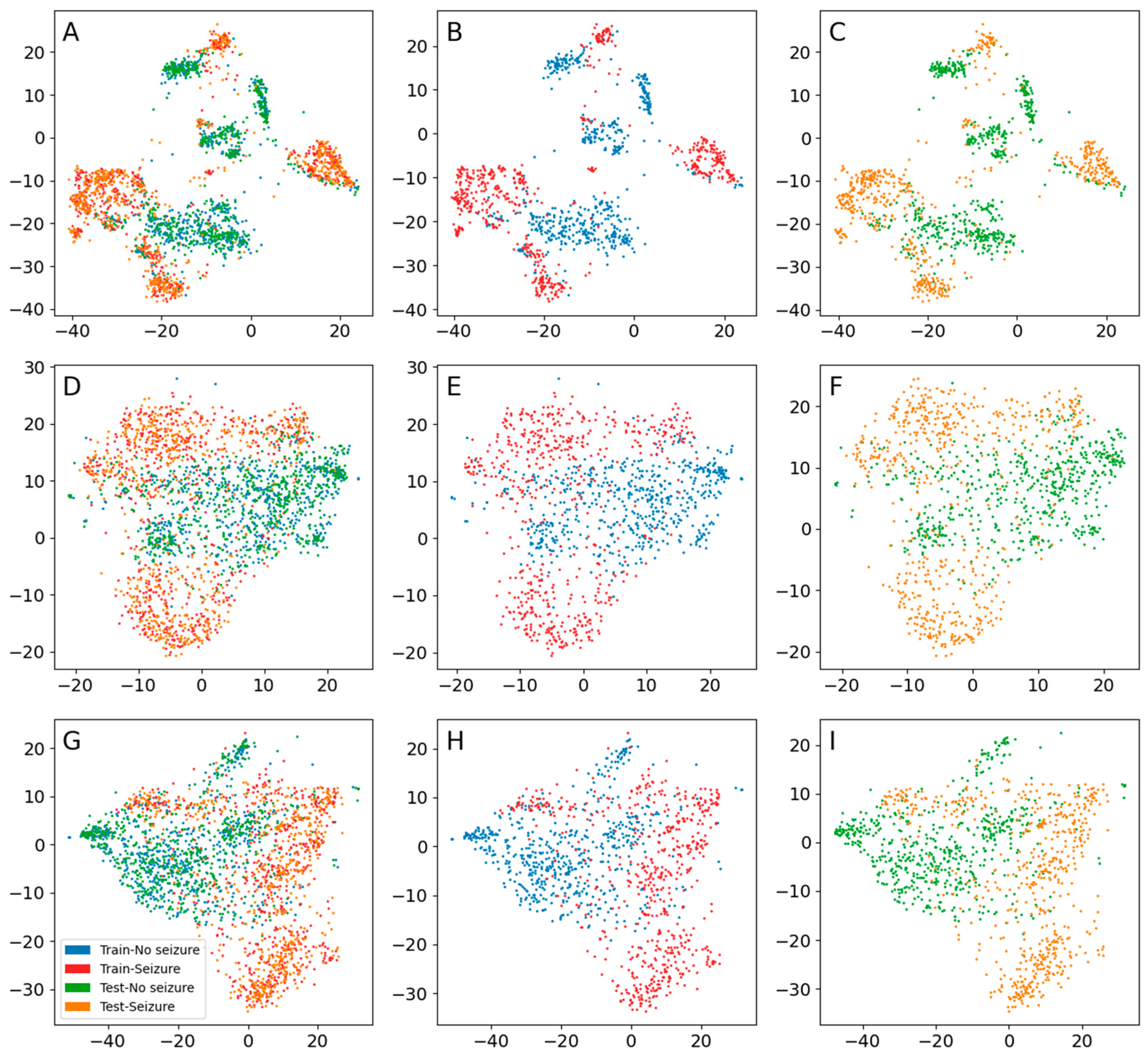
| Encoder | Input Shape | Number of Layers * | Number of Parameters | Number of Latent Dimensions | Learning Rate | Optimizer | Batch Size |
|---|---|---|---|---|---|---|---|
| CNN | 21 × 250 | 23 + 4 † | 19,084 + 1,117,970 † | 1260 | 10−4 | Adam | 500 |
| STFT | 630 | 4 | 1,823,502 | - | 10−4 | Adam | 500 |
| CWT | 2436 | 4 | 2,726,502 | - | 10−4 | Adam | 500 |
| -Train | -Test | -Train | -Test | -Train | -Test | |
|---|---|---|---|---|---|---|
| CNN | 0.80 | 0.89 | 0.93 | 0.86 | 22 | 2 |
| STFT | 0.77 | 0.86 * | 0.84 | 0.92 | 2 | 2 |
| CWT | 0.87 | 0.92 | 0.9 | 0.92 | 4 | 2 |
| -Train | -Test | -Train | -Test | -Train | -Test | |
|---|---|---|---|---|---|---|
| CNN | 0.39 | 0.33 | 0.76 | 0.69 | 18 | 17 |
| STFT | 0 | 0 | 0.43 | 0.35 | 4 | 4 |
| CWT | 0.31 | 0.29 | 0.67 | 0.56 | 2 | 2 |
| -Train | -Test | -Train | -Test | -Train | -Test | |
|---|---|---|---|---|---|---|
| CNN | 0 | 0 | 0.87 | 0.8 | 7 | 8 |
| STFT | 0 | 0 | 0.79 | 0.76 | 3 | 3 |
| CWT | 0.66 | 0.68 | 0.8 | 0.74 | 3 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svantesson, M.; Olausson, H.; Eklund, A.; Thordstein, M. Get a New Perspective on EEG: Convolutional Neural Network Encoders for Parametric t-SNE. Brain Sci. 2023, 13, 453. https://doi.org/10.3390/brainsci13030453
Svantesson M, Olausson H, Eklund A, Thordstein M. Get a New Perspective on EEG: Convolutional Neural Network Encoders for Parametric t-SNE. Brain Sciences. 2023; 13(3):453. https://doi.org/10.3390/brainsci13030453
Chicago/Turabian StyleSvantesson, Mats, Håkan Olausson, Anders Eklund, and Magnus Thordstein. 2023. "Get a New Perspective on EEG: Convolutional Neural Network Encoders for Parametric t-SNE" Brain Sciences 13, no. 3: 453. https://doi.org/10.3390/brainsci13030453
APA StyleSvantesson, M., Olausson, H., Eklund, A., & Thordstein, M. (2023). Get a New Perspective on EEG: Convolutional Neural Network Encoders for Parametric t-SNE. Brain Sciences, 13(3), 453. https://doi.org/10.3390/brainsci13030453





