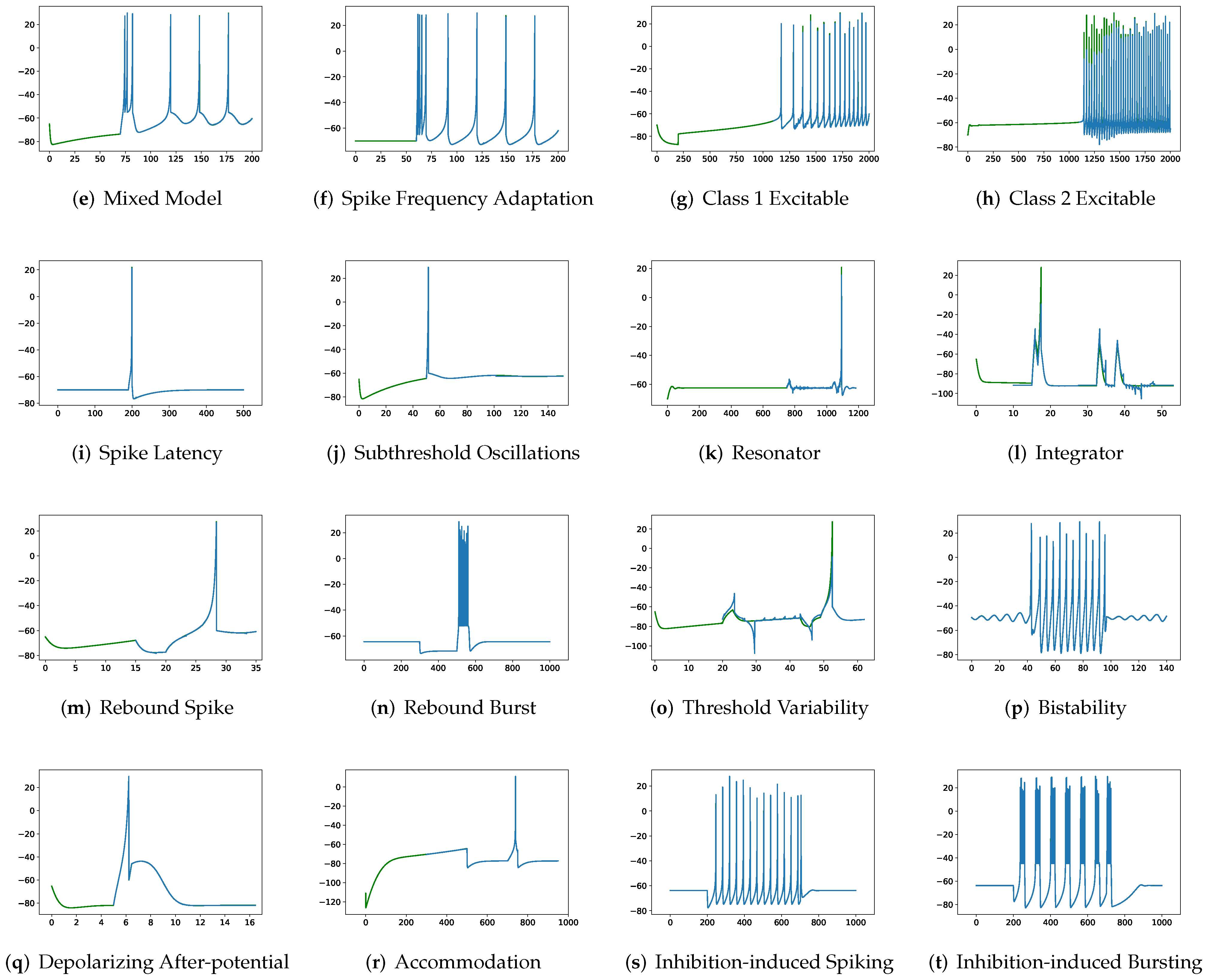
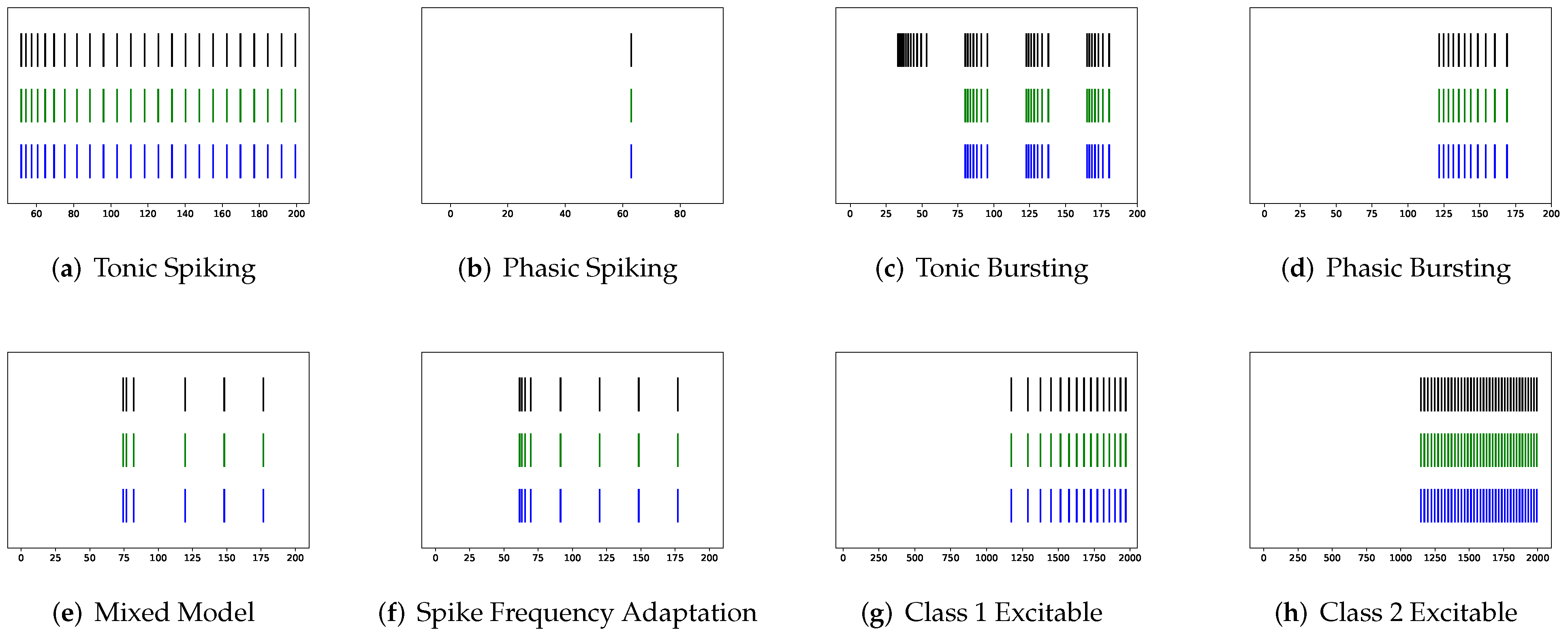
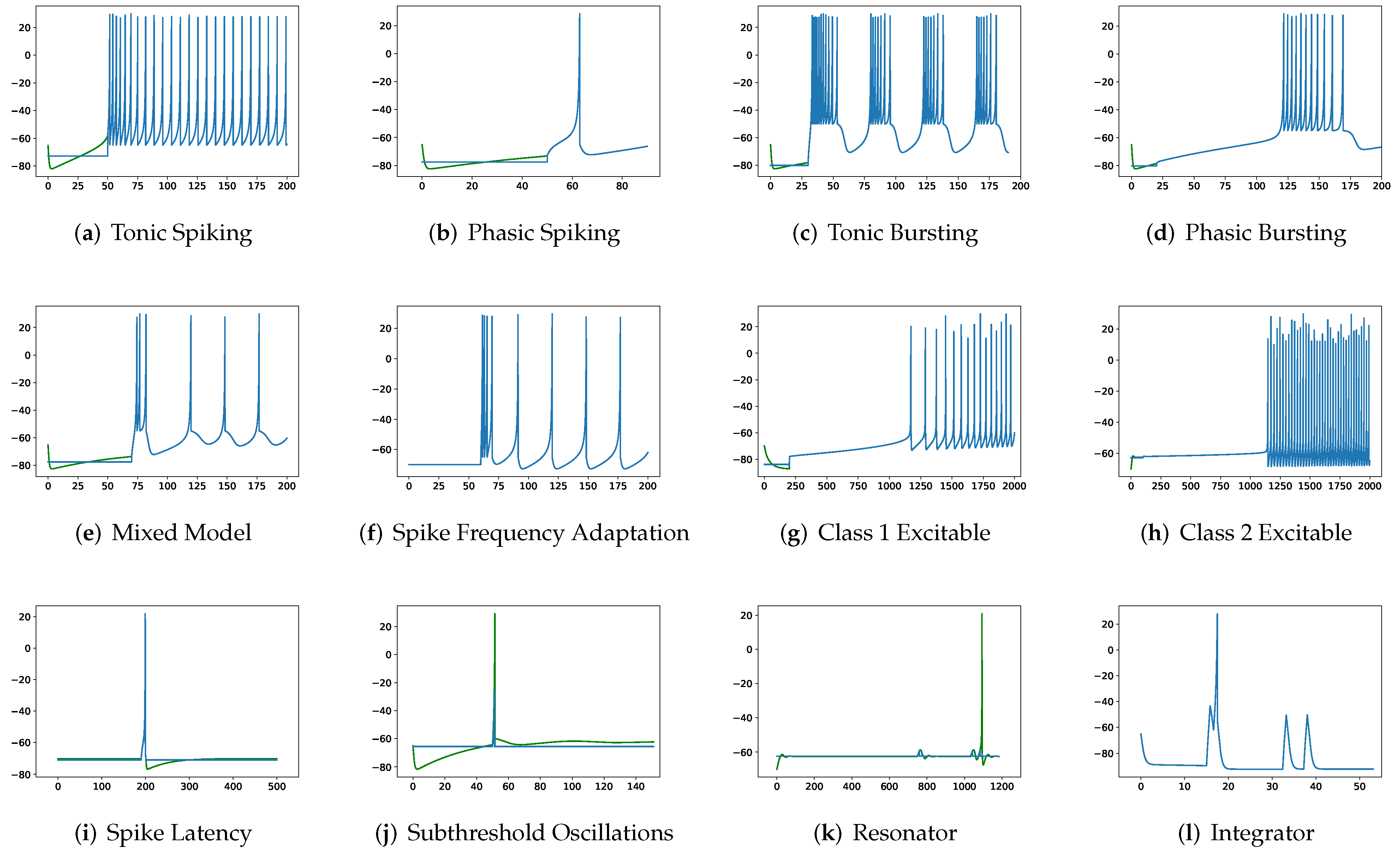
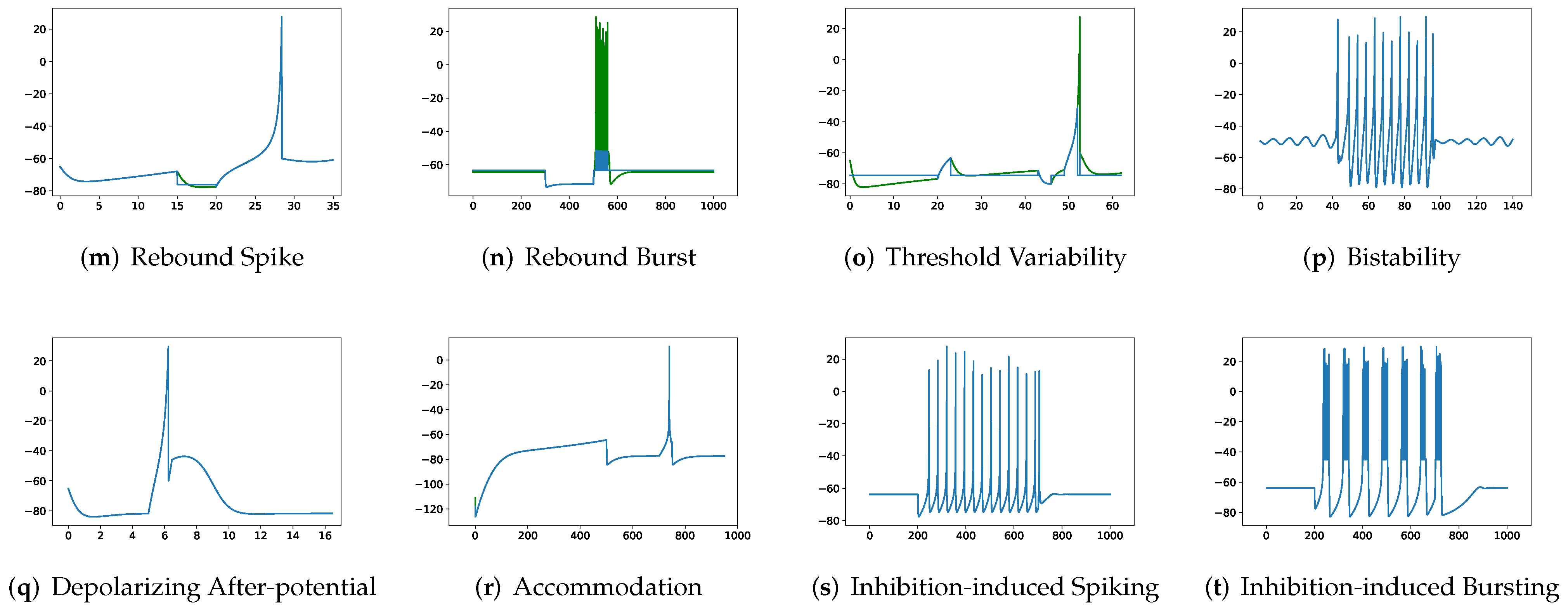
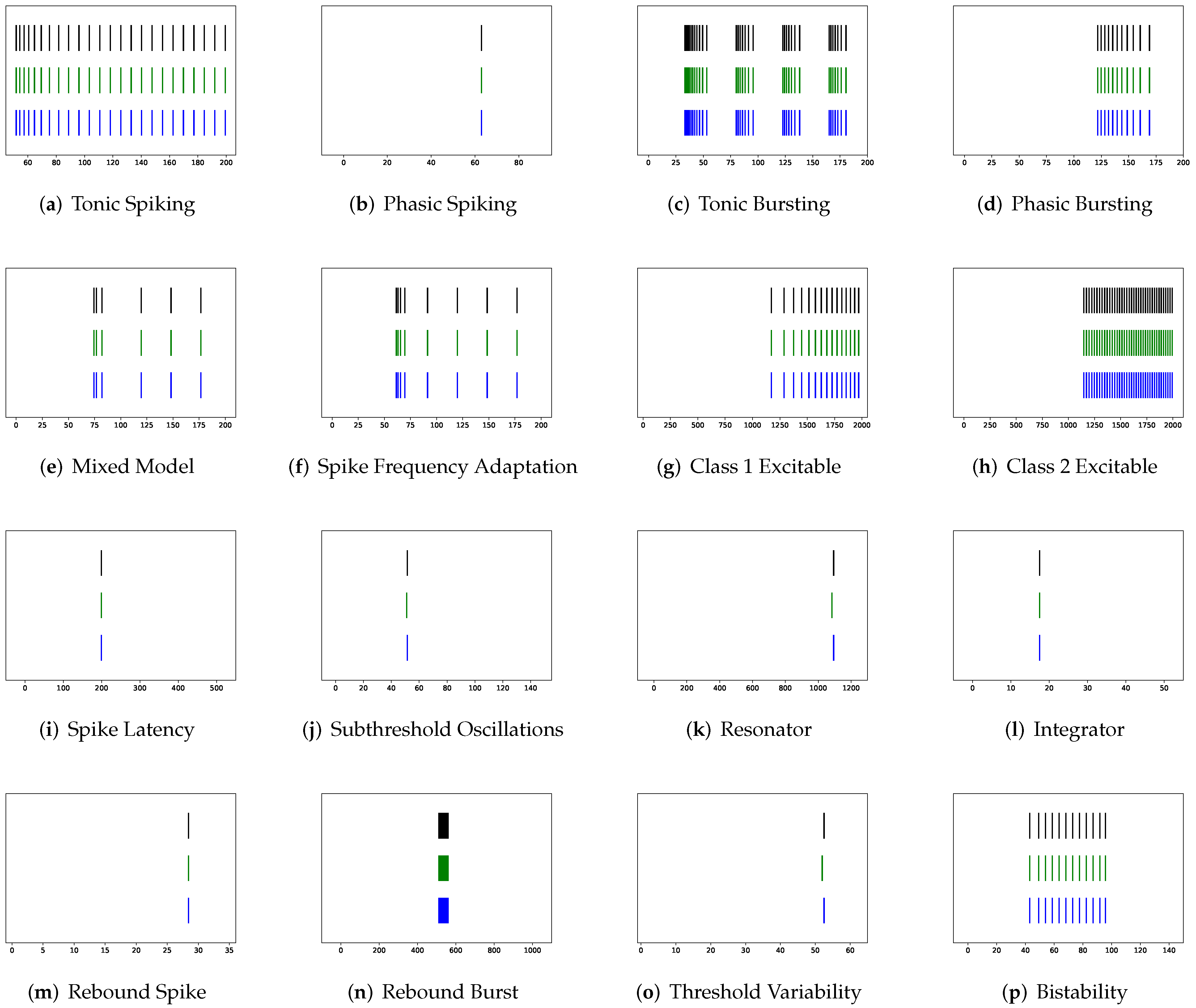
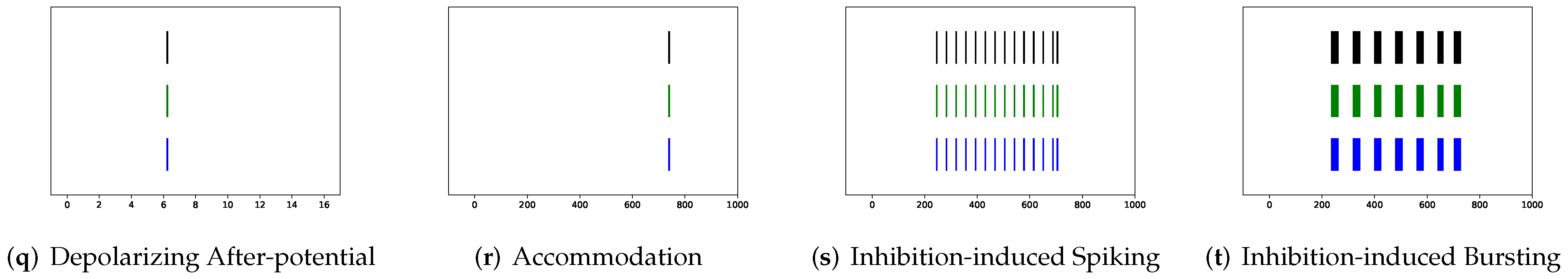
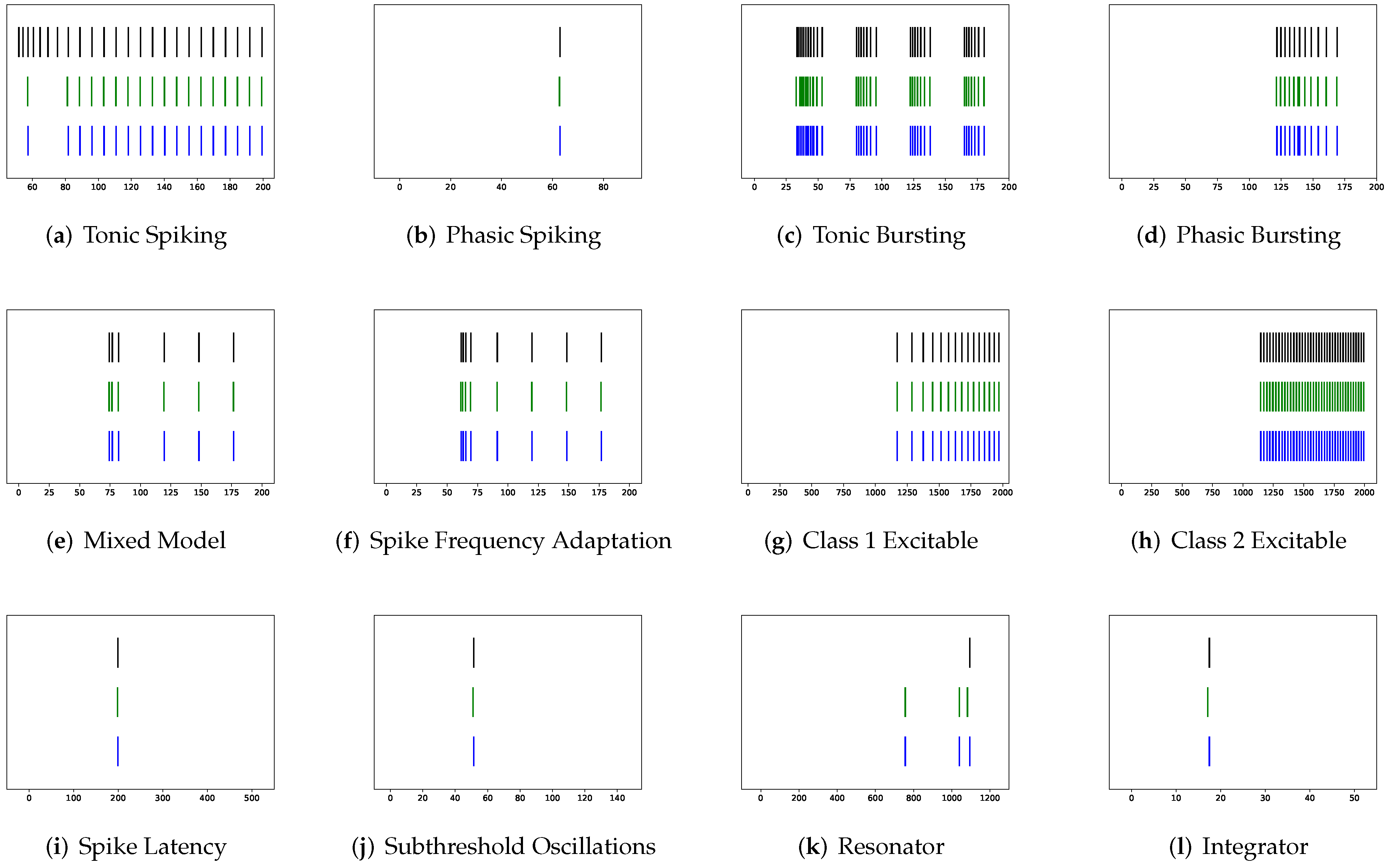
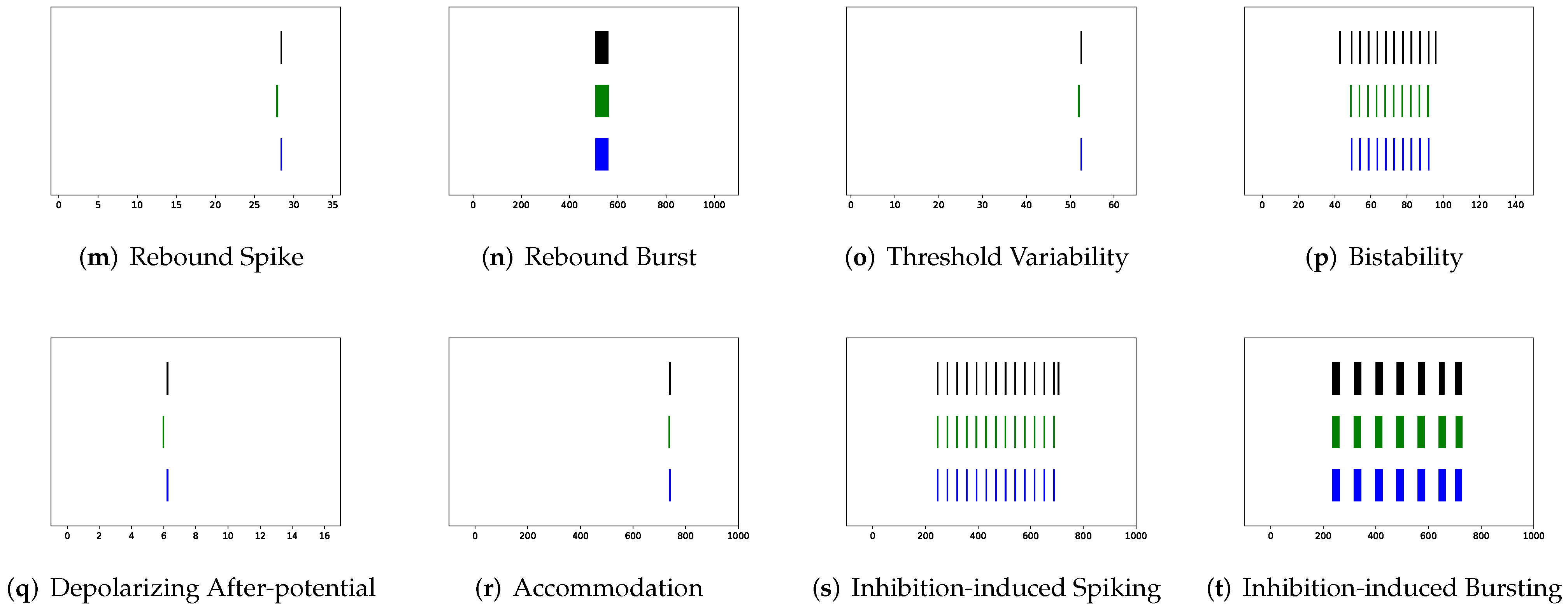
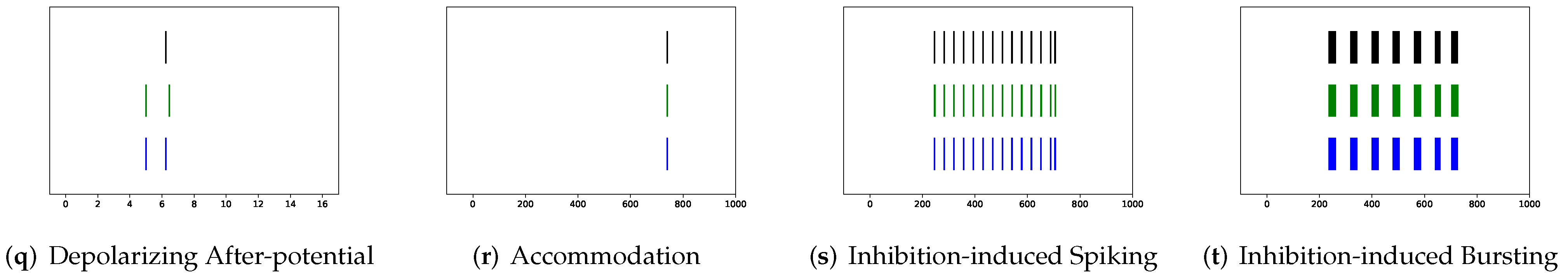
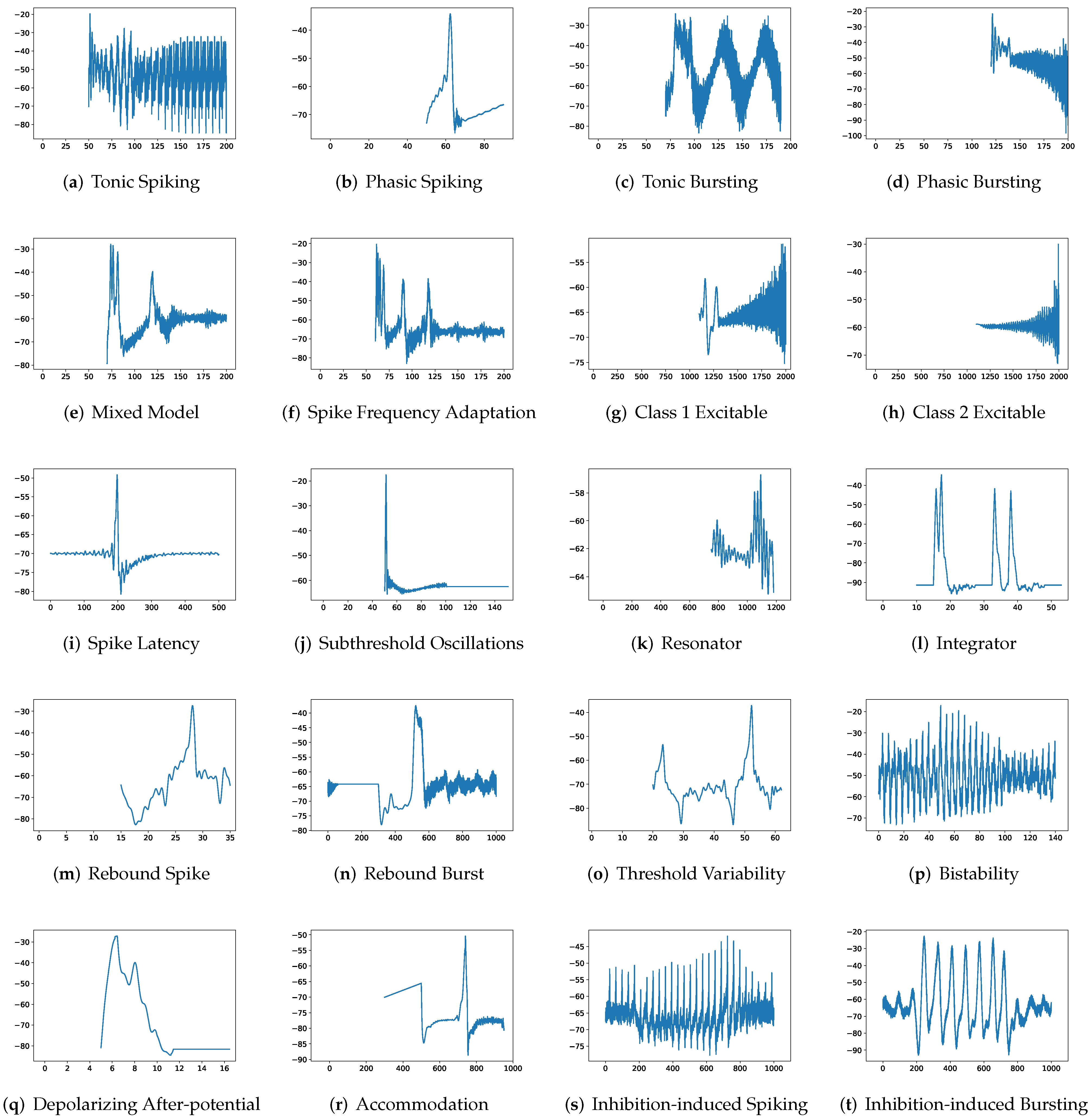
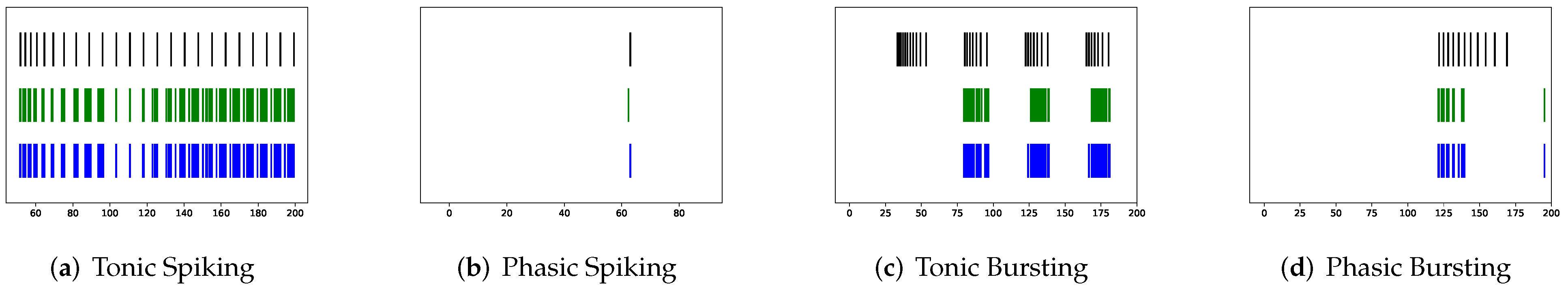
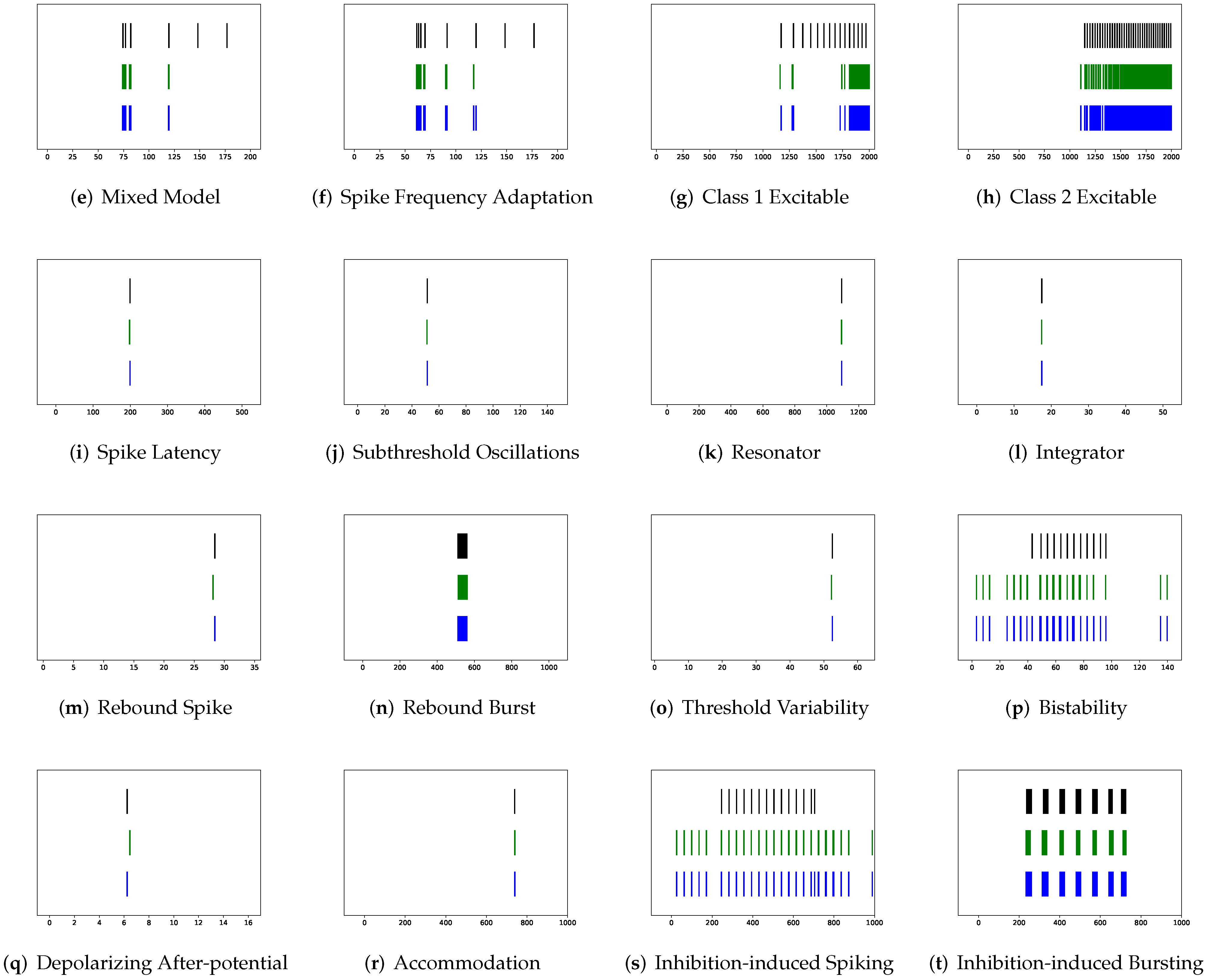
In this section, the data, including twenty classic firing behaviors of biological neurons, are generated, followed by exhaustive experiments of model fitting for membrane potentials and spike trains, respectively. Then, evaluation metrics are introduced, and comprehensive model comparisons are undertaken to evaluate the performance of each model.
3.2. Experiments
Six neuron models, the Spike Response Model (SRM), the Regularized Spike Response Model (RSRM), the Spectral Spike Response Model (SSRM), the Regularized Spectral Spike Response Model (RSSRM), the Principal Component Regression (PCR) [
28], and the Raised Cosine Basis Function Regression (RCR) [
37,
38,
39], are applied to fit the data of twenty firing behaviors. The dependent variable, membrane potentials, is shared, whereas the independent variables are versatile. Specifically, independent variables of SRM are injected currents and spike history with different amounts of time lags. SSRM takes as inputs the Fourier basis function on injected currents and the spike history with a single time lag. RSRM is SRM with L1 regularization for the parameter reduction, while RSSRM is SSRM with L1 regularization. The features of PCR are modified inputs of SRM, where the principal component analysis, a technique for dimension reduction, is performed to transform the independent variables of SRM into the principal components. Then, the first few principal components are selected as the features of PCR to decrease the dimension. RCR takes the raised cosine basis function on injected currents and spike history with a single time lag as predictors, where the raised cosine basis function captures temporal signals close to the time of a spike with relatively few hyperparameters [
39]. The details of each neuron model are shown in
Table 1.
There are two aspects of the model fitting that should be emphasized. First, parameter estimation is undertaken using the same numerical optimization method. The curse of dimensionality results from the fact that some models contain more independent variables than observations, resulting in high-dimensional input data. To fairly compare all neuron models, the L2 norm with the hyperparameter is performed in the optimization process to reduce the variability in the parameter estimation. Second, since the goal is to assess the effect of parameter reduction on neuron models, instead of splitting data of spiking patterns into training and test sets, the entire data is used for the model fitting, and the generalizability of models is not considered here.
In addition to prominent membrane potential prediction, excellent spike train forecasting is expected for neuron models. However, the relationship between estimated membrane potentials and predicted spike trains are unobvious. It is possible that undesirable estimated membrane potentials are from the prediction in the subthreshold regime, while estimated spike trains are outstanding. Three algorithms are proposed to convert estimated membrane potentials to estimated spike trains with a reasonable tolerance for mismatches between the actual and estimated spike trains.
Before discussing each algorithm, the following notations are introduced. The lowercase letters represent scalars, while the bold ones indicate vectors. In addition, calligraphic uppercase letters signify sets.
Algorithm 1 aims to convert membrane potentials to spike trains. The quantile
q in line 1 is defined as a threshold value in predicted membrane potentials, where the estimated firing ratio
is larger than the true firing proportion
r to take fluctuations in predicted membrane potentials into account. For example, given the true firing proportion
and an inflation coefficient
, the estimated firing ratio is
. The 85th quantile
q indicates that 15% of predicted membrane potentials are greater than
q. At the time
t, if the value of the predicted membrane potential is over the threshold point
q and is also a local maximum, then a predicted spike is established, which is formalized by lines 2 to 6.
| Algorithm 1: Convert membrane potentials to spike trains. |
![Brainsci 12 01008 i001 Brainsci 12 01008 i001]() |
The hyperparameter
ranges from 1 to 1.5 for SRM and SSRM, and from 1 to 270 for RSRM, RSSRM, PCR, and RCR due to their significant oscillations after parameter reduction. It is introduced because the predicted membrane potentials may be subject to substantial fluctuations so that it is difficult to differentiate between these membrane potentials and potential membrane potentials for spiking. By tuning
, potential membrane potentials for spiking are accounted for, despite the inclusion of membrane potentials with substantial fluctuations. Evaluation metrics such as F1-score is applied to to accommodate this phenomenon in
Section 3.3. Overall, the hyperparameter
is applied to capture all potential membrane potentials for spiking, despite the fact that some membrane potentials exhibit variations, which will be reflected in the evaluation metrics.
Algorithm 2 searches for the maximum tolerance of predicted spike trains, where maximum tolerance is a reasonable value such that predicted spikes within it are treated as the true spikes. The concept is introduced since it is common for the true spike time and the predicted spike time to differ by a few milliseconds [
19]. The maximum tolerance is an auxiliary variable that improves the assessment of neuron model performance. Since the performance of each neuron model in twenty firing behaviors differs, the associated maximum tolerances can vary considerably. For a certain evaluation metric, the greater the performance, the smaller the maximum tolerance. It is straightforward to transform the output vector of spike trains from Algorithm 1 into the input set of the spike times for Algorithm 2 due to the one-to-one relationship between spike trains consisting of 0 s and 1 s and spike times denoting the precise time of firings.
in line 1 is the upper bound of the maximum tolerance. For example,
and
, then
.
Within iterations of the tolerance
, a comparison is performed between the true spike time and the predicted spike time. When all elements in either set have been enumerated, indicating that the comparison is complete, the procedure terminates and further increments of
have no effect. Such a
is known as the maximum tolerance. To simulate specific firing behaviors, neuron models PCR and RCR may require large values of the maximum tolerance. Since an enormous maximum tolerance is not biologically reasonable and is inefficient in applications, the value of maximum tolerance for PCR and RCR is set to 30 ms by observing that the most considerable maximum tolerance among neuron models SRM, SSRM, RSRM, and RSSRM in twenty firing behaviors is around 30 ms. These pre-defined maximum tolerances are marked by asterisks in
Table 2.
| Algorithm 2: Find maximum tolerance. |
![Brainsci 12 01008 i002 Brainsci 12 01008 i002]() |
Algorithm 3 takes two sets of the spike time and a maximum tolerance as inputs and returns a predicted spike train with the maximum tolerance. This algorithm is similar to Algorithm 2, with the exception that when a predicted spike time falls inside the maximum tolerance, the true spike time is substituted.
| Algorithm 3: Predicted spike train with maximum tolerance. |
![Brainsci 12 01008 i003 Brainsci 12 01008 i003]() |
3.4. Model Comparison
The model comparison of neuron models is shown in
Table 3. The Izhikevich model composed of a two-dimensional dynamical system of differential equations is regarded as the ground truth, where parameters are fitted by the cortical neuron data and hyperparameters are specified. The Spike Response Model (SRM) owns 9895 parameters on average of twenty firing behaviors. In comparison, the Spectral Spike Response Model (SSRM) has an average of 29,659 parameters, which is a result of applying the Fourier basis function to injected currents and spike histories, respectively. In order to acquire 100 parameters on average in the Regularized Spike Response Model (RSRM), a parameter reduction, pruning more than 99% of parameters, is performed on SRM using L1 regularization for each firing behavior. To fairly compare different neuron models, the number of parameters for the Regularized Spectral Spike Response Model (RSSRM), the Principal Component Regression (PCR), and the Raised Cosine Basis Function Regression (RCR) is restricted and is the same as in RSRM for each firing pattern. Details are shown in
Table 2. Specifically, RSSRM applies a similar procedure to RSRM, where L1 regularization is conducted on SSRM in order to control the number of parameters in RSSRM to be the same as in RSRM by selecting hyperparameters appropriately. PCR utilizes the first
n principal components as new features [
28], where
n is the number of parameters in RSRM. The corresponding
n parameters are achieved by numerical optimization. Due to the fact that the dimensionality of the raised cosine basis is a hyperparameter, the required parameters in RCR are controlled.
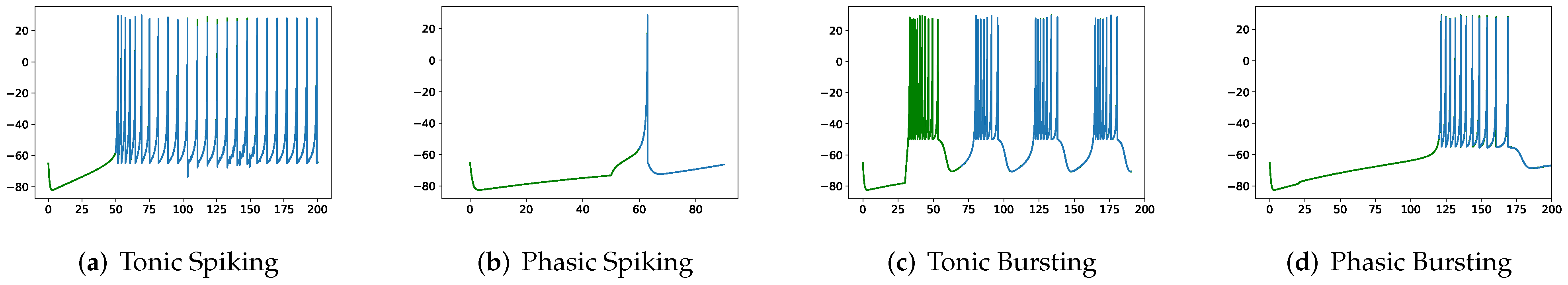
Although SRM has significantly fewer parameters than SSRM, its overall performance is superior. In particular, SRM it has a faster inference time, lower RMSE, and smaller maximum tolerance than SSRM, as well as comparable VRD and F1-score despite requiring slightly larger memory for inference, i.e., simulating firing behaviors. Both SRM and SSRM are well performed on the vast majority of firing behaviors, which are shown in
Figure 2,
Figure 3,
Figure 4 and
Figure 5. However, the parameter reduction of SRM is not robust. After pruning 99% of parameters using L1 regularization, the inference memory, RMSE, VRD, F1-score, and maximum tolerance of RSRM are inferior than those of RSSRM with the same number of parameters. Specifically, inference memory, RMSE and maximum tolerance of RSSRM are about half as tiny as those of RSRM, while VRD and F1-score of RSSRM are 72,000 times less and 8 times higher than those of RSRM, respectively. Their difference in inference time is minor, as both require approximately 1 ms, which is far faster than the Izhikevich model.
In addition, the robustness of parameter reduction in RSSRM has been demonstrated by comparing the changes in F1-score before and after the parameter pruning. RSSRM’s F1-score decreases by only 0.05 after parameter reduction, while F1-score of RSRM is reduced drastically. Furthermore, the difference in RMSE and VRD between RSSRM and SSRM is trivial when compared to the difference between RSRM and SRM. Specifically, the RMSE of RSSRM and RSRM is approximately 5 and 20 times larger than that of SSRM and SRM, whereas the VRD of RSSRM and RSRM is 47 and 3,600,000 times larger, respectively. Moreover, their changes in inference memory and inference time are noteworthy. After parameter pruning, the memory usage and runtime of RSSRM during inference are 56 times lower and 116 times faster than those of SSRM, although RSRM has 41 times larger memory and 72 times faster runtime than SRM.
Although PCR and RCR have superior overall performance than RSRM, they are not able to compete with RSSRM, which has the lowest RMSE, the lowest VRD, the highest F1-score, and the smallest maximum tolerance among these four neuron models with parameter reduction. The inadequate performance of PCR is further demonstrated by the low variance explanation. On average for twenty firing behaviors, the first 100 principal components of PCR explain only about 51% of the variance in data. In an ideal scenario, the first few principal components should explain more than 80% of variance. Since PCR assumes that principal components are obtained by a linear combination of the original data, non-linear methods are advocated. Therefore, RSSRM and RCR, obtaining nonlinearity from basis functions, perform better than PCR. RSSRM and RCR achieve the best and the second-best performance among neuron models with parameter reduction. However, considering the involvement of hyperparameters in constructing the raised cosine basis in RCR, RSSRM, consisting of the hyperparameter-free Fourier basis function, is preferable.
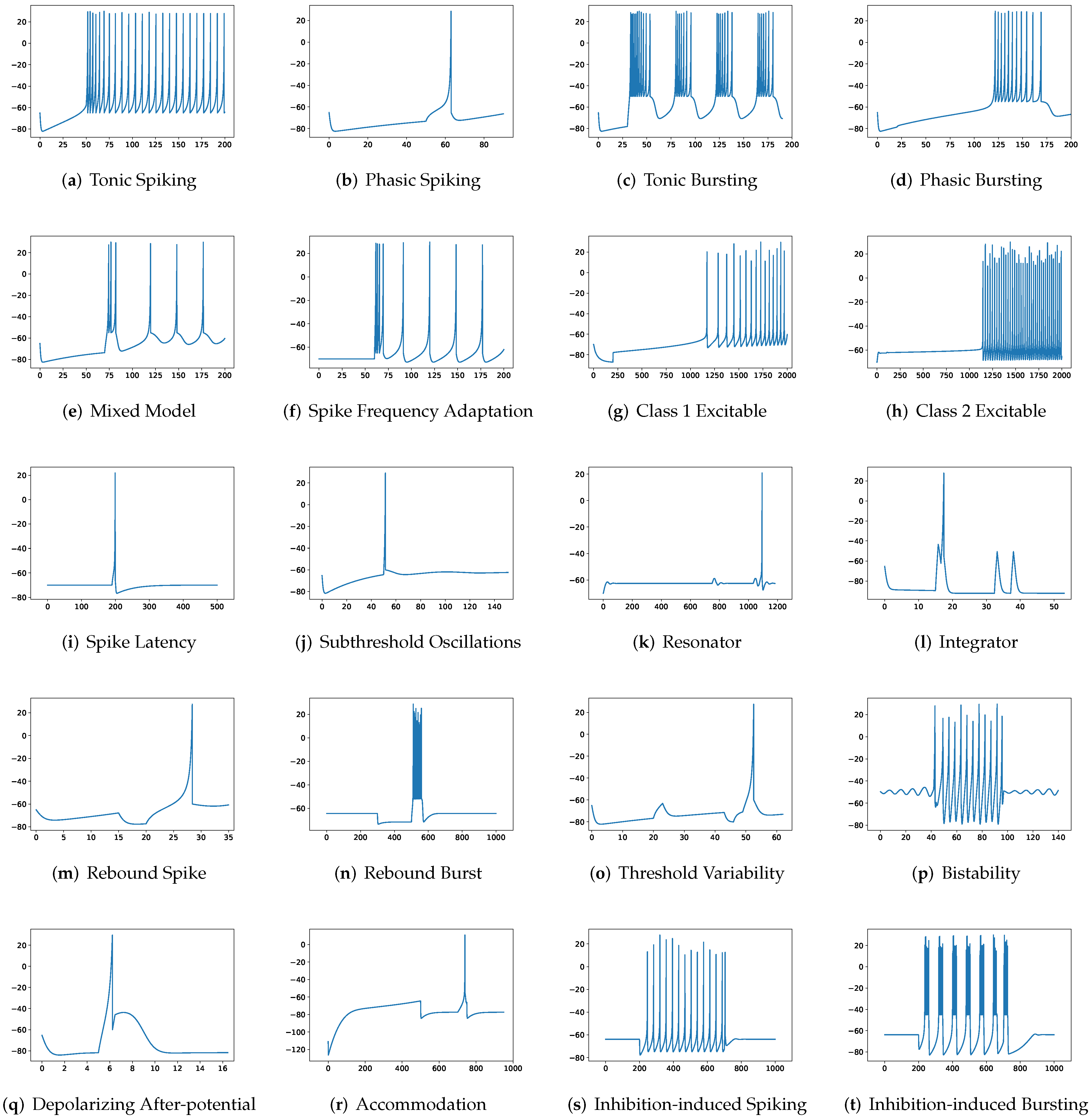
In addition to evaluating the performance of neuron models on an average of twenty firing behaviors, their capacity on different categories of spiking modes is studied, respectively. Twenty spiking patterns are classified into two types based on the number of spikes. One class, as shown in
Table 4, is the one-spike firing behavior, which includes (b) phasic spiking, (i) spike latency, (j) subthreshold oscillations, (k) resonator, (i) integrator, (m) rebound spike, (o) threshold variability, (q) depolarizing after-potential, and (r) accommodation. These nine spiking modes each produce a single spike throughout the experiment. As illustrated in
Table 5, the other class contains eleven spiking patterns that fire multiple spikes during the simulation. It consists of (a) tonic spiking, (c) tonic bursting, (d) phasic bursting, (e) mixed mode, (f) spike frequency adaptation, (g) class 1 excitable, (h) class 2 excitable, (n) rebound burst, (p) bistability, (s) inhibition-induced spiking, and (t) inhibition-induced bursting.
Table 4 demonstrates that SRM is superior to SSRM in terms of the number of parameters, inference time, RMSE, and maximum tolerance. After parameter reduction, RSSRM beats RSRM in terms of inference memory, RMSE, VRD, and F1-score. With only 50 parameters, PCR has the best performance among the neuron models with parameter reduction. It has the fastest inference time, the smallest RMSE, the lowest VRD, the highest F1-score, and the smallest maximum tolerance. Although its memory usage during inference is greater than that of other neuron models with parameter reduction, it is significantly lower than that of the Izhikevich model. PCR is the most notable model for predicting one-spike firing behaviors, whereas RSSRM is the second-best model. In
Table 5, it is challenging to distinguish between the performance of SRM and SSRM. Although SRM has fewer parameters, a faster inference time, and a smaller RMSE, SSRM shows lower memory usage, a lower VRD, a higher F1-score, and a smaller maximum tolerance. When parameter reduction is considered, RSSRM has the lowest memory consumption, the lowest RMSE, the lowest VRD, the highest F1-score, and the smallest maximum tolerance among all neuron models. Furthermore, RCR is the second most prominent neuron model though the maximum tolerance is greater than twelve times that of RSSRM. Moreover, RSRM is the most unsatisfactory neuron model. Compared with RSSRM, it has around twice the RMSE, more than seventy thousand times the VRD, one-ninth the F1-score, and more than seven times the maximum tolerance. Comparing
Table 4 and
Table 5, the Izhikevich model doubles the memory usage for inference from one-spike firing behaviors to multiple-spike firing patterns, but the other neuron models do not exhibit this increase. Moreover, RSSRM demonstrates a consistent inference time, whereas the inference time for the other neuron models increases dramatically.
Although
Table 3,
Table 4 and
Table 5 demonstrate that the overall performance of RSSRM is prominent among neuron models with parameter reduction, it is worthwhile to measure the capability of neuron models for each firing behavior. Details are shown in
Table 2.
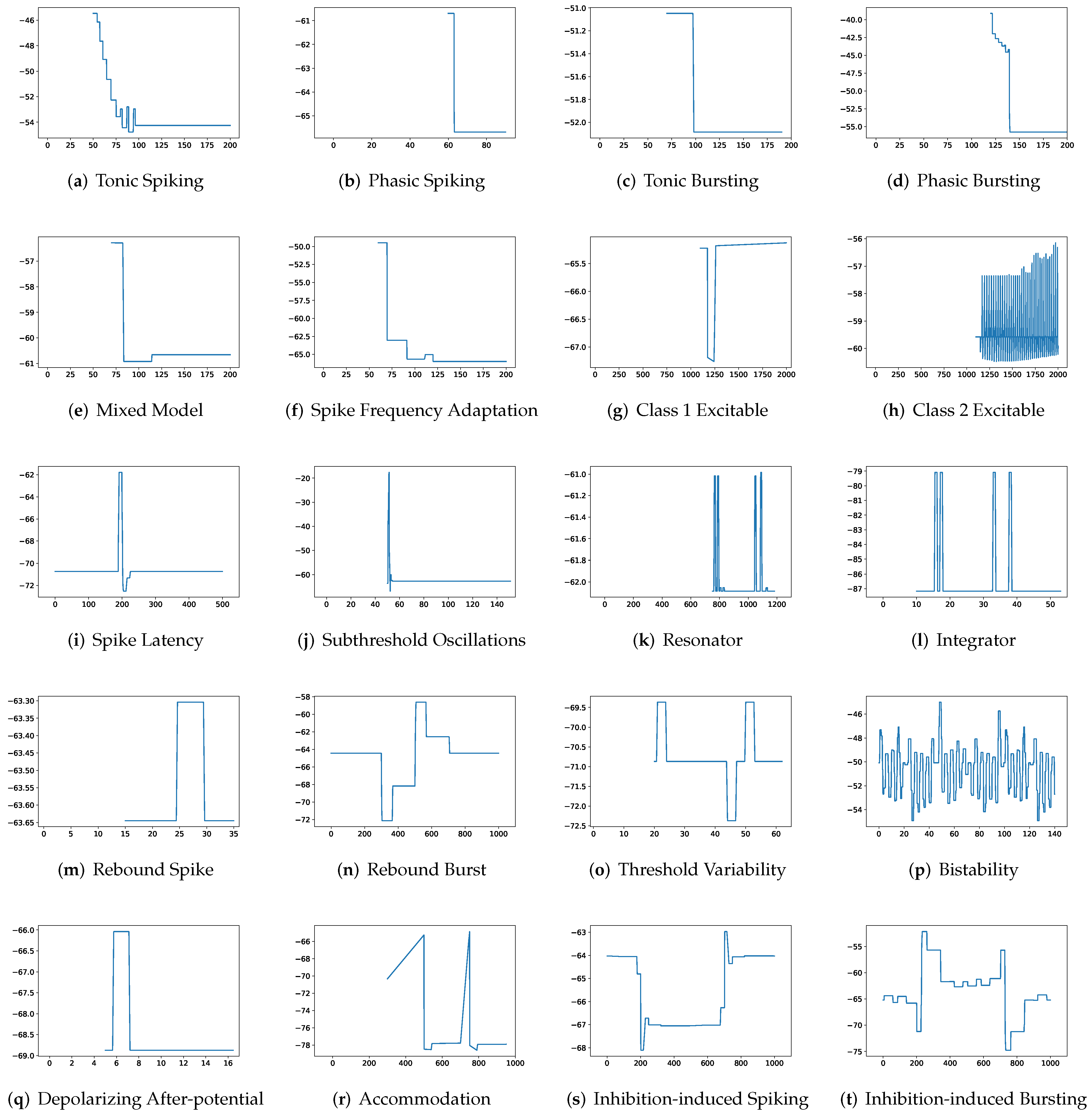
First, SRM, containing fewer parameters, exhibits faster inference time than SSRM in twenty firing patterns with the exception of (b) phasic spiking. Furthermore, RMSE of SRM for spiking behaviors (i) spike latency, (j) subthreshold oscillations, (k) resonator, (m) rebound spike, (n) rebound burst, and (o) threshold variability is smaller than that of SSRM. However, VRD and F1-score of both SRM and SSRM are nearly all 0.0 and 1.0 for twenty firing behaviors, respectively, indicating that perfect spike train prediction, the maximum tolerance of SSRM for (j) subthreshold oscillations, (k) resonator, (n) Rebound Burst, and (o) threshold variability is substantially greater than those of SRM, which remains zero, indicating perfect matches between the predicted and true spike trains. Zero maximum tolerance for SSRM is shown for (i) spike latency and (m) rebound spike due to a moderate corresponding RMSE.
In addition, in contradiction to the other firing behaviors, RMSE of RSSRM for (j) subthreshold oscillations and (n) rebound burst are larger than those of RSSRM. Specifically, the comparable RMSE of SSRM and RSSRM for (j) subthreshold oscillations indicates the Fourier basis function cannot capture the variability of membrane potentials, as depicted in
Figure 4 and
Figure 6. When it comes to (n) rebound burst,
Figure 4 and
Figure 6 reveal that neither SSRM nor RSSRM are able to reproduce the membrane potentials during spikes firing precisely.
Furthermore,
Table 2 show that none of RSRM owns a higher F1-score and a lower VRD than RSSRM though maximum tolerance of RSRM for (a) tonic spiking, (b) phasic spiking, (f) spike frequency adaptation, (i) spike latency, (j) subthreshold oscillations, and (k) resonator are all zeros. The rationale is indicated in
Figure 7 and
Figure 8, where RSRM is incapable of reproducing those firing behaviors precisely and instead provides a combination of step functions. In contrast to this circumstance, there are cases in which a considerable tolerance is required. To capture firing behaviors (g) class 1 excitable, (h) class 2 excitable, (r) accommodation, and (s) inhibition-induced spiking, the maximum tolerance values for RSRM range from 10.3 to 30.3 ms.
Moreover,
Table 2 illustrates that, among all neuron models, RSSRM takes the lowest memory consumption during inference, whereas the Izhikevich model shows the highest memory usage for twenty spiking patterns, excluding (k) resonator, (m) rebound spike, (o) threshold variability and (q) depolarizing after-potential. Compared to SRM, SSRM requires smaller memory for twenty firing behaviors despite a slower inference time. Specifically, the memory consumption of SSRM is lower for fifteen firing behaviors (b) phasic spiking, (c) tonic bursting, (e) mixed mode, (f) spike frequency adaptation, (g) class 1 excitable, (h) class 2 excitable, (i) spike latency, (k) resonator, (m) rebound spike, (n) rebound burst, (o) threshold variability, (p) bistability, (q) depolarizing after-potential, (r) accommodation, and (t) inhibition-induced bursting, while SSRM has only one faster inference time than SRM for (b) phasic spiking. Among neuron models with parameter reduction, RSRM has the second-lowest memory usage in twenty firing patterns. Although PCR shows the highest memory consumption in nine spiking modes, it consumes the largest memory on average, as shown in
Table 3. A comparison of inference time among neuron models with parameter reduction is trivial, as all of them takes about 1 ms during inference, which is over thirty times faster than the Izhikevich model.
To compare neuron models with parameter reduction,
Table 6 ranks the performance of RSRM, RSSRM, PCR, and RCR across twenty firing behaviors. RSSRM achieves the smallest RMSE for eleven spiking patterns, including (a) tonic spiking, (b) phasic spiking, (c) tonic bursting, (d) phasic bursting, (f) spike frequency adaptation, (g) class 1 excitable, (h) class 2 excitable, (i) spike latency, (p) bistability, (s) inhibition-induced spiking, and (t) inhibition-induced bursting, and the second-smallest RMSE for five firing modes, which are (e) mixed mode, (k) resonator, (l) integrator, (m) rebound spike, and (r) accommodation. Specifically, the difference between RSSRM and the best neuron model in terms of RMSE is less than 0.5. RSSRM produces the third-smallest RMSE among the other four spike behaviors, and the difference between it and the second-best model is minor, except for (q) depolarizing after-potential, where the difference is around 0.6.
Taking spike train forecasting into account, RSSRM achieves the highest F1-score for ten firing modes containing (a) tonic spiking, (b) phasic spiking, (c) tonic bursting, (d) phasic bursting, (e) mixed mode, (f) spike frequency adaptation, (h) class 2 excitable, (i) spike latency, (n) rebound burst, and (p) bistability, and the second-highest F1-score for six spiking patterns, which includes (g) class 1 excitable, (k) resonator, (m) rebound spike, (q) depolarizing after-potential, (s) inhibition-induced spiking, and (t) inhibition-induced bursting. Specifically, RSSRM maintains a perfect F1-score for (g) class 1 excitable, (m) rebound spike, and (q) depolarizing after-potential despite a slightly larger maximum tolerance. RSSRM obtains the third-highest F1-score among the other four firing modes. While the perfect F1-score remains, the corresponding maximum tolerance is insufficient. Moreover, RSSRM accomplishes the lowest VRD across twenty firing behaviors, with the exception of (d) phasic bursting, (k) resonator, (s) inhibition-induced spiking, and (t) inhibition-induced bursting, where the second-lowest VRD is obtained. Although the rankings of VRD and F1-score for RSSRM fluctuate across eight firing modes, they are nearly identical when the influence of maximum tolerance is removed.
Moreover, PCR obtains six of the smallest RMSE, nine of the lowest VRD, and six of the highest F1-score among twenty spiking behaviors, whereas RCR achieves two of the smallest RMSE, twelve of the lowest VRD, and six of the highest F1-score. Specifically, RSSRM and RCR earn the highest F1-score for (n) rebound burst, while PCR and RCR accomplish the highest F1-score for (l) integrator. Although RSRM does not attain the highest F1-score for any firing behavior, it achieves the smallest RMSE for (j) subthreshold oscillations though the corresponding F1-score is the lowest.
In
Table 6, RSSRM achieves the smallest RMSE, the lowest VRD, and the highest F1-score for eleven, sixteen, and ten firing behaviors, respectively, outperforming the other neuron models with parameter reduction.
Corresponding rankings support such an observation, where RMSE, VRD, and F1-score rankings of RSSRM are 1.65, 1.2, and 1.7, respectively. The superior performance of RSSRM is attributed to its excellent modeling strategy. Under the Fourier basis function, one time lag is imposed so that the model takes advantage of data maximally, which are shown in
Figure 6 and
Figure 9. Constructing RSRM requires an enormous number of time lags, which wastes a substantial quantity of data.
Table 1,
Figure 7 and
Figure 8 present this phenomenon. With the second-lowest VRD and the second-highest F1-score, RCR, endows the potential in spike train prediction at the expense of membrane potential forecasting.
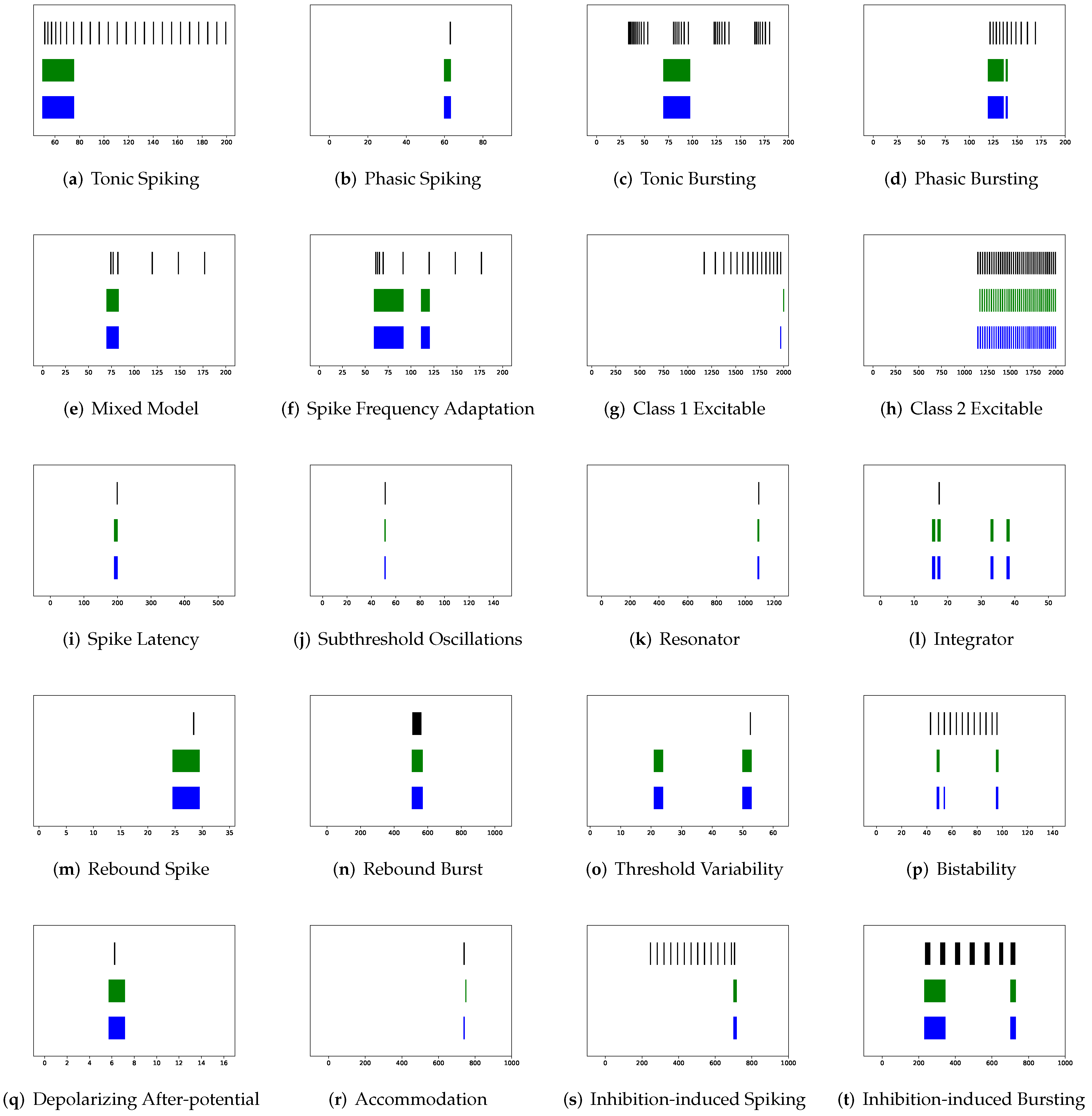
Figure 10 shows the prominent spike train prediction, whereas
Figure 11 demonstrates that predicted membrane potentials in firing behaviors such as (c) tonic bursting, (d) phasic bursting, (h) class 2 excitable, and (p) bistability are dramatically different from those of other neuron models, which use abnormal downward vertical lines to represent spikes. The performance of PCR is extreme, since the rankings of RMSE, VRD, and F1-score for the majority of firing patterns are either first or third.
Figure 12 and
Figure 13 illustrate that substantial fluctuations in the subthreshold regime contribute to its undesirable RMSE. Nevertheless, its performance exceeds expectations, considering that the modeling does not include the temporal structure.